Abstract
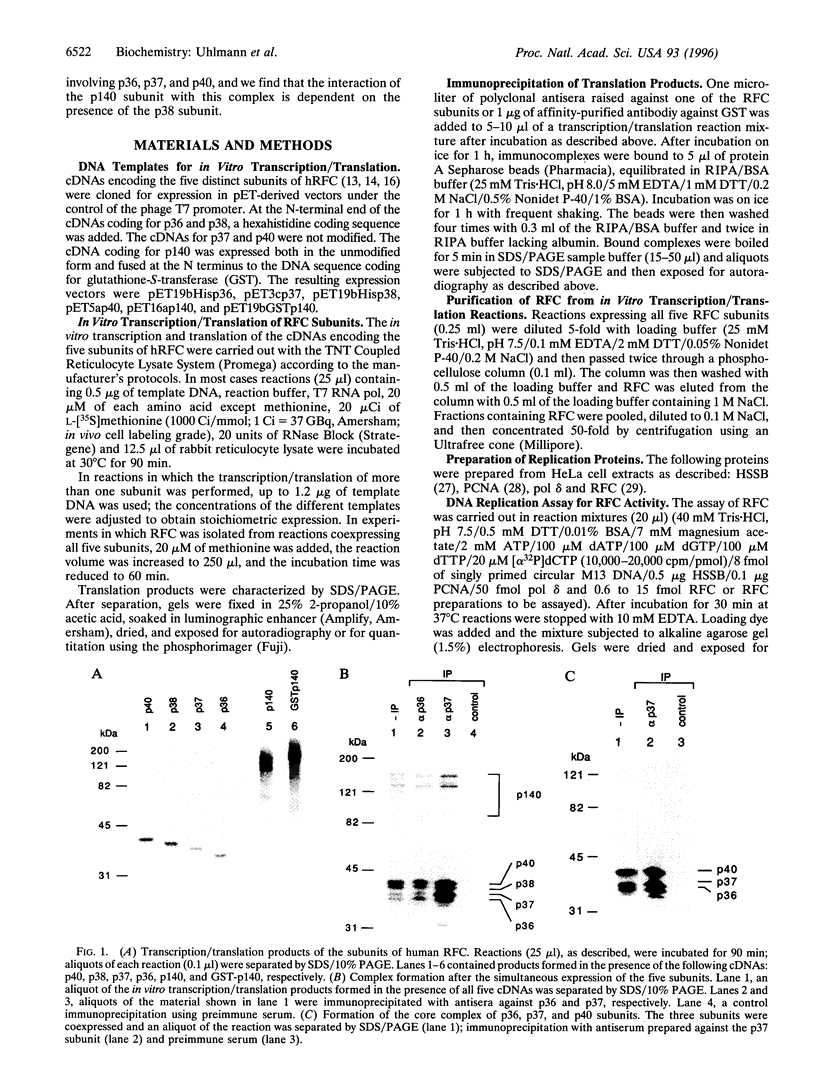
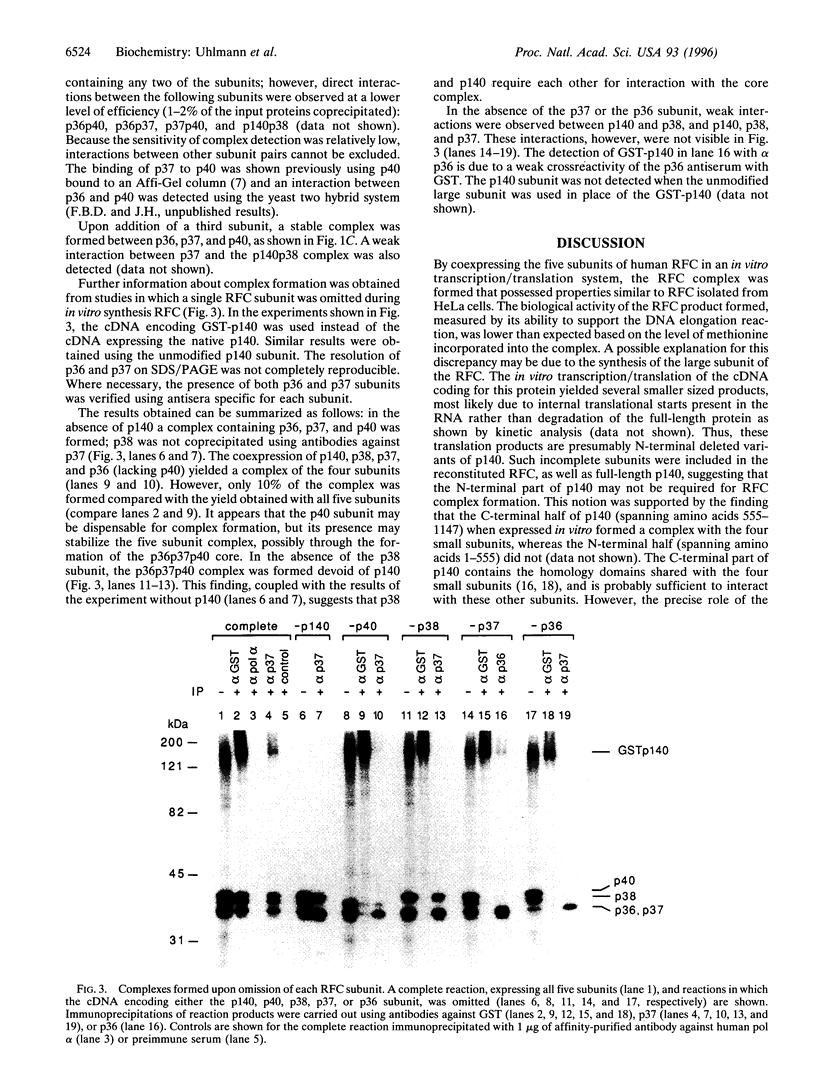
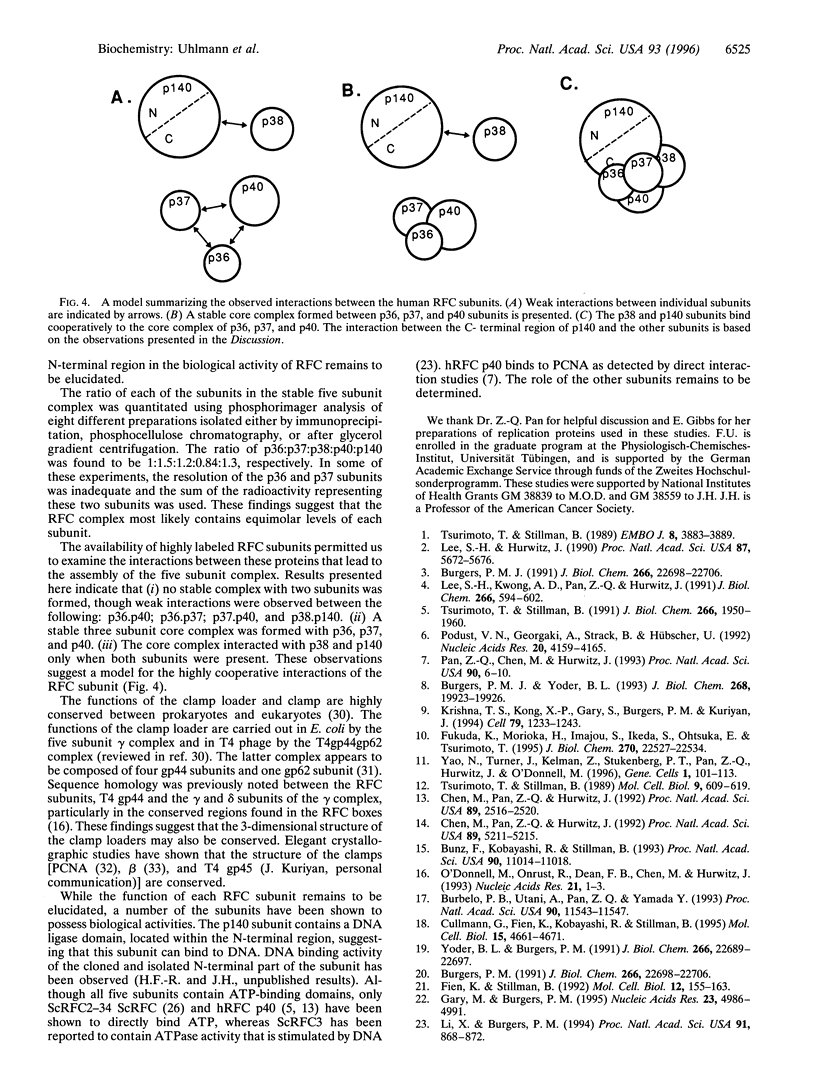
Replication factor C (RFC, also called Activator I) is part of the processive eukaryotic DNA polymerase holoenzymes. The processive elongation of DNA chains requires that DNA polymerases are tethered to template DNA at primer ends. In eukaryotes the ring-shaped homotrimeric protein, proliferating cell nuclear antigen (PCNA), ensures tight template-polymerase interaction by encircling the DNA strand. Proliferating cell nuclear antigen is loaded onto DNA through the action of RFC in an ATP-dependent reaction. Human RFC is a protein complex consisting of five distinct subunits that migrate through SDS/polyacrylamide gels as protein bands of 140, 40, 38, 37, and 36 kDa. All five genes encoding the RFC subunits have been cloned and sequenced. A functionally identical RFC complex has been isolated from Saccharomyces cerevisiae and the deduced amino acid sequences among the corresponding human and yeast subunits are homologous. Here we report the expression of the five cloned human genes using an in vitro coupled transcription/translation system and show that the gene products form a complex resembling native RFC that is active in supporting an RFC-dependent replication reaction. Studies on the interactions between the five subunits suggest a cooperative mechanism in the assembly of the RFC complex. A three-subunit core complex, consisting of p36, p37, and p40, was identified and evidence is presented that p38 is essential for the interaction between this core complex and the large p140 subunit.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bunz F., Kobayashi R., Stillman B. cDNAs encoding the large subunit of human replication factor C. Proc Natl Acad Sci U S A. 1993 Dec 1;90(23):11014–11018. doi: 10.1073/pnas.90.23.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbelo P. D., Utani A., Pan Z. Q., Yamada Y. Cloning of the large subunit of activator 1 (replication factor C) reveals homology with bacterial DNA ligases. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11543–11547. doi: 10.1073/pnas.90.24.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers P. M. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases delta and epsilon. J Biol Chem. 1991 Nov 25;266(33):22698–22706. [PubMed] [Google Scholar]
- Burgers P. M. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases delta and epsilon. J Biol Chem. 1991 Nov 25;266(33):22698–22706. [PubMed] [Google Scholar]
- Burgers P. M., Yoder B. L. ATP-independent loading of the proliferating cell nuclear antigen requires DNA ends. J Biol Chem. 1993 Sep 25;268(27):19923–19926. [PubMed] [Google Scholar]
- Chen M., Pan Z. Q., Hurwitz J. Sequence and expression in Escherichia coli of the 40-kDa subunit of activator 1 (replication factor C) of HeLa cells. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2516–2520. doi: 10.1073/pnas.89.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Pan Z. Q., Hurwitz J. Studies of the cloned 37-kDa subunit of activator 1 (replication factor C) of HeLa cells. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5211–5215. doi: 10.1073/pnas.89.12.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullmann G., Fien K., Kobayashi R., Stillman B. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol Cell Biol. 1995 Sep;15(9):4661–4671. doi: 10.1128/mcb.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fien K., Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol Cell Biol. 1992 Jan;12(1):155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K., Morioka H., Imajou S., Ikeda S., Ohtsuka E., Tsurimoto T. Structure-function relationship of the eukaryotic DNA replication factor, proliferating cell nuclear antigen. J Biol Chem. 1995 Sep 22;270(38):22527–22534. doi: 10.1074/jbc.270.38.22527. [DOI] [PubMed] [Google Scholar]
- Gary S. L., Burgers M. J. Identification of the fifth subunit of Saccharomyces cerevisiae replication factor C. Nucleic Acids Res. 1995 Dec 25;23(24):4986–4991. doi: 10.1093/nar/23.24.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell E. A., McAlear M. A., Rose D., Holm C. CDC44: a putative nucleotide-binding protein required for cell cycle progression that has homology to subunits of replication factor C. Mol Cell Biol. 1994 Jan;14(1):255–267. doi: 10.1128/mcb.14.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman Z., O'Donnell M. DNA replication: enzymology and mechanisms. Curr Opin Genet Dev. 1994 Apr;4(2):185–195. doi: 10.1016/s0959-437x(05)80044-9. [DOI] [PubMed] [Google Scholar]
- Kenny M. K., Lee S. H., Hurwitz J. Multiple functions of human single-stranded-DNA binding protein in simian virus 40 DNA replication: single-strand stabilization and stimulation of DNA polymerases alpha and delta. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9757–9761. doi: 10.1073/pnas.86.24.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X. P., Onrust R., O'Donnell M., Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992 May 1;69(3):425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Krishna T. S., Kong X. P., Gary S., Burgers P. M., Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994 Dec 30;79(7):1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Krishna T. S., Kong X. P., Gary S., Burgers P. M., Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994 Dec 30;79(7):1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Lee S. H., Eki T., Hurwitz J. Synthesis of DNA containing the simian virus 40 origin of replication by the combined action of DNA polymerases alpha and delta. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7361–7365. doi: 10.1073/pnas.86.19.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Hurwitz J. Mechanism of elongation of primed DNA by DNA polymerase delta, proliferating cell nuclear antigen, and activator 1. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5672–5676. doi: 10.1073/pnas.87.15.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Kwong A. D., Pan Z. Q., Hurwitz J. Studies on the activator 1 protein complex, an accessory factor for proliferating cell nuclear antigen-dependent DNA polymerase delta. J Biol Chem. 1991 Jan 5;266(1):594–602. [PubMed] [Google Scholar]
- Li X., Burgers P. M. Cloning and characterization of the essential Saccharomyces cerevisiae RFC4 gene encoding the 37-kDa subunit of replication factor C. J Biol Chem. 1994 Aug 26;269(34):21880–21884. [PubMed] [Google Scholar]
- Li X., Burgers P. M. Molecular cloning and expression of the Saccharomyces cerevisiae RFC3 gene, an essential component of replication factor C. Proc Natl Acad Sci U S A. 1994 Feb 1;91(3):868–872. doi: 10.1073/pnas.91.3.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov V., Maki S., Kawasaki Y., Leem S. H., Ono B., Araki H., Pavlov Y., Sugino A. The RFC2 gene encoding a subunit of replication factor C of Saccharomyces cerevisiae. Nucleic Acids Res. 1994 May 11;22(9):1527–1535. doi: 10.1093/nar/22.9.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell M., Onrust R., Dean F. B., Chen M., Hurwitz J. Homology in accessory proteins of replicative polymerases--E. coli to humans. Nucleic Acids Res. 1993 Jan 11;21(1):1–3. doi: 10.1093/nar/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z. Q., Chen M., Hurwitz J. The subunits of activator 1 (replication factor C) carry out multiple functions essential for proliferating-cell nuclear antigen-dependent DNA synthesis. Proc Natl Acad Sci U S A. 1993 Jan 1;90(1):6–10. doi: 10.1073/pnas.90.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust V. N., Georgaki A., Strack B., Hübscher U. Calf thymus RF-C as an essential component for DNA polymerase delta and epsilon holoenzymes function. Nucleic Acids Res. 1992 Aug 25;20(16):4159–4165. doi: 10.1093/nar/20.16.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T., Melendy T., Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990 Aug 9;346(6284):534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Multiple replication factors augment DNA synthesis by the two eukaryotic DNA polymerases, alpha and delta. EMBO J. 1989 Dec 1;8(12):3883–3889. doi: 10.1002/j.1460-2075.1989.tb08567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol Cell Biol. 1989 Feb;9(2):609–619. doi: 10.1128/mcb.9.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991 Jan 25;266(3):1950–1960. [PubMed] [Google Scholar]
- Yao N., Turner J., Kelman Z., Stukenberg P. T., Dean F., Shechter D., Pan Z. Q., Hurwitz J., O'Donnell M. Clamp loading, unloading and intrinsic stability of the PCNA, beta and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells. 1996 Jan;1(1):101–113. doi: 10.1046/j.1365-2443.1996.07007.x. [DOI] [PubMed] [Google Scholar]
- Yoder B. L., Burgers P. M. Saccharomyces cerevisiae replication factor C. I. Purification and characterization of its ATPase activity. J Biol Chem. 1991 Nov 25;266(33):22689–22697. [PubMed] [Google Scholar]
- Young M. C., Reddy M. K., von Hippel P. H. Structure and function of the bacteriophage T4 DNA polymerase holoenzyme. Biochemistry. 1992 Sep 22;31(37):8675–8690. doi: 10.1021/bi00152a001. [DOI] [PubMed] [Google Scholar]