Abstract
Observations of real time changes in living cells have contributed much to the field of cellular biology. Eluding the field thus far is the ability to image whole, living cells with nanometre resolution on a time scale that is relevant to dynamic cellular processes1,2. Here we investigate the kinetics of individual bacterial cell death using a novel high-speed atomic force microscope (AFM) optimized for imaging live cells in real time. The increased time resolution (13 seconds per image) allows the characterization of the initial stages of the action of the antimicrobial peptide (AmP) CM15 on individual Escherichia coli cells with nanometre resolution. Our results suggest that the killing process is a combination of a time-variable incubation phase (which takes seconds to minutes to complete) and a more rapid execution phase.
High resolution AFM images of live cells3 and cell fragments4 have resolved the nanoscale structure of cell walls5, the structural dynamics of single spores germinating6, the structural changes of bacteria treated with bactericidal enzymes7, and the location of specific binding sites8. One of the limitations that makes AFM unsuitable for studying many dynamic processes in cell biology9 is the long image acquisition time of several minutes for one high-resolution AFM image. Progress in AFM instrumentation has enabled imaging of single molecules at up to video rate, but this has been limited to small areas (hundreds of nanometres) and to flat samples10,11,12. We used a high-speed AFM specifically designed to bridge the gap between the improved AFM technology and the biologically relevant but challenging experiments interesting to cellular biology. The key benefit of this instrument is the combination of very soft but rapid imaging and a sufficient scan size to monitor multiple cells at once.
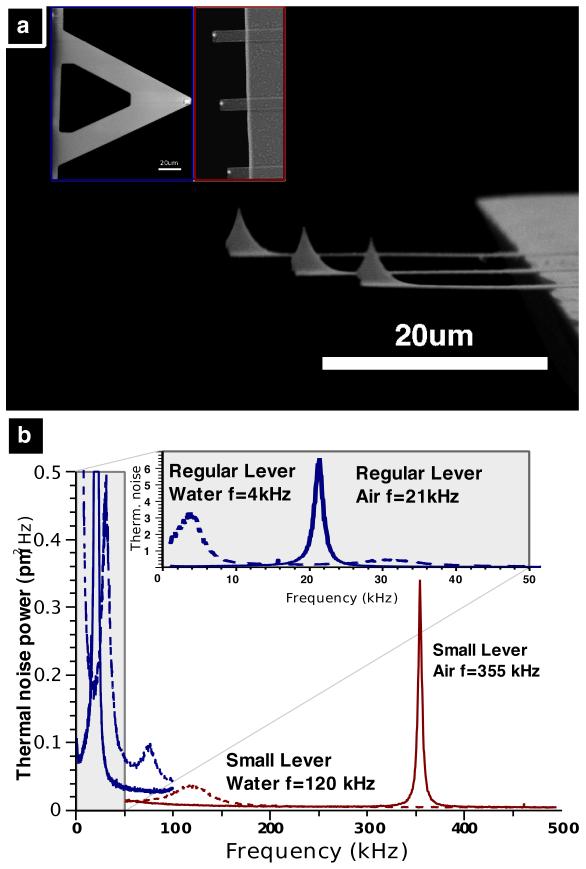
Imaging live bacteria in an aqueous environment is one of the most challenging applications for AFM13. The force interaction between the cantilever tip and the sample is critical; if the force is too high, the bacteria are damaged or disconnected from the surface, whereas if it is too low, information about the sample is lost. Maintaining this balance at high imaging speeds is achieved using prototype AFM components and micro-fabricated small cantilevers14 (~1000 times smaller mass than conventional cantilevers) with integrated tips. These cantilevers have resonance frequencies in liquid at ~100 kHz and spring constants between 300 and 1000 pN/nm. The increased resonance frequency and the reduced mass, quality factor and spring constant now allows us to increase the imaging speed in tapping mode while retaining low imaging forces on the bacteria15. Figure 1A shows an SEM image of the small cantilevers used for this research. The inset shows a comparison with a conventional cantilever (NPS-B, Veeco Metrology). Figure 1B shows the thermal spectra of the two cantilevers. The resonance frequency of the small cantilever is 17 times higher than that of the conventional cantilever in air and is 30 times higher in fluid.
Figure 1. Small AFM cantilevers for high-speed AFM.
A) SEM image of small SiN cantilevers (~10 μm wide, 100-350 nm thick, 20-30 μm long). The inset image compares the small levers (right) and a conventional lever (left) used for AFM imaging in fluid (Veeco NPS-D) at the same magnification. B) Thermal noise power spectra of regular and small cantilevers. In air (red solid line) the small cantilever’s first resonance frequency is ~350 kHz. In aqueous solution this drops to 100-120 kHz (red dashed line). The insert shows the thermal noise power spectra of an NP-S lever B with resonance frequencies of 21 kHz in air (blue solid line) and 4 kHz in aqueous solution (blue dashed line).
AmPs are a promising class of antimicrobials which have demonstrated activity against antibiotic resistant bacteria, parasites, viruses and fungi16,17,18,19. We used our high-speed AFM to measure the kinetics of the pre-death activity of a pore-forming, membrane-disrupting AmP, called CM15, on individual live E. coli cells in an aqueous solution20.
Electron microscopy and AFM experiments have demonstrated the endpoint surface morphological changes of a population of cells treated with AmPs21,22,23. Spectroscopic analyses of synthetic membranes or vesicles have provided insight into the sizes and structures of pores formed by AmPs24,25,26. However, to date the early stage kinetics of the membrane-disrupting activity of an AmP on live cells have not been reported with nanometre-spatial and seconds-temporal resolution.
In this work, bacteria were immobilized on poly-L-lysine coated cover slides. The bacteria were imaged in aqueous solution for at least 10 minutes to ensure that the cells were not altered or displaced by the AFM tip and to ensure that the poly-L-lysine did not change the cells in the timescales observed in our experiments. CM15 was added to the liquid droplet around the sample to a final concentration of 50 ug/mL or 5 times the minimum inhibitory concentration (MIC)27. Images were acquired every 13 seconds.
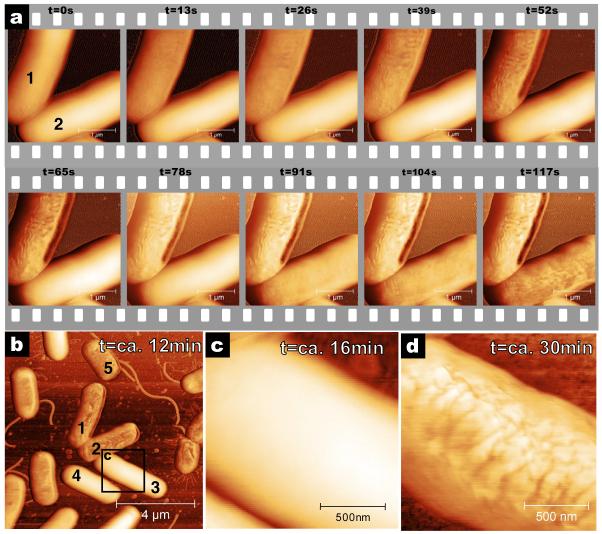
Figure 2A shows AFM phase images of the surfaces of two bacteria before and at several times after the addition of CM15. The most apparent effect of the CM15 is that the surfaces of the bacteria change from smooth to corrugated. AFM phase data is shown due to increased clarity in the image. The same changes in surface morphology are present in AFM amplitude data (supplemental figure S2). AFM height data was not suitable due to the large background variation on the surface of the bacteria. The changes in the surfaces of the bacteria are consistent with published electron microscopy data that report ultra structural damage to the outside of peptide-treated bacterial cells.21,22,23 Interestingly, there is a wide range in the time of onset of the change between individual bacteria. Bacterium 1 in figure 2A starts changing within 13 seconds after addition of CM15 and the change is completed in ~60 seconds. Bacterium 2 does not start changing until ~80 seconds and the change is not complete until ~120 seconds.
Figure 2. E. coli cell disruption induced by CM15 imaged with high-speed AFM.
A) Time series of CM15 antimicrobial action. CM15 injected at t =−6 seconds and images recorded every 13 seconds, with resolution of 1024×256 pixels and rate of 20 lines/s. The upper bacterium’s surface starts changing within 13 seconds. The lower bacterium resists changing for 78 seconds. B) Larger area view recorded 12 minutes after addition of CM15. Most bacteria are corrugated but some are still smooth. C) High-resolution image of bacterium 3 shows that this bacterium is still smooth at t=16 minutes. D) Image of the now corrugated bacterium 3 at t=30 minutes. Eventually, all bacteria in the field of view are affected by CM15. Images were recorded in liquid in tapping mode with a tapping frequency of 110 kHz. Phase images are shown here for high contrast; amplitude data is shown in figure S2. Images B, C and D were recorded with 1024×256 pixels at 2 lines/s.
Figure 2B shows a larger area of the same sample 12 minutes after addition of CM15 – bacteria 1 and 2 are in the centre of figure 2B. Some bacteria still have not changed (bacteria 3 and 4). Figure 2C is a higher resolution image of the smooth surface of bacterium 3, 16 minutes after addition of CM15. Eventually, this bacterium (figure 2D) and all bacteria in the field of view become corrugated. We considered several reasons for the cell-to-cell variation in onset time. The bacteria are all grown from a single clone, thus, we expect them to be genetically identical. They are close to each other (within 10 μm) and therefore are exposed to the same CM15 concentration at the same time. The cells are likely not all in the same stage in the cell cycle. However, in all of our experiments, we found no correlation between the time of onset of change and cell length (an indicator of stage in cell cycle28).
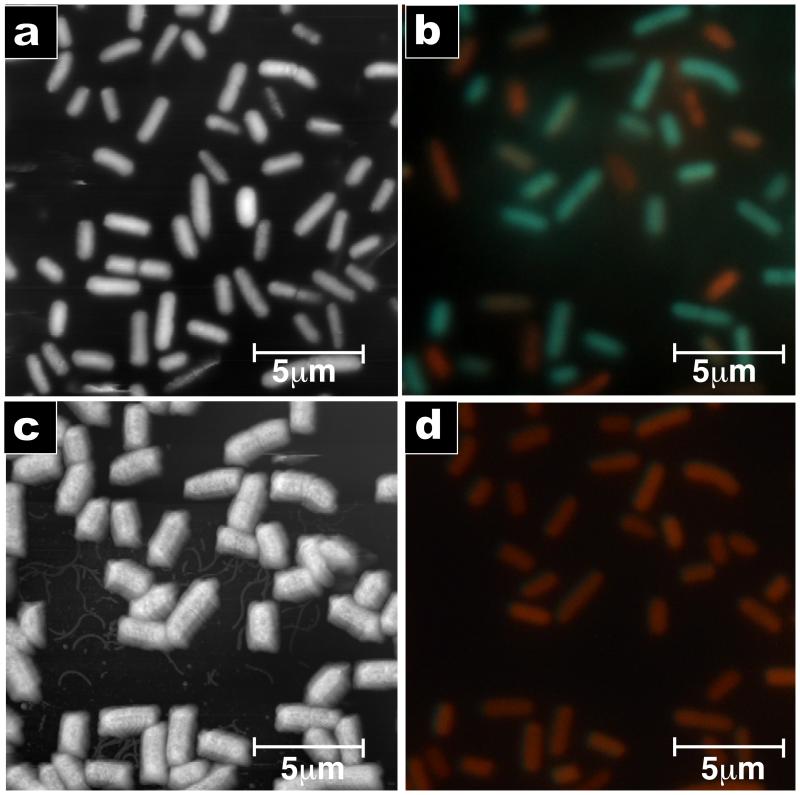
To correlate the corrugation of the cell surface with cell death, we performed combined AFM and fluorescence microscopy using a fluorescent indicator of cell viability (LIVE/DEAD BacLight). Figure 3A shows an AFM image of live cells. To minimize disruptions caused by the AFM tip, the imaging force was reduced until only the tops of the bacteria were imaged. Figure 3B shows a fluorescence image of the same cells recorded immediately after the AFM imaging. Figure 3C and 3D show the AFM and fluorescence images of the same bacteria 30 minutes after the cells were exposed to 2 times the MIC of CM15. Almost all of the bacteria have corrugated surfaces and there are no green (live) cells remaining. The apparent broadening of the cells is an imaging artefact due to the pyramidal shape of the tip29. There was a strong correlation between the fluorescence and surface variation of the cells (p-value < 0.004). To ensure that the change is induced by the CM15, we performed control experiments with a positively-charged peptide with no known antimicrobial action (figure S4) and with the conventional antibiotic ampicillin (figure S5).
Figure 3. AmP-induced surface morphology change correlates to cell death.
Combined AFM and fluorescence microscopy images recorded on the same spot before and after addition of CM15. A) Tapping mode image of bacteria before addition of CM15 (phase data). The surfaces of most bacteria are smooth. B) Fluorescence image before addition of CM15. Green represents live bacteria; red represents dead bacteria (LIVE/DEAD stain). C) AFM image 30 minutes after exposure to a 2 times the MIC solution of CM15. Nearly all the bacteria exhibit a corrugated surface. D) Fluorescence image after addition of CM15. All bacteria are red, indicating that they are dead. AFM images were taken with 512 × 256 pixels and a scan rate of 0.5 Hz.
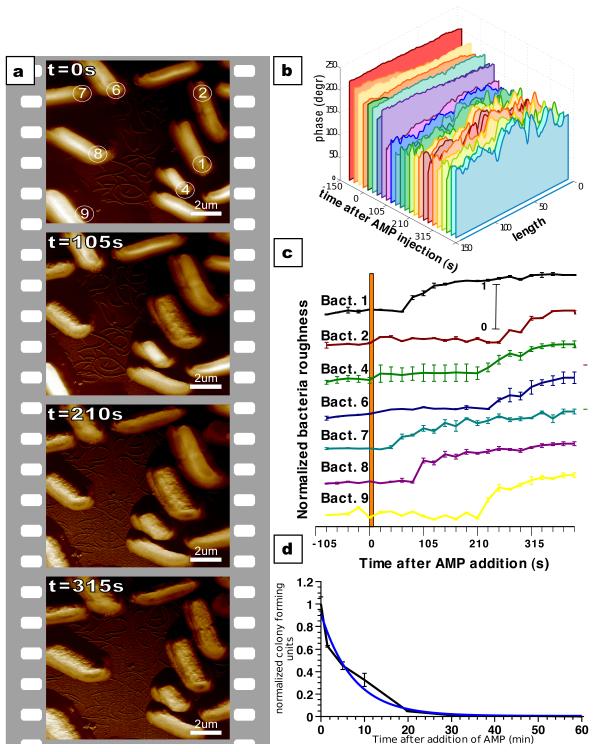
The increased time resolution of our measurements allows us to characterize the initial stages of antimicrobial action on individual live bacteria. The high-speed AFM experiment was repeated with a larger scan area to investigate relative differences in the response times of adjacent cells. At t = 0 seconds, the imaging solution was exchanged with a 2 times MIC CM15 solution using a flow-through system. One image was taken every 21 seconds, with every 5th image in figure 4A (full series in figures S6 and S7). It is evident that bacteria 1, 7 and 8 respond more quickly to the addition of CM15 than the other bacteria.
Figure 4. Early stage kinetics of CM15 action measured by AFM correlates with bulk killing activity experiment.
A) Time series of bacteria after injection of CM15. Images recorded every 21 seconds (1024×256 pixels, 12.2 lines/s) with every 5th image shown (full time series in supplemental material). B) Cross sections along the long axis of bacterium 1 showing the time progression of the surface variation. Each slice represents data extracted from one image in the full time series. C) Averaged surface variation of the bacteria as a function of time after injection of CM15 (bacteria numbers correspond to those in frame one of panel A). D) Bulk measurement of CM15 antimicrobial activity. The interpolated behaviour between 0 and 5 minutes correlates well with the single cell measurements.
To quantify the kinetics of the change, we measured, for each bacterium separately, the change in the root mean square (RMS) value of the surface corrugation along the long axis of the bacteria in every frame of the image. Figure 4B shows representative cross sections through bacterium 1 for each image. The change in surface corrugation is visible ~80 seconds after the addition of CM15. Figure 4C shows the normalized RMS corrugation values for the bacteria in figure 4A as a function of time after addition of CM15. Time of onset of change is highly variable. In this experiment it took 40 seconds (bacterium 7) to more than 4 minutes (bacterium 2) –average of 155 seconds +/− 89 seconds (mean +/− standard deviation). The time for each bacterium to complete the change (smooth to corrugated) is more consistent – 50 % of the damage is completed in 52 seconds +/−16 seconds. We chose the phase data for the analysis, since here the changes are most apparent. However, the same trends can be observed in the height and amplitude images (figure S8). We monitored the CM15-treated cells for up to 100 minutes after treatment and we did not see any additional changes. Eventually, the cells disappear, but we cannot say whether this is due to lysis or due to the detachment of cells. Figure 4D shows the results of a measurement of the kinetics of CM15 activity on a population of cells. The bulk killing rate of CM15 can be described reasonably well with a single exponential with a half-time of 4.6 minutes.
With this AFM technique, one might expect to see the pores that are formed in the outer membrane by CM15. Unfortunately, the pores formed by CM15 are reported to have diameters of 2-4 nm27 which is close to the pixel resolution of our high-speed images (4-10 nm, scan size dependent). Using a slower acquisition speed (2 lines/s), we obtained images where the pixel resolution is 1 nm for a 1 μm image (figure S3). While there are features of this size in this higher resolution image, we are hesitant to over-interpret our results.
We hypothesize that the killing of the bacteria by CM15 is a two-stage process consisting of an incubation phase, which can last seconds to minutes, followed by an execution phase, in which 50 % of the damage is completed in less than one minute. The time to complete the incubation phase varies considerably more from cell to cell than the time to complete the execution phase. This result suggests that the bulk kill rate is dominated by the time it takes to complete the incubation phase, rather than the execution phase. It also raises the question of whether the incubation and the execution phases are prolonged equally in bacteria that are more resistant to AmPs or if one particular phase is more prolonged than the other. Answering this could be important for understanding the mechanism by which bacteria can develop resistance to this class of peptide antibiotics.
The heterogeneity in the dynamics of AmP-induced bacterial cell death has an interesting parallel in eukaryotic cell biology. Mammalian cell apoptotic death recently has been demonstrated to be a two-stage process comprised of a time-variable and long-in-duration incubation phase which is followed by a more constant and relatively short execution phase30. Despite the apparent differences in the overall mechanisms, protein/peptide-induced pore formation in a membrane is a critical component of both mammalian cell apoptotic death (mitochondrial outer membrane permeabilization) and AmP-induced bacterial cell death (outer membrane permeabilization).
Our observation of this two-stage process was made possible by the use of a high-speed AFM capable of recording dynamic changes on the single cell level. These measurements demonstrate the enormous potential of high-speed AFM imaging for cellular biology. This technique can be applied to other cell types such as yeast or mammalian cells and even to eukaryotic cell organelles. The advances in AFM technology and its application reported here constitute an enabling technology for cell-biologists to explore cellular processes in real time at the nanometre level.
Materials and Methods
Antimicrobial peptide
We used a well-studied, pore-forming AmP called CM15 with the sequence KWKLFKKIGAVLKVL. The peptide was made by Genscript (Piscataway, NJ, USA) using standard solid phase peptide synthesis methods, was purified by desalting and was shipped lyophilized. The peptide was resuspended and stored in Millipore water and diluted to the appropriate concentrations in Millipore water.
Bacteria preparation
E. coli bacteria (ATCC 25922) were grown from a single colony over night in LB growth medium at 37 degrees Celsius. Cells were diluted 1:100 in fresh LB medium and were grown for 3 hours. Cells were spun down and washed three times with Millipore water.
To increase adherence of the bacteria to the glass substrate, glass cover slips were coated with poly-l-lysine before depositing the bacteria. Round glass cover slips were boiled in 2.5 M HCl solution for ten minutes to clean the glass. The cover slips were rinsed six times with Millipore water and were immersed for ten minutes in a pH 8.0 solution of 0.05 mg/mL poly-L-lysine hydrobromide (Sigma part number: P1524) and 10 mM Tris. Afterwards, they were covered and dried vertically overnight at room temperature. The coated cover slips were stored at room temperature and were used within one week.
Concentrated bacteria suspended in un-buffered Millipore water were deposited on the coated cover slips and incubated for 30 minutes. Excess cells were rinsed off with 3 washes of 1 ml Millipore water. Images were taken in Millipore water.
High speed AFM imaging
All high-speed AFM images were taken on a customized AFM instrument based on a Veeco Multimode with a Nanoscope 5 controller (Veeco Metrology, Santa Barbara, USA). The most enabling modifications are small cantilevers (SCL-Sensor.Tech. fabrication GesmbH, Vienna, Austria) and a new AFM head designed for use with the small cantilevers (See figure S1 in the supplemental materials for more details). We chose not to use a high-speed scanner for these experiments, since turnaround ripples and other distortions were small at these scanning parameters. All measurements were performed in tapping mode in fluid with an open fluid cell. Flow-through fluid exchange was achieved using a dual syringe pump. Height, amplitude and phase signals were recorded for both trace and retrace. The data was processed using ImageJ, ImageSXM and Gwyddion, using standard modification commands applied over the whole sample.
Combined AFM/Fluorescence Microscopy imaging
AFM images were recorded on a Digital Instruments Bioscope (Veeco Metrology Inc., Santa Barbara, USA) with an extended Nanoscope 3a controller, mounted on top of a Zeiss Axiovert 135 inverted microscope (Carl Zeiss Microimaging GmbH, Germany). AFM images were taken with a Veeco NPS cantilever in tapping mode (Veeco Instruments Inc., Camarillo, USA). Fluorescence images were recorded using a Zeiss 100X Plan-Neofluar oil immersion objective and a Chroma Technology Corporation (Rockingham, USA) filter set 41012 . Cells were stained with LIVE/DEAD BacLight stain (Invitrogen PN: L13152). The LIVE/DEAD stain is a mixture of SYTO® 9 green-fluorescent nucleic acid stain and propidium iodide red-fluorescent nucleic acid stain. These stains differ in their ability to penetrate healthy bacterial cells, with the green stain permeating healthy cells and the red stain permeating membrane-compromised cells.
Due to the loss in viability during the preparation process, some cells already are dead before the experiment starts (see figure 3B). During initial AFM imaging, some loosely connected cells detach from the surface and therefore not all the cells in figure 3A are present in figure 3B.
Analysis of LIVE/DEAD stain results and correlation with RMS variation of bacterial surfaces
Fluorescence images were analyzed using ImageJ with nucleus counter plug-in (downloaded from MacBiophotonics). Cells were identified and numbered using the nucleus counter plug-in. The colour images were split into red, green, and blue channels. For each cell, the mean red and the mean green values were measured and the green to red ratio for each cell was calculated. The cells were divided into two populations – those with green to red ratios less than one (dead) and those with green to red ratios greater than one (live). Using the axial RMS variation for each cell (calculation described below), a one-tailed student’s t-test was used to assess whether the live cell population and the dead cell population had statistically different RMS variation values.
Calculation of axial RMS variation of bacteria
To quantify the change in the variations on the bacterial surface, line sections were extracted from the phase data on the top of each bacterium in the longitudinal direction. The exact number of line sections per bacterium per image depended on the size of the bacterium but was always more than 20. A 3rd degree polynomial was fit through each line section and subtracted from the original data. The resulting data represented only the higher spatial frequencies from which the RMS value was calculated. The RMS values of each line were then averaged over all the lines along the bacterium. This number represents one data point in figure 4C. The error bars represent standard deviation of this calculation when varying the exact area on each bacterium and the number of longitudinal sections used for the calculation. This calculation was repeated for each frame in the time series and for each cell. The data processing was done with custom software written in Labview and NI-Vision (National Instruments, Austin, USA).
Bulk cell killing assay
Cells were grown overnight in LB from a single colony. In the morning, they were diluted 1:100 in fresh LB and grown for three hours. After three hours, they were washed three times in Millipore water. The washed cells were resuspended in Millipore water to a concentration of 2×106 cells / mL. The CM15 peptide was added to the cells to a final concentration of 20 μg / mL, which was the same peptide-to-cell ratio as that used in the AFM experiments.. Aliquots were taken from the cells at various time points and were plated in triplicate on fresh, prewarmed LB agar plates, which were incubated overnight at 37°C. The number of colonies formed is an indicator of the number of viable cells remaining in the suspension at each time point. The number of colonies formed was counted and the average and standard deviation of each time point was calculated. All data was normalized to the zero time point (before addition of peptide).
Supplementary Material
Acknowledgements
The authors would like to thank Tanguy Chau for helpful discussions about antimicrobial peptides. GEF is supported by an Erwin- Schrödinger fellowship J2778-B12. RJB is the recipient of a NIH Biotechnology Training Program Fellowship. AMB would like to thank MIT for their generous support. This work was further funded by the Army Research Office through the Institute for Soldier Nanotechnology, the National Institute of Health under Award RO1 GM065354 and by the Austrian Research Promotion Agency under award number VO156-08-BII: NSI-FABICAN.
Footnotes
Author Contributions: GEF: Experiment design and execution, writing of manuscript
RJB: Experiment design and execution, writing of manuscript
DSG: Design of methods and sample preparation.
AMB: Experimental design, supervision, editing manuscript
Additional Information: Supplementary information accompanies this paper at www.nature.com/naturenanotechonlogy. Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Dufrene YF. Using nanotechniques to explore microbial surfaces. Nat. Rev. Micro. 2004;2:451–460. doi: 10.1038/nrmicro905. [DOI] [PubMed] [Google Scholar]
- 2.Dufrene YF. Towards nanomicrobiology using atomic force microscopy. Nat. Rev. Micro. 2008;6:674–680. doi: 10.1038/nrmicro1948. [DOI] [PubMed] [Google Scholar]
- 3.Matzke R, Jacobson K, Radmacher M. Direct, high-resolution measurement of furrow stiffening during division of adherent cells. Nat. Cell. Biol. 2001;3:607–610. doi: 10.1038/35078583. [DOI] [PubMed] [Google Scholar]
- 4.Muller DJ, Baumeister W, Engel A. Controlled unzipping of a bacterial surface layer with atomic force microscopy. Proc. Natl. Acad. Sci.U.S.A. 1999;96:13170–13174. doi: 10.1073/pnas.96.23.13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller DJ, Fotiadis D, Scheuring S, Muller SA, Engel A. Electrostatically balanced subnanometer imaging of biological specimens by atomic force microscope. Biophys. J. 1999;76:1101–1111. doi: 10.1016/S0006-3495(99)77275-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plomp M, Leighton TJ, Wheeler KE, Hill HD, Malkin AJ. vitro high-resolution structural dynamics of single germinating bacterial spores. Proc. Natl. Acad. Sci.U.S.A. 2007;104:9644–9649. doi: 10.1073/pnas.0610626104. doi:10.1073/pnas.0610626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francius G, Domenech O, Mingeot-Leclercq MP, Dufrene YF. Direct Observation of Staphylococcus aureus Cell Wall Digestion by Lysostaphin. Journal of Bacteriology. 2008;190:7904–7909. doi: 10.1128/JB.01116-08. doi:10.1128/jb.01116-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinterdorfer P, Baumgartner W, Gruber HJ, Schilcher K, Schindler H. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci U.S.A. 1996;93:3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller DJ, Dufrene YF. Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nature Nanotechnology. 2008;3:261–269. doi: 10.1038/nnano.2008.100. [DOI] [PubMed] [Google Scholar]
- 10.Ando T, et al. A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl. Acad. Sci.U.S.A. 2001;98:12468–12472. doi: 10.1073/pnas.211400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi M, Sumitomo K, Torimitsu K. Real-time imaging of DNA-streptavidin complex formation in solution using a high-speed atomic force microscope. Ultramicroscopy. 2007;107:184–190. doi: 10.1016/j.ultramic.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 12.Hansma PK, Schitter G, Fantner GE, Prater C. Applied physics - High-speed atomic force microscopy. Science. 2006;314:601–602. doi: 10.1126/science.1133497. [DOI] [PubMed] [Google Scholar]
- 13.Dufrene YF. Atomic force microscopy and chemical force microscopy of microbial cells. Nat. Protocols. 2008;3:1132–1138. doi: 10.1038/nprot.2008.101. [DOI] [PubMed] [Google Scholar]
- 14.Fantner GE, et al. Components for high speed atomic force microscopy. Ultramicroscopy. 2006;106:881–887. doi: 10.1016/j.ultramic.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Viani MB, et al. Fast imaging and fast force spectroscopy of single biopolymers with a new atomic force microscope designed for small cantilevers. Rev. Sci. Instrum. 1999;70:4300–4303. [Google Scholar]
- 16.Tiozzo E, Rocco G, Tossi A, Romeo D. Wide-Spectrum Antibiotic Activity of Synthetic, Amphipathic Peptides. Biochemical and Biophysical Research Communications. 1998;249:202–206. doi: 10.1006/bbrc.1998.9114. [DOI] [PubMed] [Google Scholar]
- 17.Gottlieb CT, et al. Antimicrobial peptides effectively kill a broad spectrum of Listeria monocytogenes and Staphylococcus aureus strains independently of origin, sub-type, or virulence factor expression. BMC Microbiology. 2008;8 doi: 10.1186/1471-2180-8-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hancock REW, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nature Biotechnology. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 19.Loose C, Jensen K, Rigoutsos I, Stephanopoulos G. A linguistic model for the rational design of antimicrobial peptides. Nature. 2006;443:867–869. doi: 10.1038/nature05233. [DOI] [PubMed] [Google Scholar]
- 20.Andreu D, et al. Shortened Cecropin-a Melittin Hybrids - Significant Size-Reduction Retains Potent Antibiotic-Activity. FEBS Letters. 1992;296:190–194. doi: 10.1016/0014-5793(92)80377-s. [DOI] [PubMed] [Google Scholar]
- 21.Kalfa VC, et al. Congeners of SMAP29 kill ovine pathogens and induce ultrastructural damage in bacterial cells. Antimicrobial Agents and Chemotherapy. 2001;45:3256–3261. doi: 10.1128/AAC.45.11.3256-3261.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meincken M, Holroyd DL, Rautenbach M. Atomic Force Microscopy Study of the Effect of Antimicrobial Peptides on the Cell Envelope of Escherichia coli. Antimicrobial Agents and Chemotherapy. 2005;49:4085–4092. doi: 10.1128/AAC.49.10.4085-4092.2005. doi:10.1128/aac.49.10.4085-4092.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangoni ML, et al. Effects of the Antimicrobial Peptide Temporin L on Cell Morphology, Membrane Permeability, and Viability of Escherichia Coli. Journal of Peptide Science. 2004;10:178–178. doi: 10.1042/BJ20031975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bechinger B. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochimica Et Biophysica Acta-Biomembranes. 1999;1462:157–183. doi: 10.1016/s0005-2736(99)00205-9. [DOI] [PubMed] [Google Scholar]
- 25.Ladokhin AS, Selsted ME, White SH. Sizing membrane pores in lipid vesicles by leakage of co-encapsulated markers: Pore formation by melittin. Biophys. J. 1997;72:1762–1766. doi: 10.1016/S0006-3495(97)78822-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee MT, Hung WC, Chen FY, Huang HW. Mechanism and kinetics of pore formation in membranes by water-soluble amphipathic peptides. Proc. Natl. Acad. Sci.U.S.A. 2008;105:5087–5092. doi: 10.1073/pnas.0710625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato H, Feix JB. Osmoprotection of bacterial cells from toxicity caused by antimicrobial hybrid peptide CM15. Biochemistry. 2006;45:9997–10007. doi: 10.1021/bi060979m. [DOI] [PubMed] [Google Scholar]
- 28.Ferullo DJ, Cooper DL, Moore HR, Lovett ST. Cell cycle synchronization of Escherichia coli using the stringent response, with fluorescence labeling assays for DNA content and replication. Methods. 2009;48:8–13. doi: 10.1016/j.ymeth.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alsteens D, et al. Organization of the mycobacterial cell wall: a nanoscale view. Pflug. Arch. Eur. J. Phy. 2008;456:117–125. doi: 10.1007/s00424-007-0386-0. [DOI] [PubMed] [Google Scholar]
- 30.Albeck JG, et al. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol. Cell. 2008;30:11–25. doi: 10.1016/j.molcel.2008.02.012. doi:10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.