Abstract
Multidrug resistance (MDR) caused by ATP-binding cassette (ABC) transporter P-glycoprotein (P-gp) through extrusion of anticancer drugs from the cells is a major cause of failure to cancer chemotherapy. Previously, selenazole containing cyclic peptides were reported as P-gp inhibitors and these were also used for co-crystallization with mouse P-gp, which has 87% homology to human P-gp. It has been reported that human P-gp, can simultaneously accommodate 2-3 moderate size molecules at the drug binding pocket. Our in-silico analysis based on the homology model of human P-gp spurred our efforts to investigate the optimal size of (S)-valine-derived thiazole units that can be accommodated at drug-binding pocket. Towards this goal, we synthesized varying lengths of linear and cyclic derivatives of (S)-valine-derived thiazole units to investigate the optimal size, lipophilicity and the structural form (linear and cyclic) of valine-derived thiazole peptides that can accommodate well in the P-gp binding pocket and affects its activity, previously an unexplored concept. Among these oligomers, lipophilic linear- (13) and cyclic-trimer (17) derivatives of QZ59S-SSS were found to be the most and equally potent inhibitors of human P-gp (IC50 = 1.5 μM). Cyclic trimer and linear trimer being equipotent, future studies can be focused on non-cyclic counterparts of cyclic peptides maintaining linear trimer length. Binding model of the linear trimer (13) within the drug-binding site on the homology model of human P-gp represents an opportunity for future optimization, specifically replacing valine and thiazole groups in the non-cyclic form.
Keywords: ABC transporter, P-glycoprotein, molecular modeling, multidrug resistance, peptide mimics
Introduction
P-glycoprotein (P-gp) or multidrug resistance protein 1 (MDR1) encoded by ABCB1, is a plasma membrane bound ATP-binding cassette (ABC) transporter. P-gp catalyzes the ATP-dependent efflux of a highly diverse set of compounds such as amphipathic, neutral or weakly basic with their molecular weights ranging from less than 200 to 2000 Da.[1-3] The 1280 amino acid long human P-gp consists of two transmembrane domains (TMDs) each with six α-helices and two nucleotide-binding domains (NBDs). The drug/substrate-binding sites are predicted to be located in the TMDs.[4,5] It has been well established that the drug-binding pocket is capable of even binding to two to three molecules simultaneously.[4,6-9] Drug transport by P-gp is driven by hydrolysis of ATP at NBDs. The close conformation of NBDs generated by ATP-binding/hydrolysis mediates substrate translocation from the drug-binding sites in TMDs, thus triggering the release of the substrate to the extracellular face of the membrane.[10,11]
In cancer patients, resistance to chemotherapy is a critical issue and can be due to various mechanisms. Overexpression of multidrug resistance (MDR) proteins such as P-gp is one of the well studied mechanisms of drug resistance.[12] Up-regulation of MDR proteins has been demonstrated in a variety of cancer types and has been shown to result in reduced intracellular concentration of chemotherapeutic drugs. P-gp is largely recognized for its role in enabling cancer cells to evade response to treatment via the efflux of chemotherapeutic agents. This multidrug resistance impedes the clinical efficiency of chemotherapy. Therefore, many researchers have been engaged in developing human P-gp inhibitors to reverse the chemotherapeutic drug resistance. Since there is no high-resolution crystal structure of human P-gp available yet, homology model of human P-gp based on the crystal structure of P-gp from mouse[13] serve as a tool for structure-based drug design of P-gp inhibitors. Although various strategies such as random and focused screening, systematic chemical modifications and combinatorial chemistry has been practiced so far to develop first three generations of P-gp inhibitors, their toxicity and drug interaction profiles are still undesirable. Therefore, new strategies leading to the development of fourth generation P-gp inhibitors (isolation of natural products or chemically modified natural product analogs) with high P-gp selectivity and potency seems to be a novel approach.[14]
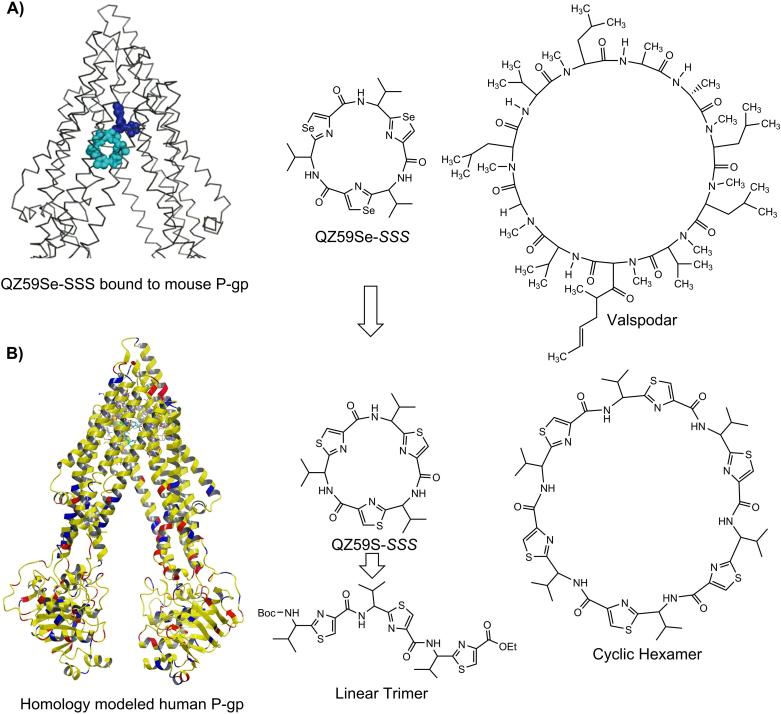
Analysis of QZ59-selenazole cyclic peptides (QZ59Se-SSS and QZ59Se-RRR)[5] within the drug binding cavity of homology model of human P-gp (built previously using mouse P-gp)[13] led to the following observations: a) two copies of QZ59Se-SSS were found at QZ59Se-SSS binding site[5] (Figure 1); moreover, it has been reported that human P-gp, can simultaneously accommodate 2-3 molecules at drug binding transmembrane domain.[4,6-9] b) P-gp can distinguish between the stereoisomers of cyclic peptides such that the QZ59Se-SSS forms extensive hydrophobic contacts with the hydrophobic residues of drug binding site and four-fold potent than corresponding QZ59Se-RRR isomer. Therefore, in our design strategy described below, we choose to maintain S-stereochemistry at all chiral centers. c) Comparison of inhibitory activity of QZ59S-SSS for Chinese hamster or mouse P-gp (IC50 = 2.7 μM)[15] with that of potent inhibitor valspodar[16] suggest the role of macrocyclic peptidic nature of valspodar for its high potency as compared to QZ59Se-SSS (Figure 1). During ongoing Phase III clinical trials, valspodar has shown very restricted oral bioavailability which along with its efficacy and safety concerns further suggested the need of optimization.[17]
Figure 1. Design Strategy.
A) Schematic structure of mouse P-gp crystal structure bound to QZ59Se-SSS,[5] chemical structures of QZ59Se-SSS and Valspodar, B) Human P-gp homology model generated from mouse P-gp (PDB ID: 3G60), cyclic trimer (QZ59S-SSS) and cyclic hexamer.
Unlike valspodar and cyclosporine A, both macrocyclic undecamer peptides, our peptidomimetics oligomers incorporate insertion of thiazole ring between the Cα-carbon and carbonyl carbon making them non-natural peptides which may promise them to be potentially devoid of immunosuppressant activity as well as resistant to hydrolytic cleavage by proteases. The constrained five-member thiazole ring imposes conformational restrictions leading to entropically favorable binding at the drug-binding site of P-gp. With this backdrop, herein we report the effect of structural form, length and flexibility of the designed (S)-valine thiazole-derived peptidomimetic oligomers on human P-gp function.
Co-crystal structure of QZ59Se-SSS-mouse P-gp has been previously reported.[5] The sulfur analogue was also shown to have IC50 value of 2.7 μM against mouse P-gp;[15] however, our current study reported here illustrate inhibitory activity of QZ59S-SSS against human P-gp. Replacement of selenium with sulfur could be effective strategy for designing natural product analogues, which are better tolerated by human cells. Thiazole structures are abundant in natural products and have direct applications in drug discovery.[18] Furthermore, we performed docking experiments on these analogs at all the possible binding sites of homology modeled human P-gp.
Results and Discussion
Chemistry
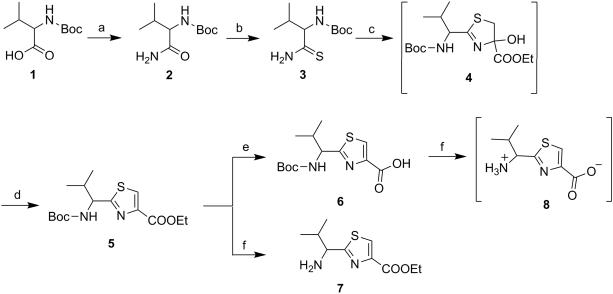
The thiazole derivatives were synthesized with diminutive variations in the procedures reported by Bertram et al and as shown in Scheme 1 to 6.[19,20] The compounds, linear as well as cyclic, with one thiazole unit are referred to as monomer derivatives, those with two thiazole units are called as dimer and so on and so forth as trimer, tetramer and hexamer. Scheme 1 shows the synthesis of monomer derivatives starting with commercially available N-Boc-(S)-valine 1 which was converted to amide 2 by general mixed anhydride/aqueous ammonia method in quantitative yield (Scheme 1). The amide 2 was then transformed to thioamide 3 by Holpfazel–Hantzsch procedure[21] using Lawesson's reagent in THF.[22,23] Compound 5 was synthesized by modified Hantzsch method.[24,25] The procedure involves reaction between compound 3 and ethyl bromopyruvate to give cyclocondensed intermediate 4,5-dihydrothiazole 4, that upon treatment with trifluoroacetic anhydride (TFAA) furnished aromatized thiazole product 5, in low yields.[21] Here, highly acidic reaction condition in the later step presents a possibility of N-trifluoroacetylation of amide ‘NH’. For this reason, the crude product obtained was further treated with freshly prepared sodium ethoxide in ethanol to produce the desired product 5 in appreciable yields without affecting the stereocenter adjacent to the 2-position of thiazole ring. Next, saponification of compound 5 using sodium hydroxide produced the monomer acid (6) and deprotection of ‘Boc-NH’ in compound 5 using trifluoroacetic acid (TFA) produced the monomer amine TFA salt (7). Further, Boc-deprotection of carboxylic acid derivative 6 yielded zwitterion product 8 (Scheme 1).
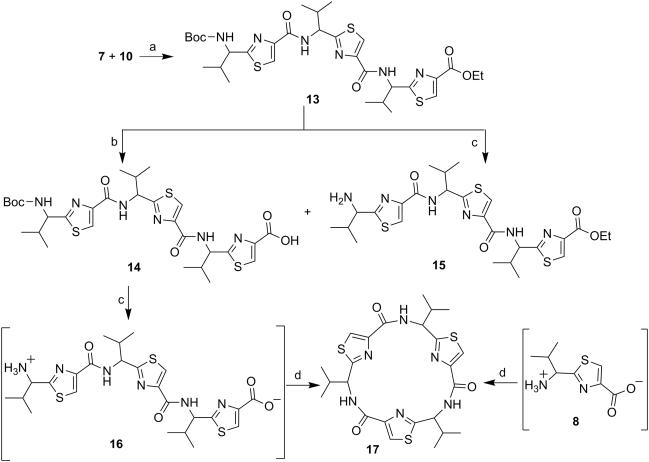
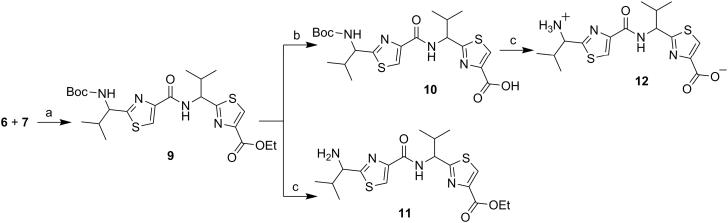
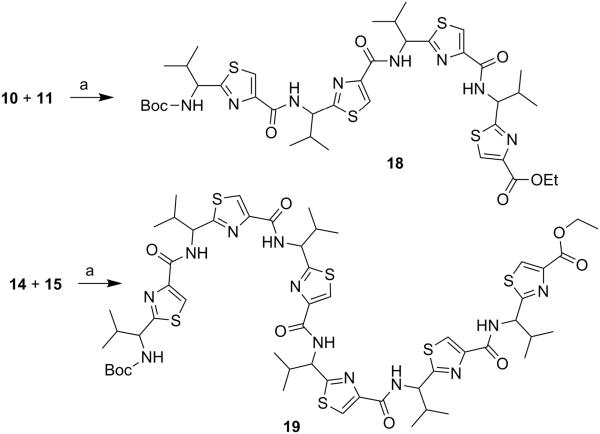
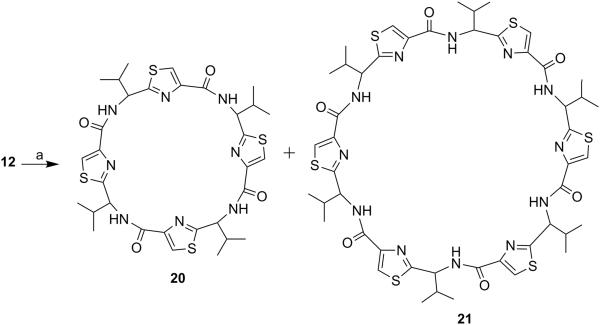
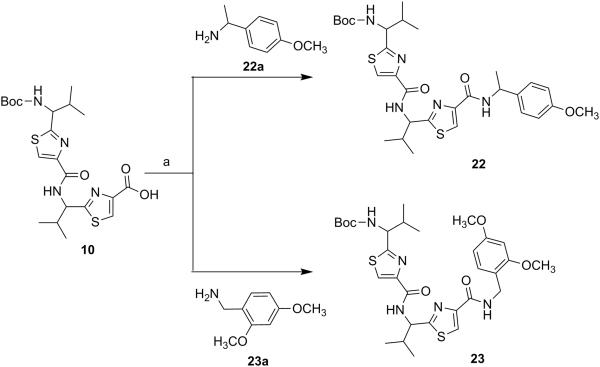
Next, through quick initial experiments we optimized the coupling of valine-derived monomer thiazole acid (6) via activation using BOP (benzotriazol-1-yl-oxy)-tris(dimethylamino)-phosphonium hexafluorophosphate reagent), HOBt and DIEA in CH2Cl2: DMF (4:1) with excess of monomer thiazole amine (7) to produce the required dimer (9) in moderate yield as shown in Scheme 2. This may be attributed to sterically demanding isopropyl group adjacent to the amine functionality.[26,27] Compounds 10 and 11 were synthesized by basic and acidic hydrolysis of dimer 9, respectively. Further, the deprotected compound 12 was made by treating dimer acid 10 with TFA in DCM (Scheme 2). The synthetic route to trimer derivatives is portrayed in Scheme 3. Monomer amine (7) and dimer acid (10) were coupled using BOP-HOBt method to obtain the desired linear trimer (13). Compound 13 was hydrolyzed by sodium hydroxide and TFA, respectively, to give trimer acid 14 and trimer amine 15. The acid 14 was again treated with TFA in DCM to obtain intermediate trimer zwitterion (16), which was carried forward without further purification. Intermediate 16 was cyclized in presence of pentafluorophenyl diphenylphosphinate (FDPP), anhydrous ZnCl2 and DIEA in CH2Cl2: DMF (4:1) to provide the target cyclic trimer (17) (QZ59S-SSS).[28] Alternatively, cyclic trimer 17 was also prepared by coupling and subsequent cyclization of three molecules of monomer zwitterion compound (8) using the above mentioned procedure. The above compounds were found to be optically pure (ee≥95%) based on analysis by chiral HPLC (single peak using 85% acetonitrile: 15% water: 0.1% TFA). After exploring a number of different conditions to prepare linear peptides, we found that coupling reactions of thiazole acid in presence of 1.5 equiv of BOP, 1.5 equiv of HOBt and 1.5 equiv of N,N-diisopropylethylamine (DIEA), with valine-derived thiazole amine-TFA salt in excess amount (1.3-1.5 equiv) in 4:1 CH2Cl2: DMF worked satisfactorily. Linear derivatives of tetramer (18) and hexamer (19) were prepared from corresponding thiazole starting materials (Scheme 4). Compound 18 was prepared by coupling 10 and 11 while linear hexamer (19) was made beginning with 14 and 15. The cyclic tetramer (20) and cyclic hexamer (21) were synthesized starting from dimer zwitterion (12) giving both the products in one step (Scheme 5). The tetramer and hexamer derivatives were produced in very low yields (Scheme 4 and 5), therefore the desired products were purified by means of preparative HPLC with subsequent electron spray ionization-mass spectrometry (ESI-MS) analysis. In our further investigation, carboxyl terminus of valine-derived bis-thiazole acid (10) was coupled with commercially available arylalkyl amines (22a and 23a) using coupling reagents HCTU, HOBt and DIEA in DMA to obtain compounds 22 and 23 in moderate yield as shown in Scheme 6. The activity of the synthesized analogs was assessed for P-gp modulation using Calcein-AM and Bodipy-FL-Prazosin efflux assays, inhibition of photolabeling of P-gp with [125I]-IAAP and ATPase activity assay.
Structure-activity relationship
The thiazole containing peptides were studied for their modulatory effect on the transport function of human P-gp. Herein, we wanted to investigate the optimal size, lipophilicity and the structural form (linear and cyclic) of these peptides that can accommodate well in the P-gp drug-binding pocket and affects its activity.
The synthesized (S)-valine-derived thiazole products (5-15, 17-23) were evaluated for their effect on human P-gp activity utilizing multiple assays. Table 1 represents the percent inhibition values for all thiazole derivatives obtained from Calcein-AM and Bodipy-FL-Prazosin efflux assays in HeLa cells expressing human P-gp. In Table 1, the inhibition of the P-gp transport activity by the synthesized compounds was expressed as relative to the percent of the inhibition caused by 1 μM tariquidar, a known P-gp third generation inhibitor.[29-31] All the synthesized derivatives were tested for their effect on photoaffinity labeling of P-gp with [125I]-iodoarylazidoprazosin ([125I]-IAAP), which is a transport substrate for P-gp and the results are given in Table 2. For comparison of inhibitory potency of our derivatives, we used tariquidar as a standard inhibitor of photolabeling of P-gp [125I]-IAAP) and 5 μM tariquidar gave complete (100%) inhibition (see legend to Table 2).
Table 1.
Effect of (S)-valine based thiazole derivatives on the transport function of P-gp in HeLa cells
| Compd | Common Name | Tested conc. | Calcein-AM efflux % Inhibition[a] |
Bodipy-Prazosin efflux % Inhibition[a] |
|---|---|---|---|---|
| 5 | Monomer | 20 μM | <5 | 10 |
| 6 | Monomer acid | 20 μM | <5 | 10 |
| 7 | Monomer amine | 20 μM | <5 | <5 |
| 8 | Monomer zwitterion | 20 μM | <5 | ND[c] |
| 9 | Dimer | 20 μM | 55 | 20 |
| 10 | Dimer acid | 20 μM | <5 | 12 |
| 11 | Dimer amine | 20 μM | 55 | 13 |
| 12 | Dimer zwitterions | 20 μM | <5 | 9 |
| 13 | Linear trimer | 10 μM | 98[b] | 75 |
| 14 | Trimer acid | 10 μM | <5 | 10 |
| 15 | Trimer amine | 10 μM | 55 | 67 |
| 17 | Cyclic trimer (QZ59S-SSS) | 10 μM | 97[b] | 89 |
| 18 | Linear tetramer | 10 μM | 59 | 81 |
| 19 | Linear hexamer | 10 μM | <5 | 12 |
| 20 | Cyclic tetramer | 10 μM | 62 | 79 |
| 21 | Cyclic hexamer | 10 μM | <5 | 15 |
| 22 |
R-1-(4-methoxy- phenyl)ethyl-dimer acid |
10 μM | 97[b] | 88 |
| 23 | 2,4-Dimethoxy benzyl amine-dimer acid |
10 μM | 91[b] | 87 |
BacMam-P-gp virus transduced HeLa cells were incubated with 0.5 μM calcein-AM for 10 min or Bodipy-FL-Prazosin for 45 min at 37 °C under dark in the presence and absence of 10 μM (S)-valine based thiazole derivatives. Cells were washed once with IMDM medium and data acquired in FL-1 channel in flow cytometer. Percentage transport inhibition was derived by taking the level of inhibition obtained with the standard inhibitor, tariquidar at 1 μM equal to 100% and the values shown are an average of two independent experiments done in triplicate.
The IC50 values of inhibition of calcein-AM efflux for selected compounds: #13 linear trimer = 1.5 μM; #17 cyclic trimer = 1.5 μM; #22 R-1-(4-methoxyphenyl)ethyl-dimer acid = 2. 0 μM and #23 2, 4-Dimethoxybenzyl amine-dimer acid = 2.0 μM. These values were determined by taking the average of at least two independent experiments done in triplicate.
ND, not determined.
Table 2.
Effect of (S)-valine based thiazole derivatives on photolabeling of P-glycoprotein with [125I]-IAAP
| Compd | Common Name | Tested conc. |
[125I]-IAAP % Inhibition[a] |
|---|---|---|---|
| 5 | Monomer | 20 μM | 20% |
| 6 | Monomer acid | 20 μM | <10% |
| 7 | Monomer amine | 20 μM | <10% |
| 8 | Monomer zwitterion | 20 μM | <10% |
| 9 | Dimer | 20 μM | 87% |
| 10 | Dimer acid | 20 μM | <10% |
| 11 | Dimer amine | 20 μM | 87% |
| 12 | Dimer zwitterion | 20 μM | 23% |
| 13 | Linear trimer | 10 μM | 91% |
| 14 | Trimer acid | 10 μM | 60% |
| 15 | Trimer amine | 10 μM | 91% |
| 17 | Cyclic trimer(QZ59S-SSS) 10 μM | 87% | |
| 18 | Linear tetramer | 10 μM | 99% |
| 19 | Linear hexamer | 10 μM | 94% |
| 20 | Cyclic tet ramer | 10 μM | 95% |
| 21 | Cyclic hexamer | 10 μM | 91% |
| 22 |
R-1-(4-methoxy- phenyl)ethyl-Dimer acid |
10 μM | 100% |
| 23 | 2,4-Dimethoxybenzyl amine-Dimer acid |
10 μM | 95% |
Crude membranes (50-75 μg protein) from P-gp expressing High-Five insect cells were incubated in the presence and absence of 10 μM thiazole-derivatives in 50 mM Tris-HCl pH (7.5) and 4-6 nM [125I]-IAAP (2200 Ci/mmol). The samples were then photocross-linked by exposure to 366 nm UV light for 10 min and incorporation of [125I]-IAAP was determined using phosphorimager as described previously.[45] 5 μM Tariquidar (positive control) inhibited IAAP incorporation into P-gp by 100%. The results are given as % inhibition of [125I]-IAAP incorporation into P-gp by taking the average of two independent experiments.
The monomer compounds comprising of single thiazole unit (5-8) were devoid of P-gp inhibitory activity as indicated by the data obtained from the efflux assays (<5-10% inhibition). These results were also corroborated by the [125I]-IAAP % inhibition. Ineffective inhibition of P-gp efflux function by monomers (5-8) may possibly be due to their smaller size and limited hydrophobic surface area. In this regard, increasing the number of thiazole units to two in compound 5, thus resulting compound 9 was found to possess substantial improvement in P-gp inhibition. Dimer compound 9 inhibited the Calcein-AM efflux by 55% and the [125I]-IAAP labeling by 87% as compared to <5% and 20%, respectively, for monomer compound 5. On the contrary, the Bodipy-FL-Prazosin efflux analysis (20% inhibition) showed modest improvement for compound 9. However, compounds 10 and 12, the acid and zwitterion derivatives correspondingly of 9 exhibited complete loss in inhibitory activity (<5% Calcein-AM efflux inhibition; 12% and 9% Bodipy-Prazosin efflux inhibition). On the other hand, amine 11 maintained similar inhibition potency as that of compound 9 (55% Calcein-AM efflux inhibition and 13% Bodipy-FL-Prazosin efflux inhibition). Among dimers, free carboxy group proved unfavorable which may be due to poor cell penetration. However, dually protected dimer 9 as well as its Boc hydrolyzed analog 11 both showed identical inhibition of P-gp efflux function as compared to acid 10 and zwitterion 12. The [125I]-IAAP labeling inhibition results for these dimer derivatives (10, <10%; 11, 87%; 12, 23%) were corresponding to the data from the efflux assays. These results suggest that the free acid group is not accepted; however the free amino group is rather tolerated in the P-gp binding pocket as frequently seen in P-gp modulators.[32] Next, further increasing the length to tripeptide, compound 13 showed impressive activity with 98% inhibition of Calcein-AM efflux by P-gp along with 75% inhibition of Bodipy-FL-Prazosin transport. The dose response curve for this analog against calcein-AM efflux function revealed an IC50 of 1.5 μM. In affirmation, the [125I]-IAAP labeling was inhibited by a significant extent of 91%. Next to this substantial enhancement, deprotection of compound 13 was effected at both ends. As observed for compound 10, the acid 14 was devoid of Calcein-AM or Bodipy-FL-Prazosin efflux inhibition potential (<10%) but showed moderate (60%) inhibition of [125I]-IAAP labeling of P-gp. From these results it is evident that larger molecular size coupled with extended hydrophobic surface area and lipophilic characteristics are the key features contributing towards increased inhibition of P-gp efflux function. The increase in the [125I]-IAAP labeling inhibition by compound 14 may be due to increase in the hydrophobic segment as compared to dimer acid 10 and lack of transport function inhibitory activity may be attributed to the fact that these being cell based assays requires compound to permeate through lipophilic cell bilayer. Persistently, the amine derivative 15 showed moderate inhibitory activity of 55% and 67% for the transport, yet 91% inhibition of [125I]-IAAP labeling. Subsequently, the trimer (13) was cyclized to obtain compound (17) i.e., QZ59S-SSS and evaluated against human P-gp efflux function. The observed results suggest that the linear and cyclic forms of the tripeptide structure are equipotent based on the inhibition of Calcein-AM efflux (% inhibition = 97%, IC50 = 1.5 μM), Bodipy-FL-Prazosin assay (89%) and [125I]-IAAP labeling assay (87%). The results obtained from trimer analogs further reinforce the notion that increased molecular size and hydrophobicity contribute towards inhibition of P-gp efflux function. Since increasing molecular size and hydrophobicity proved beneficial to inhibit P-gp efflux activity, hereon, we decided to synthesize only the lipophilic linear structure protected at both ends and the cyclic form for the higher analogues such as tetramer and hexamer. In this direction, four units of thiazole containing linear tetramer (18) was tested to find that the cell based inhibitory activity (59% and 81%), though decreased by 1.6-fold as compared to that of homolog 13 despite increase in [125I]-IAAP labeling inhibition (99%). Surprisingly, the linear hexamer (19) lacked any significant P-gp efflux inhibitory activity (5% and 12%) in transport assays. Additionally, cyclic tetramer (20) which showed moderate inhibition of efflux transports (62% and 79%) demonstrated appreciable inhibition of [125I]-IAAP labeling (95%) of P-gp. Similar to its linear analogue, cyclic hexamer (21) also proved to be ineffective in transport assays (<5% and 15%). It was interesting to note that linear and cyclic hexamers (19 and 21) did not inhibit Calcein-AM or Bodipy-FL-Prazosin transport significantly, although these derivatives significantly inhibited photoaffinity labeling of [125I]-IAAP (94% for 19 and 91% for 21). This suggests that the hexamer compounds interact with P-gp contrary to the transport assay results. These discrepancies may be ascribed to decrease in ability of the higher analogues of the trimer compounds to cross the cell membrane in the short-term transport assays.
Functionally, P-gp possesses ATPase activity that is stimulated by many substrates.[33,34] ATPase stimulation activity for selected derivatives (13-15, 17-23) was determined using membranes of P-gp expressing Hive-Five insect cells (Table 3). Here as well, the linear (13, fold stimulation = 2.80) and the cyclic (17, fold stimulation = 2.81) trimer analogs were found to stimulate the ATPase activity equivalently. Corresponding to the above transport assays, the trimer acid (14, fold stimulation = 1.47) was less effective whereas increase in the ATPase activity by the amine derivative (15, fold stimulation = 2.16) was comparable to that of compound 13. For the higher analogues, the ATPase activity fold stimulation data was consistent with the results from the labeling assay which suggest that these compounds interact with P-gp. For comparison, we used a well established substrate verapamil,[35] which is also a good stimulator of ATPase activity[33,34] and found that 50 μM verapamil gave 2.5-fold stimulation of P-gp ATPase activity (see legend to Table 3). Nonetheless, the results here suggest that the cell membrane crossing ability decreases with increase in the size of the molecule above that of the trimer length structures. Furthermore, to understand the binding mechanism of oligomers to the drug-binding pocket of homology model of human P-gp[13,36] at molecular level, we performed glide docking using ABCB1-QZ59Se-RRR (site-1), ABCB1-QZ59Se-SSS (site-2), ABCB1-verapamil (site-3), site common to above three sites (site-4)[5] and ATP-binding site following the protocol mentioned in our previous studies.[13,36] The binding energy data for the docked poses of oligomers were compared at all sites and QZ59Se-RRR binding site of P-gp i.e., site-1, was found to be the most favorable site for binding of linear trimer analog (13) which was also supported by the photoaffinity labeling assay with substrate [125I]-IAAP (Table 2).
Table 3.
Effect of selected thiazole derivatives on vanadate-sensitive ATPase activity of P-glycoprotein
| Compd | ATPase activity (nmoles Pi/min/mg protein)[a] |
Fold stimulation |
|---|---|---|
| 13 | 78 | 2.80 |
| 14 | 41.40 | 1.47 |
| 15 | 60.70 | 2.16 |
| 17 | 78.75 | 2.81 |
| 18 | 83.70 | 2.98 |
| 19 | 54.30 | 1.93 |
| 20 | 42.15 | 1.50 |
| 21 | 77.20 | 2.75 |
| 22 | 89.4 | 3.31 |
| 23 | 92.3 | 3.42 |
Crude membranes (100 μg protein/ml) from P-gp expressing High-Five cells were incubated at 37 °C with 2.5 μM of selected derivatives in the presence and absence of 0. 3 mM sodium orthovanadate in ATPase assay buffer for 5 min, and the vanadate-sensitive ATPase activity of P-gp was determined as described.[44] The vanadate-sensitive ATPase activity was calculated as nmoles Pi/min/mg protein and expressed as fold stimulation with respect to basal ATPase activity (in the presence of solvent DMSO) taken as 1.0. With the same membranes 50 μM verapamil stimulated the ATPase activity of P-gp by 2.5-fold (70 nmoles Pi/min/mg protein).The values are an average of two independent experiments.
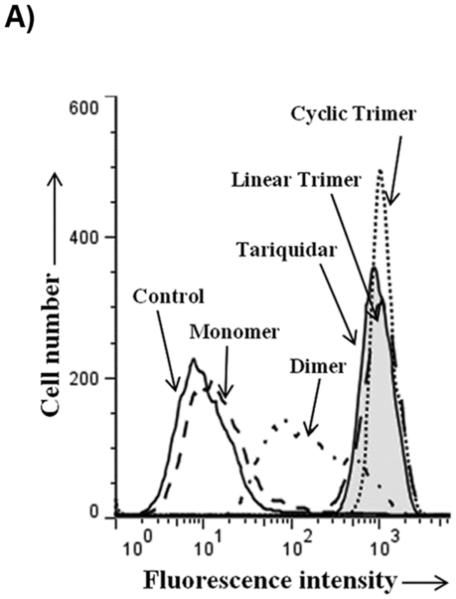
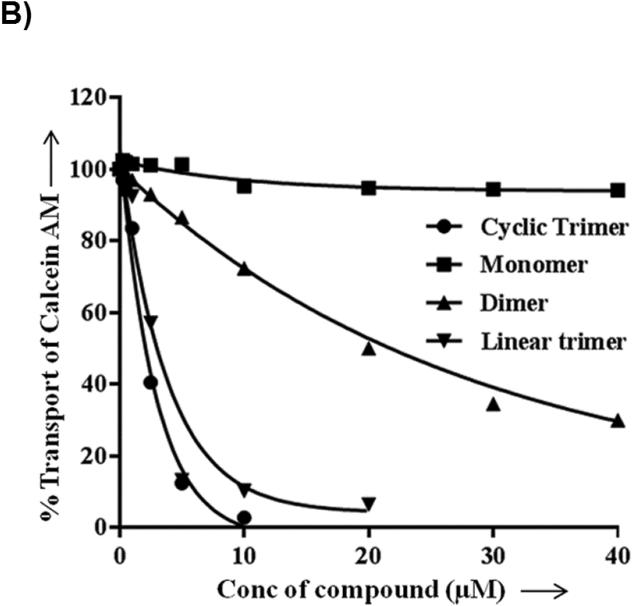
For testing the potency of synthesized derivatives, the flow cytometer based transport assay using fluorescent substrates was used. The Figure 2A shows a typical histogram of effect of selected compounds on Calcein-AM efflux by P-gp. It is clear from the Figure that linear trimer (13) is as effective as cyclic form (17) at 5 μM, whereas dimer (9) is moderately and monomer (5) is least effective. For comparison, inhibition of calcein-AM efflux with 1 μM tariquidar is also shown in Figure 2A (tainted filled trace). The Figure 2B depicts concentration-dependent effect of these three compounds and from the inhibition curves, IC50 (concentration required for 50% inhibition) were determined. The IC50 value for the linear (13) and cyclic (17) trimers was similar (1.5 μM; see legend to Table 1), whereas for the dimer (9), it was much higher (20 μM; legend to Figure 2B).
Figure 2. Inhibition of P-gp-mediated Calcein-AM transport by various derivatives.
HeLa cells transduced with wild-type P-gp BacMam virus were evaluated for transport function using calcein-AM (0.5 μM) as described in Experimental Section. A) The reversal of calcein-AM transport was carried out in the presence of solvent DMSO (control, solid line), or in the presence of various synthesized derivatives at 5 μM. The reversal by monomer (5) (dashed line), dimer (9) (complex line), linear trimer (13) (long dashed line), and cyclic trimer (QZ59S-SSS, 17) (dotted line) are compared with tariquidar (1 μM), which is a known inhibitor of P-gp (tinted filled trace). B) BacMam P-gp transduced HeLa cells were assayed for calcein-AM transport in the presence of increasing concentrations of monomer (5) (filled squares), dimer (9) (filled triangles), linear trimer (13) (filled inverted triangles) and cyclic trimer (QZ59S-SSS, 17) (filled circles). The transport in the absence of these compounds was taken as 100%. The results are represented as an average of two independent experiments. The IC50 value for the dimer (9) was 20 μM and that for both linear trimer (13) and cyclic trimer (17) was 1.5 μM.
The data obtained so far suggests the trimer length, linear as well as cyclic, to be optimum for blocking P-gp efflux activity. To this end, we synthesized two derivatives (22 and 23) of dimer acid (10) by coupling with corresponding methoxy substituted arylalkyl amines to mimic the trimer length. A critical role of methoxy substituted arylalkyl amines in tariquidar and elacridar, towards P-gp inhibition prompted us to choose these pharmacophores. Further, we chose dimer acid in spite of its inactivity because protected dimer 9 was found to be active which indicates a possible role of carboxyl terminal extension in enhanced binding. Also, we wanted to limit the size of the resulting chain to trimer because higher than trimer analogs proved ineffective. Indeed, these compounds showed significant inhibition of efflux of both Calcein-AM and Bodipy-FL-Prazosin (22, % inhibition = 97% and 88% and 23, % inhibition = 91% and 87%) as well as [125I]-IAAP labeling (22, % inhibition = 100% and 23, % inhibition = 95%) of P-gp. The IC50 values for inhibition of Calcein-AM efflux for both 22 and 23 were found to be 2.0 μM (see legend to Table 1). Both 22 and 23 also stimulated ATPase activity of P-gp by 3.31 and 3.42-fold, respectively.
In general, logP values of resulting compounds may have major role in the activity of compounds as observed in various QSAR analyses on P-gp inhibitors.[37-39] It is evident that lipophilicity contributes significantly for high P-gp inhibitory activity. These studies have also described that molecules with logP values between 3 and 6 have been shown to exhibit higher mean efflux ratios.[37-39] Interestingly the QikProp derived clogP of potent trimer analogues (13, 17, 22 and 23) were found to be within the range of 3 and 6. However, the higher oligomers namely tetramer and hexamer derivatives 18-21 (clogP = 6-9) were in the higher range of clogPs which could have been a possible barrier with the inhibitory activity of P-gp, whereas inactive zwitterion compounds were found to be in negative clogP values. This is also evident from the graphs illustrated in the Supplementary Figure S1 A-D (Supporting information). There is a clear correlation between size and potency of derivatives till ~850 Dalton for inhibition of transport but for inhibition of [125I]-IAAP incorporation and stimulation of ATPase activity, which is not a transport assay, the same does not hold true.
Molecular docking
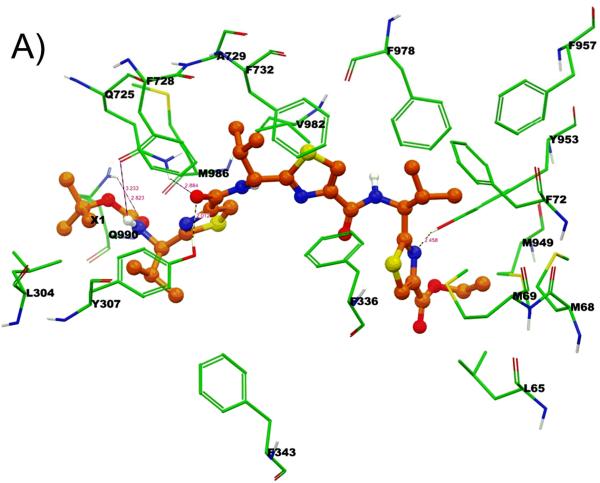
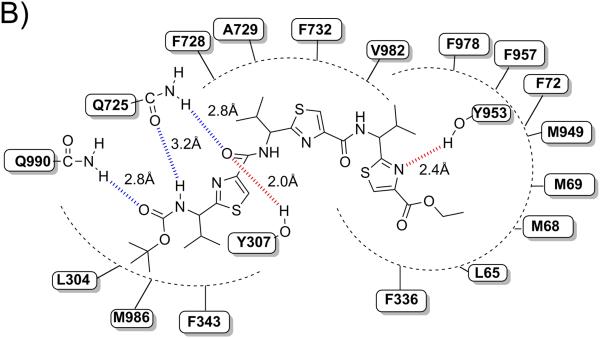
Binding model of linear trimer (13) (labeled as X1) is shown in Figure 3 (A and B). The tert-butyl group of Boc is involved in hydrophobic contacts with the side chains of residues L304 and M986, whereas the carbamate group of Boc enter into electrostatic interactions with Q725 (-NH---O(H2N)C-Q725, 3.2 Å) and Q990 (-C(H3C)3-O---H2N(O)C-Q990, 2.8 Å). The thiazole ring nitrogen atom next to the carbamate group is involved in electrostatic interaction with Q725 (N---H2N(O)C-Q725, 2.2 Å). Further, carbonyl oxygen atom attached to the same thiazole ring was shown to have critical hydrogen bonding interaction with Y307 (-CO---HO-Y307, 2.0 Å). Thiazole rings and the isopropyl groups are responsible for the significant interaction with M69, F72, F336, F343, F728, A729, F732, F957, F978 and V982. The ethyl group of ester function is located in the vicinity of hydrophobic residues L65, M68 and M949. Thiazole ring nitrogen atom near to the ester group is shown to have another hydrogen bonding interaction with Y953 (-CO---HO-Y953, 2.4 Å).
Scheme 3.
Reagents and conditions: (a) BOP, HOBt, DIEA, CH2Cl2:DMF (4:1), rt, 18 h; (b) NaOH, THF:Methanol:H2O (10:2:3), rt, 4 h; (c) TFA, CH2Cl2, rt, 12 h; (d) FDPP, DIEA, anhydrous ZnCl2, CH2Cl2:DMF (4:1), rt, 5 days
Among these residues, Y307, Q725, F728 and V982 may have significant role in interactions, as these residues were found to be involved with most of the ligands during docking at different drug binding sites of human P-gp. These residues were also shown to be critical by labeling with methanethiosulfonate-verapamil (MTS-verapamil).[40]
Conclusion
We synthesized a series of (S)-valine-derived thiazole containing cyclic and non-cyclic peptidomimetic oligomers to identify the optimal structural requirements for potent inhibition of human P-gp. Based on a set of 17 compounds from monomer to hexamer, it was revealed that linear- (13) and cyclic trimer (17) oligomers of (S)-valine-derived thiazole units were found to be the most potent inhibitors of human P-gp (IC50 = 1.5 μM) and their potency is comparable to third generation inhibitors (e.g., tariquidar) of this transporter. In addition, the analysis also indicated the importance of the molecular size, lipophilicity and hydrophobic contact surface area of the synthesized compounds. Cyclic trimer and linear trimer being equipotent, future studies can focus on non-cyclic versions of various known cyclic peptides. Binding model of compound (13) within the drug-binding site on the homology model of human P-gp represents an opportunity for future optimization with linear trimer based on the present interactions. Two representative dimer analogs 22 and 23 were also synthesized to verify the hypothesis that maintaining the length up to trimer will improve inhibition of P-gp efflux function. Indeed, 22 and 23 were shown to have significant inhibitory capacity on human P-gp efflux transport. Therefore, future SAR will be mainly devoted to dimer derivatives modified to mimic trimer length to obtain highly active inhibitors specific for human P-gp.
Experimental Section
Chemistry-General
Chemicals were purchased from Aldrich Chemical Co. (Milwaukee, WI), AK scientific (Union City, CA), Oakwood Products (West Columbia, SC), Alfa Aesar (Ward Hill, MA) and TCI America (Portland, OR) and were used as received. All compounds were checked for homogeneity by TLC using silica gel as stationary phase. Melting points were determined on Thomas Hoover capillary melting point apparatus and are uncorrected. NMR spectra were recorded on a Bruker 400 Avance DPX spectrometer (1H at 400 MHz) outfitted with a z-axis gradient probe. The chemical shifts for 1H NMR are reported in parts per million (δ ppm) downfield from tetramethylsilane (TMS) as an internal standard. The 1H NMR data are reported as follows: chemical shift, multiplicity s (singlet), d (doublet), t (triplet), dd (doublet of doublets), m (multiplet) and bs (broad singlet), respectively. Flash chromatography was performed using silica gel (0.060–0.200 mm) obtained from Dynamic adsorbents. High pressure liquid chromatography (HPLC) was performed using Agilent 1260 Infinity system employing Dynamax silica gel cartridge column (30 cm × 10 mm internal diameter). The elutions were monitored at UV 254 nm. Optical rotations of the chiral compounds were measured using PerkinElmer 241 Polarimeter with chloroform as solvent; concentration (c) is expressed as g/100 mL. Chiral HPLC analysis was performed using Dionex Ultimate 3000 Series instrument. The compounds were dissolved in methanol and injected (20 μL) into the chiralpak 1A column (Daicel Corp., Fort Lee, NJ) with stationary phase as amylose tris (3,5-dimethylphenylcarbamate) immobilized on 5 μm silica-gel. The chiral homogeneity was ensured using an isocratic mobile phase (1:1 n-hexane:ethyl acetate) eluting at a flow rate of 1 mL/min and monitored at UV 370 nm.
Synthesis
Method A. General procedures for ester hydrolysis of thiazole amino acid derivatives
Thiazole ethyl ester derivatives (0.01 M) were added in a mixture of solvents [THF: Methanol: Water (10:2:3)], and cooled to 0°C. Sodium hydroxide (10 equiv) was added and the mixture was stirred at RT for 12 h. The reaction mixture was then concentrated in vacuo and partitioned between ethyl acetate (30 mL) and water (20 mL). The aqueous phase containing compound was collected and acidified to pH 4 with 10% KHSO4 and then extracted with ethyl acetate (3 × 20 mL). The organic fractions were dried over sodium sulfate (Na2SO4) and concentrated under reduced pressure to yield the carboxylic acid derivatives.
Method B. General procedures for Boc-N-deprotection of thiazole amino acid derivatives
Trifluoroacetic acid (42 equiv) was added drop wise to a solution of N-Boc protected thiazoles in CH2Cl2 (1 mL/mmol) at 0 °C and the solution was stirred at RT under nitrogen atmosphere for 12 h. After completion of the reaction, the solvent was removed in vacuo followed by co-evaporation of the residual solvent with toluene (3 times). The remaining crude mass was added to water (20 mL) acidified with 2 M HCl and extracted with ethyl acetate (30 mL). To the aqueous phase containing amine salt was added saturated aqueous NaHCO3 and extracted with ethyl acetate (3 × 30 mL). The organic layers were washed with brine, dried over Na2SO4 and evaporated. The concentrate obtained was triturated with diethyl ether and dried in vacuo to afford free amine derivatives.
Method C. General procedures for peptide coupling to obtain linear thiazole derivatives
Di-isopropylethylamine (DIEA) (1.5 equiv) was added to the stirred, suspension of the carboxylic acid derivatives (1.0 equiv), in 4:1 CH2Cl2: DMF (0.25 M) or di-methyl acetamide (DMA) (0.10 M). The mixture was cooled to 0°C, and then BOP reagent (1.5 equiv) in DMF or HCTU (1.5 equiv.) in DMA, followed by HOBt (1.5 equiv) were added. The solution was stirred at 0°C for 10 min, then a pre-cooled solution of the thiazole amine TFA salt (1.3-1.5 equiv) in DMF (or DMA) and DIEA (1.5 equiv) were added and stirred at rt for 12-15 h. The reaction mixture was then concentrated using vacuum rotary evaporator followed by partitioning of the residual mass between ethyl acetate and aq. citric acid (10% w/v). The aqueous layer was repeatedly extracted with ethyl acetate (4 times). The combined organic fractions were washed sequentially with saturated aq. NaHCO3 and brine. This was then dried over Na2SO4 with subsequent evaporation under reduced pressure. The residue was purified by flash chromatography on silica gel using n-hexane:ethyl acetate (1:1) as eluent to give the required compounds.
Method D. General procedures for cyclization through peptide coupling of thiazole amino acids
The zwitterion derivatives were suspended in a mixture of anhydrous CH2Cl2: DMF (4:1) (20 mL/mmol) under inert atmosphere with subsequent addition of DIEA (3 equiv), pentafluorophenyl diphenylphosphinate (FDPP; 3 equiv) and anhydrous ZnCl2 (1 equiv) (to induce cyclization to trimer).[28] The reaction mixture was stirred at RT and monitored by TLC. Upon disappearance of the starting compound (5 days), the reaction was quenched with sat. aq. NaHCO3 and solvent was concentrated in vacuo. The residual mixture was partitioned between dichloromethane (50 mL) and water (25 mL). The aqueous fraction was back extracted with CH2Cl2 (2 × 50 mL) and combined organic layers were washed successively with 10% aq. citric acid (2 × 25 mL), water (25 mL) and brine (50 mL), dried over Na2SO4 followed by evaporation under reduced pressure to leave a mixture of cyclic peptide products, which were separated by silica gel column chromatography (ethyl acetate: n-hexane 7:3) or by preparative HPLC using gradient solvent system 60-90 % acetonitrile in mixture of 0.1% TFA in water with the flow rate of 1 mL/min to obtain the pure cyclic peptides.
(S)-(1-Carbamoyl-2-methylpropyl)carbamic acid tert-butyl ester (2)
To a stirred solution of N-Boc-(S)-valine (1) (3.00 g, 13.80 mmol) in anhydrous tetrahydrofuran (THF) (20 mL), was added isobutyl chloroformate (2.26 g, 16.56 mmol) and N-methylmorpholine (NMM) (1.68 g, 16.56 mmol) under nitrogen atmosphere and was allowed to stir for 4 h at −20°C. Afterwards, excess (20 mL) of aqueous ammonia (30%) was added and the resulting biphasic mixture was stirred at room temperature for 4 h. After completion of reaction, THF was removed under reduced pressure. The residual mixture was extracted with ethyl acetate (EtOAC) (3 × 30 mL). The combined organic fraction was washed with 1N KHSO4 and brine and then dried over anhydrous Na2SO4. After evaporation of the solvent, the product was recrystallized by trituration with EtOAC: n-hexane (3:8) to provide the amide (2) as a white solid (2.87 g, 96%); mp 156-159°C (Lit. 160-161°C);[25] 1H NMR (400 MHz; DMSO-d6; TMS) δ 7.29 (s, 1H), 7.03 (s, 1H), 6.54 (d, 1H, J = 8.9 Hz), 3.73 (t, 1H, J = 8.9 Hz), 1.91-1.89 (m, 1H), 1.38 (s, 9H), 0.85 (d, 3H, J = 7.1 Hz), 0.81 (d, 3H, J = 7.1 Hz).
(S)-(2-Methyl-1-thiocarbamoylpropyl)carbamic acid tert-butyl ester (3)
Compound 2 (2.50 g, 11.56 mmol) was dissolved in dry THF (30 mL) under nitrogen atmosphere. Lawesson's reagent (9.35 g, 23.12 mmol) was added under a well-ventilated fume hood, and the resulting suspension was stirred at 50°C for 16 h. After cooling to RT, the reaction was quenched with saturated aqueous NaHCO3 (20 mL) and then diluted with EtOAc (40 mL), and stirred at RT for additional 2 h. The layers were separated and the organic layer was washed with brine (20 mL), dried over Na2SO4 and concentrated in vacuo. Purification by flash column chromatography (5-25% EtOAc:n-hexane), yielded title compound (3) as a yellow solid (2.15 g, 80%); mp 72-76°C (Lit. 84-85°C);[21] Rf = 0.6 (EtOAc: n-hexane 2:3); 1H NMR (400 MHz; DMSO-d6; TMS) δ 9.63 (s, 1H), 9.18 (s, 1H), 6.52 (d, 1H, J = 9.1 Hz), 3.55-3.53 (m, 1H), 1.90-1.88 (m, 1H), 1.43 (s, 9H), 0.87 (d, 3H, J = 7.1 Hz), 0.84 (d, 3H, J = 7.1 Hz).
(S)-2-(1-tert-Butoxycarbonylamino-2-methylpropyl)thiazole-4-carboxylic acid ethyl ester (5)[23]
The solution of 3 (2.00 g, 8.61 mmol) in dry DME (30 mL) was cooled to −15°C with subsequent addition of KHCO3 (7.76 g, 77.49 mmol) under nitrogen atmosphere. The resulting suspension was vigorously stirred for 15 min, followed by dropwise addition of ethyl bromopyruvate (2.01 g, 10.33 mmol) under inert atmosphere. The reaction was then continued at −15°C for 1 h after which it was allowed to warm gradually to RT and stirred for 12 h. Then, the mixture was filtered through celite pad. The filtrate was concentrated in vacuo at 40°C to yield a dark colored crude mass of hydroxythiazoline intermediate 4. The crude mass was further dissolved per se in dry DME (20 mL) under nitrogen and the resulting solution was cooled to −15°C. To this, a solution of 2,6-lutidine (7.38 g, 68.88 mmol) and TFAA (9.04 g, 43.05 mmol) in dry DME (10 mL) was added dropwise over a period of 40 min (large amounts of CO2 was released). The solution was stirred at −15°C for 3 h and then allowed to stir at RT for additional 12 h, afterwards; the reaction mixture was concentrated and then stirred with freshly prepared sodium ethoxide (3 equiv) in ethanol (20 mL). After 6 h, ethanol was evaporated under reduced pressure. The residual product was added with saturated aqueous NaHCO3 (10 mL) solution. The resulting mixture was extracted with ethyl acetate (3 × 30 mL). The combined organic layer was washed with citric acid (10%) and brine, dried over Na2SO4, and concentrated in vacuo. Purification of the residue by column chro-matography (5-35% EtOAc:n-hexane), afforded the title compound (5) as a white solid (1.04 g, 37%); mp 112-114°C (Lit. 114-116°C);[21] Rf = 0.5 (EtOAc: n-hexane 2:3); purity (HPLC, UV 254 nm) 95%; 1H NMR (400 MHz, DMSO-d6; TMS) δ 8.40 (s, 1H), 7.72 (d, 1H, J = 8.4 Hz), 4.61 (t, 1H, J = 7.5 Hz), 4.31-4.27 (m, 2H), 2.21-2.19 (m, 1H) 1.39 (s, 9H), 1.29 (t, 3H, J = 7.1 Hz), 0.88 (d, 3H, J = 6.7 Hz), 0.84 (d, 3H, J = 6.7 Hz).
(S)-2-(1-tert-Butoxycarbonylamino-2-methylpropyl)thiazole-4-carboxylic acid (6)
Compound 5 (0.90 g, 2.74 mmol) was hydrolyzed to obtain compound 6 according to method A as a white solid (0.75 g, 91%); mp 131-133°C; Rf = 0.20 (MeOH: CH2Cl2 5:95); purity (HPLC, UV 254 nm) 97%; 1H NMR (400 MHz; DMSO-d6; TMS) δ 12.34 (1H, s), 8.32 (1H, s), 7.70 (d, 1H, J = 8.4 Hz), 4.60 (t, 1H, J = 7.5 Hz), 2.21-2.19 (m, 1H) 1.39 (s, 9H), 0.88 (d, 3H, J = 6.7 Hz), 0.84 (d, 3H, J = 6.7 Hz).
(S)-2-(1-Amino-2-methylpropyl)thiazole-4-carboxylic acid ethyl ester (7)
Compound 5 (0.80 g, 3.50 mmol) was used to obtain compound 7 as a yellow solid (0.47 g, 85%) by using method B; mp 38-39°C; Rf = 0.30 (MeOH: CH2Cl2 5:95); purity (HPLC, UV 254 nm) 95%; 1H NMR (400 MHz; DMSO-d6; TMS) δ 8.50 (s, 1H), 5.10-4.80 (m, 1H), 4.39 (q, 2H, J = 7.0 Hz), 2.44-2.42 (m, 1H), 2.1 (bs, 2H), 1.38 (t, 3H, J = 7.0 Hz), 0.89 (d, 3H, J = 6.7 Hz), 0.86 (d, 3H, J = 6.8 Hz).
(S)-2-(1-Amino-2-methylpropyl)thiazole-4-carboxylic acid (8)
Compound 6 (0.50 g, 1.66 mmol) was first dissolved in CH2Cl2 (15 mL) in nitrogen atmosphere and cooled to 0°C. Trifluoroacetic acid (42 equiv.) was added dropwise and the solution was stirred at RT under nitrogen atmosphere for 12 h. After removal of dichloromethane, the residual mixture was co-evaporated with toluene (3 × 10 mL) to obtain salt form of compound 8 which was used further without purification.
(S)-2-(1-{[2-(1-tert-Butoxycarbonylamino-2-methylpropyl) thiazole-4-carbonyl]amino}-(S)-2-methylpropyl)thiazole-4-carboxylic acid ethyl ester (9)
Dimer (9) was obtained using compound 6 (0.45 g, 1.99 mmol) and 7 (0.50 g, 1.66 mmol) following method C as a white solid (0.45 g, 53%); mp 90-92°C (lit 91-93°C);[20] Rf = 0.40 (EtOAc: n-hexane 2:3); purity (HPLC, UV 254 nm) 98%; 1H NMR (400 MHz; DMSO-d6; TMS) δ 8.73 (d, 1H, J = 8.4 Hz), 8.46 (s, 1H), 8.23 (s, 1H), 7.74 (t, 1H, J = 6.9 Hz), 5.15–5.06 (m, 1H), 4.70 (bs, 1H), 4.32 (q, 2H, J = 7.0 Hz), 2.47-2.45 (m, 1H), 2.24-2.22 (m, 1H), 1.40 (s, 9H), 1.30 (t, 3H, J = 7.1 Hz), 0.97 (d, 3H, J = 6.8 Hz), 0.90-0.88 (m, 9H); ESI-MS(+) m/z = 533.18 (C23H34N4O5S2Na, [M+Na]+).
(S)-2-(1-{[2-(1-tert-Butoxycarbonylamino-2-methylpropyl) thiazole-4-carbonyl]amino}-(S)-2-methylpropyl)thiazole-4-carboxylic acid (10)
Dimer acid (10) was obtained by hydrolyzing dimer (9) (0.40 g, 0.78 mmol) according to method A as a white solid (0.33 g, 79%); mp 101-103°C [lit 101-106°C];[20] Rf = 0.20 (MeOH: CH2Cl2 5:95); purity (HPLC, UV 254 nm) >99%; 1H NMR (400 MHz; DMSO-d6; TMS) δ 8.69 (d, 1H, J = 8.4 Hz), 8.38 (s, 1H), 8.23 (s, 1H), 7.74 (d, 1H, J = 8.1 Hz), 5.11 (t, 1H, J = 8.4 Hz), 4.70 (t, 1H, J = 7.6 Hz,), 2.47-2.45 (m, 1H), 2.25-2.23 (m, 1H), 1.40 (s, 9H), 0.97 (d, 3H, J = 6.8 Hz), 0.92-0.85 (m, 9H) (acid peak was missing); ESI-MS(+) m/z = 505.15 (C21H30N4O5S2Na, [M+Na]+).
(S)-2-(1-{[2-(1-Amino-2-methylpropyl)thiazole-4-carbonyl]amino}-(S)-2-methylpropyl)thiazole-4-carboxylic acid ethyl ester (11)
Dimer amine (11) was obtained using dimer (9) (0.40 g, 0.78 mmol) following method B as a yellowish-white solid (0.12 g, 36%); mp 119-122°C [lit 120-123°C];[20] Rf = 0.20 (MeOH: CH2Cl2 5:95); purity (HPLC, UV 254 nm) 95%; 1H NMR (400 MHz; CD3OD; TMS) δ 8.62 (d, 1H, J = 8.7 Hz), 8.42 (s, 1H), 8.21 (s, 1H), 5.15-5.11 (m, 2H), 4.32 (q, 2H, J = 7.1 Hz), 2.47–2.40 (m, 2H), 1.30 (t, 3H, J = 7.1 Hz), 1.16 (m, 12H) (amine peak was missing); ESI-MS(+) m/z = 433.18 (C18H26N4O3S2Na, [M+Na]+).
(S)-2-(1-{[2-(1-Amino-2-methylpropyl)thiazole-4-carbonyl]amino}-(S)-2-methylpropyl)thiazole-4-carboxylic acid (12)
To the suspension of Dimer acid (10) (0.30 g, 0.62 mmol) in CH2Cl2 (15 mL) at 0°C was added trifluoroacetic acid (42 equiv) dropwise. After completion of the reaction, solvent was co-evaporated with toluene (3 vol). The crude mass was dissolved in ethyl acetate and washed with 10% aq. NaHCO3 (2 × 10 mL) and brine (2 × 10 mL). The organic extract was dried over Na2SO4, and then concentrated in vacuo. The residual mass was triturated with ether and hexane to afford the title compound 12 as a white solid (0.21 g, 86%); Rf = 0.20 (MeOH: CH2Cl2 5:95); purity (HPLC, UV 254 nm) 98%; 1H NMR (400 MHz; DMSO-d6; TMS) δ 8.69 (t, 1H, J = 8.9 Hz), 8.45 (d, 1H, J = 3.3 Hz), 8.40 (s, 1H), 5.18–5.12 (m, 1H), 4.70 (t, 1H, J = 7.6 Hz), 2.47-2.45 (m, 1H), 2.25-2.3 (m, 1H), 1.01-0.90 (m, 12H) (amine and acid peaks were missing). ESI-MS (+) m/z = 382.11 (C16H22N4O3S2Na, [M+Na] +)
(S)-2-(1-{[2-(1-{[2-(1-tert-Butoxycarbonylamino-2-methylpropyl)thiazole-4-carbonyl]amino}-(S)-2-methylpropyl)thiazole-4-carbonyl]amino}-(S)-2-methylpropyl) thiazole-4-carboxylic acid ethyl ester (13)
Linear trimer (13) was obtained using Dimer acid (10) (0.30 g, 0.62 mmol) and monomer amine (7) (0.17 g, 0.74 mmol) following method C as a white solid (0.16 g, 36%); purity (HPLC, UV 254 nm) 98%; Rf = 0.40 (EtOAc: n-hexane 1:1); 1H NMR (400 MHz; CDCl3; TMS) δ 8.07 (s, 1H), 8.06 (s, 1H), 8.04 (s, 1H), 7.95 (d, 1H, J = 9.2 Hz), 7.82 (d, 1H, J = 9.2 Hz), 5.39-5.32 (m, 2H), 5.11 (d, 1H, J = 8.7 Hz,), 4.89 (bs, 1H), 4.43 (q, 2H, J = 7.1 Hz), 2.71–2.60 (m, 2H), 2.41–2.30 (m, 1H), 1.45 (s, 9H), 1.40 (t, 3H, J = 7.1 Hz), 1.08-1.00 (m, 15H), 0.94-0.87 (d, 3H, J = 6.8 Hz); ESI-MS(+) m/z = 693.26 (C31H45N6O6S3, [M+H]+).
(S)-2-(1-{[2-(1-{[2-(1-tert-Butoxycarbonylamino-2-methylpropyl)thiazole-4-carbonyl]amino}-(S)-2-methylpropyl)thiazole-4-carbonyl]amino}-(S)-2-methylpropyl)thiazole-4-carboxylic acid (14)
Trimer acid (14) was obtained using linear trimer (13) (0.30 g, 0.43 mmol) following method A as a white solid (0.24 g, 83%); Rf = 0.20 (MeOH: CH2Cl2 5:95); purity (HPLC, UV 254 nm) 98%; 1H NMR (400 MHz; DMSO-d6; TMS) δ 12.95 (s, 1H), 8.73 (s, 2H), 8.38 (s, 1H), 8.28 (d, 1H, J = 5.6 Hz), 8.23 (bs, 1H), 7.74 (d, 1H, J = 9.2 Hz), 5.18 (d, 2H, J = 8.7 Hz), 4.68 (bs, 1H), 2.47–2.31 (m, 2H), 2.24-2.21 (m, 1H), 1.41 (s, 9H), 1.04-0.86 (m, 18H); ESI-MS(+) m/z = 687.21 (C29H40N6O6S3Na, [M+Na]+).
(S)-2-(1-{[2-(1-{[2-(1-Amino-2-methylpropyl)thiazole-4-carbonyl]amino}-(S)-2-methylpropyl)thiazole-4-carbonyl]amino}-(S)-2-methylpropyl)thiazole-4-carboxylic acid ethyl ester (15)
Trimer amine (15) was obtained using linear trimer (13) (0.40 g, 0.58 mmol) following method B as a white solid (0.11 g, 31%); Rf = 0.20 (MeOH: CH2Cl2 5:95); purity (HPLC, UV 254 nm) 95%; 1H NMR (400 MHz; DMSO-d6; TMS) δ 8.74-8.68 (m, 2H), 8.62 (bs, 2H), 8.47 (s, 2H), 8.30 (s, 1H), 5.21-5.10 (m, 2H), 4.74 (bs, 1H), 4.29 (q, 2H, J = 7.1 Hz), 2.47–2.31 (m, 2H), 2.29-2.27 (m, 1H), 1.28 (t, 3H, J = 7.1 Hz), 1.32-0.87 (m, 18H); ESI-MS(+) m/z = 593.00 (C26H37N6O4S3, 593.20, [M+H]+).
(S)-2-(1-{[2-(1-{[2-(1-Amino-2-methylpropyl)thiazole-4-carbonyl]amino}-(S)-2-methylpropyl)thiazole-4-carbonyl]amino}-(S)-2-methylpropyl)thiazole-4-carboxylic acid (16)
Trifluoroacetic acid (42 equiv) was added dropwise to a solution of trimer acid (14) in CH2Cl2 (15 mL) at 0°C and the solution was stirred at RT under nitrogen atmosphere for 15 h. After completion of the reaction, the solvent was co-evaporated with toluene (3 times). The obtained product was used further without purification. ESI-MS (+) m/z = 565.18 (C24H33N6O4S3, [M+H] +).
Cyclic Tris-thiazole amino acid (QZ59S-SSS) (17)
Compound 17 was obtained using compound 16 (0.80 g, 1.42 mmol) following method D as a white solid (0.25 g, 32%); purity (HPLC, UV 254 nm) ≥98%; mp 243-247°C; Rf = 0.45 (EtOAc:n-hexane 1:1); [α]D25= -102.64° (c 5.3, CHCl3); 1H NMR (400 MHz; CDCl3; TMS) δ 8.47 (d, 3H, J = 9.1 Hz), 8.09 (s, 3H), 5.43 (dd, 3H, J = 9.3 Hz, 5.8 Hz), 2.34-2.27 (m, 3H), 1.10 (d, 9H, J = 6.8 Hz), 1.05 (d, 9H, J = 6.8 Hz); ESI-MS(+) m/z = 547.15 (C24H31N6O3S3, [M+H+]).
Preparation of 17 through coupling and cyclization of 8
Alternatively, compound 17 was obtained using compound 8 (0.75 g, 2.45 mmol) following method D as a white solid (0.11 g, 24%).
(S)-2-(1-{[2-(1-{[2-(1-{[2-(1-tert-Butoxycarbonylamino-2-methylpropyl)thiazole-4-carbonyl]amino}-(S)-2-methylpropyl)thiazole-4-carbonyl]amino}-(S)-2-methylpropyl)thiazole-4-carbonyl]amino}-(S)-2-methylpropyl)thiazole-4-carboxylic acid ethyl ester (18)
The dimer acid (10) (0.40 g, 0.46 mmol) and the dimer amine (11) (0.23 g, 0.55 mmol) were coupled according to general procedure C to give the linear tetramer (18) as a white solid (0.23 g, 32%); purity (HPLC, UV 254 nm) 98%; 1H NMR (400 MHz; DMSO-d6; TMS) δ 8.77-8.70 (m, 3H), 8.45 (s, 1H), 8.27 (bs, 2H), 8.23 (s, 1H), 7.76 (d, 1H, J = 7.9 Hz), 5.19-5.07 (m, 3H), 4.70 (bs, 1H), 4.28 (q, 2H, J = 7.2 Hz), 2.47–2.42 (m, 3H), 2.27-2.23 (m, 1H), 1.39 (s, 9H), 1.31 (t, 3H, J = 7.1 Hz), 1.10–0.95 (m, 9H), 0.94-0.82 (m, 15H); ESI-MS(+) m/z = 897.2871 (C39H54N8O7S4Na, [M+Na]+).
Linear Hexamer (19)
The trimer acid (14) (0.10 g, 0.15 mmol) and the trimer amine (15) (0.11 g, 0.18 mmol) were coupled according to general procedure C and the crude product was purified by preparative HPLC using gradient solvent system of 60-90% acetonitrile in water with 0.1% TFA. Peak separated at 51 minute was characterized to be linear hexamer (19) by ESI-MS analysis. Compound was obtained as a white solid (0.002 g, 0.60 %); purity (HPLC, UV 254 nm) ≥99%; ESI-MS(+) m/z = 1261.48 (C55H74N12O9S6Na, [M+Na]+).
Cyclic Tetramer (20) and cyclic hexamer (21)
Cyclic tetramer (20) and cyclic hexamer (21) were obtained using dimer zwitterion (0.90 g, 2.74 mmol) following method D and were separated by preparative HPLC using gradient solvent system of 60-90% acetonitrile in water with 0.1% TFA at a flow rate of 1 mL/min. Both compounds were confirmed by single peaks in ESI-MS spectrum. The extremely low yield of cyclic hexamer permitted us to do ESI-MS and biological activity only.
Cyclic Tetramer (20)
Peak separated at 19 minute was cyclic tetramer (20) (0.001 g, 0.11%). mp. 150-153 °C; purity (HPLC, UV 254 nm) 99%; Rf = 0.45 (EtOAc:n-hexane 2:1); 1H NMR (400 MHz; CDCl3; TMS) δ 8.66 (d, 1H, J = 8.9 Hz), 8.31 (d, 1H, J = 8.6 Hz), 8.18 (s, 1H), 8.15 (bs, 2H), 8.13 (s, 1H), 8.01 (d, 1H, J = 7.3 Hz), 7.89 (d, 1H, J = 10.0 Hz), 5.52-5.48 (m, 1H), 5.36 (t, 1H, J = 9.8 Hz), 5.25 (t, 1H, J = 7.8 Hz), 5.03 (t, 1H, J = 7.8 Hz), 2.69-2.67 (m, 1H), 2.43-2.41 (m, 2H), 2.29-2.27 (m, 1H), 1.23 (d, 3H, J = 6.7 Hz) 1.17 (d, 3H, J = 6.7 Hz), 1.12 (d, 3H, J =6.52 Hz), 1.07 (d, 3H, J = 6.8 Hz), 1.04-0.90 (m, 9H), 0.87 (d, 3H, J = 6.7 Hz); ESI-MS(+) m/z = 729.00 (C32H41N8O4S4, [M+H+])
Cyclic Hexamer (21)
Peak separated at 55 minutes was cyclic hexamer (21) (0.001 g, 0.11%). Purity (HPLC, UV 254 nm) ≥98%; ESI-MS (+) m/z = 1094, (C48H60N12O6S6, [M+H+]).
{1-[4-(1-{4-[(R)-(+)-1-(4-Methoxyphenyl)ethylcarbamoyl]thiazol-2-yl}-(S)-2-methylpropylcarbamoyl)thiazol-2-yl]-(S)-2-methylpropyl}carbamic acid tert-butyl ester (22)
Compounds 10 (0.10 g, 0.21 mmol) and 22a (0.06 g, 0.42 mmol) were reacted together by following general method C using HCTU to obtain compound 22 as a yellowish white solid (0.08 g, 63%); mp 64-66°C; Rf = 0.55 (EtOAc: n-hexane 1:1); purity (HPLC, UV 254 nm) 96%; 1H NMR (400 MHz; CDCl3; TMS) δ 8.66-8.61 (m, 1H), 8.43-8.36 (m, 1H), 8.22 (s, 1H), 8.16 (s, 1H), 7.74 (d, 1H, J = 7.8 Hz), 7.32 (d, 2H, J = 8.4 Hz), 6.88 (d, 2H, J = 8.4 Hz), 5.18-5.08 (m, 2H), 4.70-4.66 (m, 1H), 3.72 (s, 3H), 2.47-2.44 (m, 1H), 2.27-2.21 (m, 1H), 1.48 (d, 3H, J = 6.9 Hz), 1.40 (s, 9H), 0.97 (d, 3H, J = 6.2 Hz), 0.90-0.86 (m, 9H); ESI-MS(+) m/z = 638.2430 (C30H41N5O5S2Na, [M+Na]+).
[1-(4-{1-[4-(2, 4-Dimethoxybenzylcarbamoyl)thiazol-2-yl]-(S)-2-methylpropylcarbamoyl}thiazol-2-yl)-(S)-2-methylpropyl]carbamic acid tert-butyl ester (23)
Compounds 10 (0.10 g, 0.21 mmol) and 23a (0.07 g, 0.42 mmol) were reacted together by following general method C using HCTU to obtain compound 23 as a white solid (0.09 g, 68%); mp 70-74°C; Rf = 0.55 (EtOAc: n-hexane 1:1); purity (HPLC, UV 254 nm) 98%; 1H NMR (400 MHz; DMSO-d6; TMS) δ 8.68 (bs, 1H), 8.39 (d, 1H, J = 5.8 Hz), 8.23 (s, 1H), 8.19 (d, 1H, J = 3.6 Hz), 7.75 (bs, 1H), 7.08 (d, 1H, J = 8.2 Hz), 6.56 (s, 1H), 6.47 (d, 1H, J = 8.4 Hz), 5.17-5.07 (m, 1H), 4.69-4.67 (m, 1H), 4.37 (d, 2H, J = 5.8 Hz), 3.80 (s, 3H), 3.73 (s, 3H), 2.48-2.46 (m, 1H), 2.25-2.22 (m, 1H), 1.40 (s, 9H), 1.12-1.05 (m, 12H); ESI-MS(+) m/z = 654.2399 (C30H41N5O6S2Na, [M+Na]+).
Biological procedures
Cell lines
HeLa cells were cultured in DMEM media supplemented with 10% fetal bovine serum (FBS), 1% glutamine, and 1% penicillin as described earlier.[3]
Cloning and amplification of BacMam-P-gp virus and transduction of HeLa cells
The expression clones for P-gp were generated in pDest-625, as described previously.[3] These were then transformed into E. coli DH10Bac cells (Invitrogen, Carlsbad, CA), and plated on gentamycin, kanamycin, tetracycline, IPTG, and X-gal selective media as per the manufacturer’s protocols. White colonies were then selected from these plates and bacmid DNA was prepared by alkaline lysis method, which was further verified by PCR amplification across the bacmid junctions.
HeLa cells (2.5 million) were transduced with BacMam WT-P-gp virus at a titer of 50-60 viral particles per cell in 3 mL DMEM medium.[3] The DMEM medium (17 ml) was added after an hour and the cells were incubated further at 37°C in 5% CO2. After 3-4 h, 10 mM butyric acid was added and the cells were grown overnight at 37°C. After 24 h, the cells were trypsinized, washed, counted and analyzed by flow cytometry for testing potency of various derivatives to inhibit transport function of P-gp.
Fluorescent substrate transport assay
The ability of various derivatives to inhibit the transport function of human P-gp was checked with fluorescent substrates using flow cytometry as described previously.[41,42] Briefly, the baculovirus transduced cells were trypsinized and resuspended in IMDM medium containing 5% FBS. Cells were incubated with indicated concentration of the compounds or 1 μM tariquidar followed by calcein-AM (0.5 μM) for 10 min or Bodipy-FL-Prazosin (0.5 μM) for 45 min at 37°C as described previously.[3] The cells were washed with cold PBS, re-suspended in 300 μL of PBS with 0.1% Bovine Serum Albumin and analyzed. Fluorescence of Calcein or Bodipy-FL-Prazosin was measured with a FACSort flow cytometry equipped with a 488 nm argon laser and 530 nm bandpass filter. Percentage transport inhibition was derived when compared with inhibition obtained with the standard inhibitor, tariquidar at 1 μM. The results are plotted as an average of two experiments. The IC50 values for inhibition of calcein-AM efflux by selected derivatives were calculated using GraphPad Prism 5.0.
Photoaffinity labeling of P-gp with [125I]-IAAP
Crude membranes (1 mg protein/mL) from P-gp-expressing High-Five insect cells were photo labeled with [125I]-IAAP (2200 Ci/mmol) in the absence or presence of tested compounds, as described previously.[43] Briefly, crude membranes from P-gp expressing High-Five insect cells were incubated in the presence and absence of 10 μM thiazole-derivatives or 5 μM tariquidar in 50 mM Tris-HCl pH (7.5) and 6 nM [125I]-IAAP (2200 Ci/mmol). The samples were then photocross-linked and images were developed using phosphorimager. The density of P-gp band intensity was quantified and % inhibition of IAAP incorporation was calculated. The results are given as % inhibition as average of two independent experiments.
ATPase assay
Crude membrane protein (100 μg protein/ml) from High-Five insect cells expressing P-gp was incubated at 37 °C with 2.5 μM of thiazole-derivatives or 50 μM verapamil in the presence and absence of 0.3 mM sodium orthovanadate and the ATP hydrolysis was measured as described previously.[44] The vanadate-sensitive ATPase activity is expressed as nanomoles Pi/min/mg protein.
Molecular modeling
Ligand preparation
The structures of (S)-valine based thiazole derivatives were built using the fragment dictionary of Maestro v9.0 and subsequent energy minimization was performed by Macromodel program v9.7 (Schrödinger, Inc., New York, NY, 2009). The low-energy 3D structures of compounds were generated by LigPrep v2.3 as described previously.[13]
Protein preparation
The X-ray crystal structure of mouse P-gp in apo state (PDB ID: 3G5U) and in complex with inhibitors QZ59Se-RRR (PDB ID: 3G60), QZ59Se-SSS (PDB ID: 3G61)[5] and ATP bound (PDB ID: 1MV5) obtained from the RCSB Protein Data Bank were used to build the homology model of human ABCB1. The protocol for homology modeling is essentially same as reported before.[13] Refined human P-gp homology model was further used to generate different receptor grids by selecting QZ59Se-RRR (site-1) and QZ59Se-SSS (site-2) bound ligands, all amino acid residues known to contribute to verapamil binding (site-3), two residues (Phe728 and Val982) known to be common to sites 1-3 (site-4) and ATP binding site. Derivatives were docked on all the mentioned sites for comparison.
Docking protocol
Conformational library of ligands were docked at each of the generated grids (site-1 to site-4 and ATP binding site of P-gp using the “Extra Precision” (XP) mode of Glide program v5.0 (Schrödinger, Inc., New York, NY, 2009) with the default functions. The top scoring ligands conformation was used for graphical analysis. All computations were carried out on a Dell Precision 470n dual processor with the Linux OS (Red Hat Enterprise WS 4.0).
Supplementary Material
Figure 3. XP-Glide predicted binding mode of compound 13 with homology modeled P-gp.
A) 3D-model of binding mode of linear trimer (13) in homology modeled human P-gp drug binding cavity. Important amino acids present within 4 Å from the ligand are depicted as sticks with the atoms colored as carbon – green, hydrogen – white, nitrogen – blue, oxygen – red, sulfur – yellow) whereas the inhibitor is shown as ball and stick model with the same color scheme as above except carbon atoms are represented in orange. Electrostatic and hydrogen bonding interactions are shown in Å with the distances in dotted lines. B) Schematic representation diagram of the 3D model with important interactions observed in the complex of liner trimer (13) with the drug binding site residues of human P-gp. Electrostatic (blue dotted lines) and hydrogen bonding (red dotted lines) interactions are shown with the distances in Å.
Scheme 1.
Reagents and conditions: (a) (i) Isobutyl chloroformate, N-Methyl morpholine, THF, −20°C, 4 h; (ii) 30% NH4OH in excess, −20°C to rt, 4 h; (b) Lawesson's reagent, THF, 50°C, 16 h; (c) Ethyl bromopyruvate, KHCO3, DME, −15°C to rt, 12 h; (d) (i) TFAA, 2,6-Lutidine, DME, −15°C to rt, 15 h; (ii) NaOEt, Ethanol, −15°C to rt, 6 h; (e) NaOH, THF:Methanol:H2O (10:2:3), rt, 4 h; (f) TFA, CH2Cl2, rt, 12 h
Scheme 2.
Reagents and conditions: (a) BOP, HOBt, DIEA, CH2Cl2: DMF (4:1), RT, 18 h; (b) NaOH, THF: Methanol: H2O (10:2:3), RT, 4 h; (c) TFA, CH2Cl2, RT, 12 h
Scheme 4.
Reagents and conditions: (a) BOP, HOBt, DIEA, CH2Cl2: DMF (4:1), RT, 18 h
Scheme 5.
Reagents and conditions: a) FDPP, DIEA, CH2Cl2: DMF (4:1), RT, 5 days
Scheme 6.
Reagents and conditions: (a) HCTU, HOBt, DIEA, DMA, RT, 18 h
Acknowledgements
This research was supported by the Department of Pharmaceutical Sciences of St. John’s University and St. John’s University Seed Grant No. 579-1110 to T.T.T. Drs. NRP, KP, EEC and SVA were supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Financial support from Indian Council of Medical Research, New Delhi in the form of International Fellowship for Young Indian Biomedical Scientists to Dr. Nagarajan Rajendra Prasad is gratefully acknowledged (Indo/FRC/452(Y-19)/2012-13IHD). SS and TTT are thankful to Dr. Sanjai Kumar and Dibyendu Dana for assisting in preparative HPLC, ESI-MS and chiral HPLC analyses.
Footnotes
Dr. N. R. Prasad Present address: Department of Biochemistry and Biotechnology Annamalai University Annamalai 608002, Tamilnadu, India
Supporting information for this article is available on the WWW under http://www.chembiochem.org
References
- 1.Ambudkar SV, Dey S, Hrycyna CA, Ramachandra M, Pastan I, Gottesman MM. Annu. Rev. Pharmacol. Toxicol. 1999;39:361–398. doi: 10.1146/annurev.pharmtox.39.1.361. [DOI] [PubMed] [Google Scholar]
- 2.Schinkel AH, Jonker JW. Adv. Drug Deliv. Rev. 2003;55:3–29. doi: 10.1016/s0169-409x(02)00169-2. [DOI] [PubMed] [Google Scholar]
- 3.Shukla S, Schwartz C, Kapoor K, Kouanda A, Ambudkar SV. Drug Metab. Dispos. 2012;40:304–312. doi: 10.1124/dmd.111.042721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambudkar SV, Kim I, Sauna ZE. Eur. J. Pharm. Sci. 2006;27:392–400. doi: 10.1016/j.ejps.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 5.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dey S, Ramachandra M, Pastan I, Gottesman MM, Ambudkar SV. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10594–10599. doi: 10.1073/pnas.94.20.10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loo TW, Bartlett MC, Clarke DM. J. Biol. Chem. 2003;278:39706–39710. doi: 10.1074/jbc.M308559200. [DOI] [PubMed] [Google Scholar]
- 8.Lugo MR, Sharom FJ. Biochemistry. 2005;44:14020–14029. doi: 10.1021/bi0511179. [DOI] [PubMed] [Google Scholar]
- 9.Orlowski S, Mir LM, Belehradek Jr J, Garrigos M. Biochem. J. 1996;317:515–522. doi: 10.1042/bj3170515. Pt 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins CF. Nature. 2007;446:749–757. doi: 10.1038/nature05630. [DOI] [PubMed] [Google Scholar]
- 11.Kapoor K, Sim H, Ambudkar SV. Multidrug Resistance in Cancer: A Tale of ABC Drug Transporters, Vol. 1. Springer; New York: 2013. pp. 1–34. [Google Scholar]
- 12.Sekine I, Shimizu C, Nishio K, Saijo N, Tamura T. Int. J. Clin. Oncol. 2009;14:112–119. doi: 10.1007/s10147-008-0813-z. [DOI] [PubMed] [Google Scholar]
- 13.Shi Z, Tiwari AK, Shukla S, Robey RW, Singh S, Kim IW, Bates SE, Peng X, Abraham I, Ambudkar SV, Talele TT, Fu LW, Chen ZS. Cancer Res. 2011;71:3029–3041. doi: 10.1158/0008-5472.CAN-10-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shukla S, Wu CP, Ambudkar SV. Expert Opin. Drug Metab. Toxicol. 2008;4:205–223. doi: 10.1517/17425255.4.2.205. [DOI] [PubMed] [Google Scholar]
- 15.Tao H, Weng Y, Zhuo R, Chang G, Urbatsch IL, Zhang Q. ChemBioChem. 2011;12:868–873. doi: 10.1002/cbic.201100048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boesch D, Gavériaux C, Jachez B, Pourtier-Manzanedo A, Bollinger P, Loor F. Cancer Res. 1991;51:4226–4233. [PubMed] [Google Scholar]
- 17.Loor F. Expert Opin. Investig. Drugs. 1999;8:807–835. doi: 10.1517/13543784.8.6.807. [DOI] [PubMed] [Google Scholar]
- 18.Bagley MC, Dale JW, Merritt EA, Xiong X. Chem. Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 19.Bertram A, Hannam JS, Jolliffe KA, Gonzalez-Lopez Felix d. T., Pattenden G. Synlett. 1999:1723–1726. [Google Scholar]
- 20.Bertram A, Blake AJ, Turiso F. G. d., Hannam JS, Jolliffe KA, Pattenden G, Skae M. Tetrahedron. 2003;59:6979–6990. [Google Scholar]
- 21.Merritt EA, Bagley MC. Synthesis. 2007;2007:3535–3541. [Google Scholar]
- 22.Bredenkamp MW, Holzapfel CW, van Zyl WJ. Synth. Commun. 1990;20:2235–2249. [Google Scholar]
- 23.Aguilar E, Meyers AI. Tetrahedron Lett. 1994;35:2473–2476. [Google Scholar]
- 24.Boden CDJ, Pattenden G, Ye T. Synlett. 1995:417–19. [Google Scholar]
- 25.Moody C, Bagley M. J. Chem. Soc., Perkin Trans. 1. 1998:601–608. [Google Scholar]
- 26.Ramamoorthy PS, Gervay J. J. Org. Chem. 1997;62:7801–7805. [Google Scholar]
- 27.Hudson D. J. Org. Chem. 1988;53:617–24. [Google Scholar]
- 28.Blake AJ, Hannam JS, Jolliffe KA, Pattenden G. Synlett. 2000:1515–1518. [Google Scholar]
- 29.Martin C, Berridge G, Mistry P, Higgins C, Charlton P, Callaghan R. Br. J. Pharmacol. 1999;128:403–411. doi: 10.1038/sj.bjp.0702807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mistry P, Stewart AJ, Dangerfield W, Okiji S, Liddle C, Bootle D, Plumb JA, Templeton D, Charlton P. Cancer Res. 2001;61:749–758. [PubMed] [Google Scholar]
- 31.Fox E, Bates SE. Expert Rev. Anticancer Ther. 2007;7:447–459. doi: 10.1586/14737140.7.4.447. [DOI] [PubMed] [Google Scholar]
- 32.Priebe W, Van NT, Burke TG, Perez-Soler R. Anticancer Drugs. 1993;4:37–48. doi: 10.1097/00001813-199302000-00005. [DOI] [PubMed] [Google Scholar]
- 33.Sarkadi B, Price EM, Boucher RC, Germann UA, Scarborough GA. J. Biol. Chem. 1992;267:4854–4858. [PubMed] [Google Scholar]
- 34.Ambudkar SV, Lelong IH, Zhang J, Cardarelli CO, Gottesman MM, Pastan I. Proc. Natl. Acad. Sci. U. S. A. 1992;89:8472–8476. doi: 10.1073/pnas.89.18.8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Omote H, Al-Shawi MK. J. Biol. Chem. 2002;277:45688–45694. doi: 10.1074/jbc.M206479200. [DOI] [PubMed] [Google Scholar]
- 36.Tiwari AK, Sodani K, Dai CL, Abuznait AH, Singh S, Xiao ZJ, Patel A, Talele TT, Fu L, Kaddoumi A, Gallo JM, Chen ZS. Cancer Lett. 2013;328:307–317. doi: 10.1016/j.canlet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klopman G, Shi LM, Ramu A. Mol. Pharmacol. 1997;52:323–334. doi: 10.1124/mol.52.2.323. [DOI] [PubMed] [Google Scholar]
- 38.Crivori P, Reinach B, Pezzetta D, Poggesi I. Mol. Pharm. 2006;3:33–44. doi: 10.1021/mp050071a. [DOI] [PubMed] [Google Scholar]
- 39.Pajeva IK, Globisch C, Wiese M. ChemMedChem. 2009;4:1883–1896. doi: 10.1002/cmdc.200900282. [DOI] [PubMed] [Google Scholar]
- 40.Loo TW, Clarke DM. J. Biol. Chem. 2001;276:14972–14979. doi: 10.1074/jbc.M100407200. [DOI] [PubMed] [Google Scholar]
- 41.Tiberghien F, Loor F. Anticancer Drugs. 1996;7:568–578. doi: 10.1097/00001813-199607000-00012. [DOI] [PubMed] [Google Scholar]
- 42.Robey RW, Steadman K, Polgar O, Morisaki K, Blayney M, Mistry P, Bates SE. Cancer Res. 2004;64:1242–1246. doi: 10.1158/0008-5472.can-03-3298. [DOI] [PubMed] [Google Scholar]
- 43.Shukla S, Robey RW, Bates SE, Ambudkar SV. Drug Metab. Dispos. 2009;37:359–365. doi: 10.1124/dmd.108.024612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ambudkar SV. Methods Enzymol. 1998;292:504–514. doi: 10.1016/s0076-6879(98)92039-0. [DOI] [PubMed] [Google Scholar]
- 45.Sauna ZE, Ambudkar SV. Proc. Natl. Acad. Sci. U. S. A. 2000;97:2515–2520. doi: 10.1073/pnas.97.6.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.