Abstract
Analysis of natural host-parasite relationships reveals the evolutionary forces that shape the delicate and unique specificity characteristic of such interactions. The accessory long gland-reservoir complex of the wasp Leptopilina heterotoma (Figitidae) produces venom with virus-like particles. Upon delivery, venom components delay host larval development and completely block host immune responses. The host range of this Drosophila endoparasitoid notably includes the highly-studied model organism, Drosophila melanogaster. Categorization of 827 unigenes, using similarity as an indicator of putative homology, reveals that approximately 25% are novel or classified as hypothetical proteins. Most of the remaining unigenes are related to processes involved in signaling, cell cycle, and cell physiology including detoxification, protein biogenesis, and hormone production. Analysis of L. heterotoma’s predicted venom gland proteins demonstrates conservation among endo- and ectoparasitoids within the Apocrita (e.g., this wasp and the jewel wasp Nasonia vitripennis) and stinging aculeates (e.g., the honey bee and ants). Enzyme and KEGG pathway profiling predicts that kinases, esterases, and hydrolases may contribute to venom activity in this unique wasp. To our knowledge, this investigation marks the first functional genomic study for a natural parasitic wasp of Drosophila. Our findings will help explain how L. heterotoma shuts down its hosts’ immunity and shed light on the molecular basis of a natural arms race between these insects.
Keywords: venom, venom gland, transcriptome, parasitoid, Leptopilina heterotoma, Drosophila, host, functional genomics, co-evolution
1.0 INTRODUCTION
The order Hymenoptera comprises approximately 130,000 insect species, with as many as 20% of these estimated to be parasitoid wasps in the Apocrita (Pennacchio, 2006). The reproductive strategies within this group target host development and viability, and contribute to community structure and ecology. Venom protein bioactivity has been studied since the early twentieth century, when the first snake (Noguchi, 1909) and scorpion venoms were investigated (Todd, 1909). The venom studies for pain-inflicting social insects such as bees, bumblebees, yellow jackets, and ants, have clarified the ontology of venom proteins and provided treatment applications (Hoffman, 1977; Peiren, 2005; deGraaf, 2009). In contrast to social insects, parasitoid wasps must apprehend and physiologically control their hosts to assure the success of their offspring. Early indications suggest that the venom pharmacopeia of these insects will prove to be richer (Danneels, 2010), paralleling the specific demands of host-parasite interactions.
Venom factors provide the armament for success in the host/parasitoid arms race. Venom proteins target host physiology and development to provide the developing parasitoid with a secure and nutrient-rich environment that will optimize its consumption of host resources (Rivers, 1994; Rivers, 1995). Hosts often are subdued through neuro-active venom components that may cause prolonged paralysis, particularly in ectoparasitic wasp attack (Rivers, 2002). Additionally, parasitic wasps protect their progeny either by passively evading the host immune system (e.g., Asobara tabida, (Prevost, 2009)) or by actively suppressing host immunity (e.g., Leptopilina spp. (Dubuffet, 2009; Lee, 2009)). Many studies in D. melanogaster have found that the cellular and humoral responses are predominantly under the control of Toll/NF-kappa B and JAK-STAT signaling pathways. Melanization of wasp egg also contributes to the host defense response (Lemaitre, 2007; Schlenke, 2007; Govind, 2008). These molecular mechanisms appear to be active in other insects as well (Bitra, 2012), and are targets of inhibitors arising from venoms, polydnavirus gene expression, and calyx fluid (Nappi, 2009; Strand, 2012).
Leptopilina heterotoma (Lh), a member of a moderately sized genus (Schilthuizen, 1998; Allemand, 2002), successfully parasitizes most Drosophila species tested (Carton, 1986; Schlenke, 2007). It has been known for over fifty years that Lh strains must produce venom factors (Walker, 1959). The majority of the virulence activity is attributed to the action of virus-like particles (VLPs) that are produced and assembled in the long gland-reservoir complex (Rizki, 1992; Morales, 2005; Chiu, 2006; Ferrarese, 2009). The long gland is a simple cylindrical organ lined peripherally with large, polyploid secretory cells. Internal and concentric to this cell layer is a single-celled layer of intimal cells, which lines the long gland lumen. A supracellular canal system of individual secretory units, one per secretory cell, feeds into the long gland lumen (Ferrarese, 2009). Antibody staining experiments have revealed that some VLP proteins are produced in the secretory cells; they enter the long gland lumen via secretory units and appear associated with small membranous structures. These structures undergo morphogenesis and assemble 3–6 spikes to assume unique stellate morphologies. Stellate VLPs and their constituent proteins block hemocyte-mediated wasp egg encapsulation by inducing cell lysis and apoptosis (Rizki, 1992; Chiu, 2002; Morales, 2005; Chiu, 2006; Ferrarese, 2009).
Leptopilina heterotoma attack delays larval host development (Schlenke, 2007). The biological activities of venom components that contribute to the alteration of Drosophila development and immunity are largely unknown. We are interested in understanding not only the nature of bioactive molecules in the venom and those associated with VLPs, but also the process of VLP assembly and morphogenesis that occurs in the unique long gland-reservoir environment. We also want to know if the venom factors can contribute to immune suppression via an activating or adjuvant-type role, and whether VLPs have a viral origin.
To address these questions, we have initiated a cDNA-based transcriptome analysis of the venom gland. The enzymatic profile and KEGG terms of our Blast-based protein predictions suggest that in addition to conserved signaling, cell cycle, and housekeeping proteins, the Lh venom gland expresses hypothetical and unknown proteins that may help maintain the glandular environments for VLP and venom activities. Many enzymes with predicted biological activities that have been reported in studies of other parasitoid wasps, and in the stinging Aculeata, also appear to be utilized by Lh. Given the conservation among immune pathways in insects, of which Drosophila has been the classic model (Schmid-Hempel, 2005; Tanji, 2005; Cherry, 2006; Govind, 2008), we predict that Lh venom factors with inhibitory functions in the D. melanogaster host will also modulate immune physiologies of other Drosophila species. A comprehensive understanding of the molecular strategies underlying the success of this natural Drosophilia parasitoid can potentially be used to target economically significant insect pests and pathogens.
2.0 METHODS
2.1 Insect stocks
L. heterotoma strain New York USA (Chiu, 2006; Schlenke, 2007) were raised in house at 25 °C on the y w strain of D. melanogaster on standard corn meal and yeast diet.
2.2 Transcriptome library preparation and sequencing
500 Lh females were anaesthetized by CO2 and washed with 70% alcohol. Their long gland-reservoir-ovipositor complexes (called venom glands here), were removed simply by pulling the ovipositor, and frozen at −70°C. Eight μg of total RNA were extracted and used to prepare a standard cDNA library (Evrogen) in the pAL17.3 vector using the SMART approach (Zhu, 2001). The library was amplified by PCR. SMART-Sfi1A oligonucleotide 5′-AAGCAGTGGTATCAACGCAGAGTGGCCATTACGGCCrGrGrG-3′ CDS-Sfi1B primer 5′-AAGCAGTGGTATCAACGCAGAGTGGCCGAGGCGGCCd(T)20-3′ SMART PCR primer 5′-AAGCAGTGGTATCAACGCAGAGT-3′ pAL 17 dir primer 5′-CCAGGGTTTTCCCAGTCACGA-3′ pAL 17 rev primer 5′-CACAGGAAACAGCTATGACCA–3′ More than 950 randomly selected clones in ten 96-well plates were sequenced by Sanger method (Genewiz, New Jersey).
2.3 Sequencing confirmation
A dozen clones were re-sequenced. Transformed E. coli were grown for 12 hours at 37°C in 5 ml of Luria Broth-ampicillin cultures. Approximately 500 ng of the associated pAL 17.3 plasmids were obtained from 1 ml Luria Broth-ampicillin cultures grown for 12 hrs at 37°C. QIAprep Spin Miniprep Kit (http://www.qiagen.com) procedure was followed to obtain the cloned inserts that were then sequenced using a T7 sequencing primer (Genewiz, New Jersey). T7 Universal 20mer Primer: 5′-TAA TAC GAC TCA CTA TAG GG-3′ The sequences were compared to the originals using EBI (http://www.ebi.ac.uk/Tools/) Needleman pairwise alignment (Needleman, 1970). The average percent identity of the nucleotide sequences was 98.8%, calculated as the number of indels and mismatches.
2.4 Raw EST processing
The raw Sanger nucleotide sequences were processed with the standard methodologies of (1) phred/phrap/consed (Ewing, 1998b; Ewing, 1998a) and (2) Cap3 (Huang, 1999). For phredPhrap, base calls and quality assignments were made; cloning elements and terminal N’s were trimmed, and sequence assemblies were compiled with the highest stringency (phrap 1.090518 http://phrap.org): (1) Minimum of 40 bp in common (minmatch 40); (2) Minimum of 95% sequence identity (penalty 95); (3) 95% identity within joint overlaps (repeat stringency 0.95). This analysis of 960 unigenes resulted in 90 contigs assembled from 223 clones.
The results were validated by submitting the original singlet unigene sequences to Cap3 via the Mobyle Pasteur webserver (http://mobyle.pasteur.fr). 65 contigs (72% of total) were identified by both phrap and Cap3. Individual clones from contigs assembled from phrap but not confirmed by Cap3 were Blasted. In all cases, the individual Blasts supported the assembled Blast results. The E values of the unique contig Blasts were significant, averaging 10−41, supporting their quality. In addition six randomly chosen phrap-identified contigs were selected and manually aligned. Overlapping regions were 99% identical. These alignments confirmed the phrap-assembled results in addition to manual consed reviews. The assemblies are referred to simply by a contig number while singlet unigenes are referred to by their plate number.
2.5 Characterizations and annotations of sequences based on similarities and potential homologies
Clean, base-called nucleotide sequences and contigs were submitted to the NCBI website (http://blast.ncbi.nlm.nih.gov/) BlastX algorithm (S. Altschul, 1997). Default parameters were utilized (Alignment scoring: Word length = 3; Expect threshold = 10; BLOSUM62; Existence = 1; Extension = 1) and searches were conducted against the RefSeq nr database (Pruitt, 2004). An E-value of 10−5 was applied as criterion for the identification of the most distant similarity and putative homology for consideration. Alignments were inspected for sufficient length of 75 contiguous residues or 25% of the putative best homolog. Further investigations were conducted as necessary by translation to the appropriate reading frame and BlastP or PSI-Blast (Altschul, 1997) using the default parameters. Results are presented in Supplementary Tables S1 and S2. The San Diego Supercomputer (SDSC) Biology Workbench 3.2 webserver (http://workbench.sdsc.edu/) was used for ORF analysis and translations. Rarely identified similarities with higher level eukaryotic sequences did not surpass those with insect species and likely arise due to extreme conservation in sequences that are not necessarily specific only to insects.
Alignments were created using Needleman pairwise (Needleman, 1970), ClustalOmega (Sievers, 2011), and/or MUSCLE (Edgar, 2004b; Edgar, 2004a) algorithms with default parameters via the EBI webserver. Domain annotation was used when the evolutionary relationship was not fully resolved and limited to motifs and/or folds. The NCBI Conserved Domains Database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) (Marchler-Bauer, 2004; Marchler-Bauer, 2010), SMART (http://smart.embl-heidelberg.de/) (Letunic, 2012), and PFAM 26.0 (http://pfam.sanger.ac.uk/) (Finn, 2010) were utilized. Criteria for the domain identification included primarily an E-value of no more than 10−5. E-values of 10−3 were accepted only with support from an additional source that provided concurrent sequence groupings within motifs, domains, and/or superfamilies. Annotations found in UniProt (http://www.uniprot.org/) (Magrane, 2011) were frequent starting points for transcript annotation.
Sequence characterizations include the terms “novel” and “hypothetical.” A sequence was considered novel if blast searches yielded no significant alignments at Evalues of less than 1. Sequences were defined as hypothetical when their most similar significant blast results were annotated as hypothetical in the nr database.
2.6 KEGG and EC number annotations
WebMGA (http://weizhong-lab.ucsd.edu/metagenomic-analysis/) (Wu, 2011), KAAS (http://www.genome.jp/tools/kaas/) (Moriya, 2007), and PRIAM (http://priam.prabi.fr/REL_JUL06/index_jul06.html, database profil_ENZYME_SEP12) (Claudel-Renard, 2003) webservers were utilized to collect the Enzyme Commission (E.C.) and KEGG classifications. EC/KEGG annotations were collected to supplement and organize the primary sequence-specific assignments from the NCBI Blast analyses. A significance criterion of a maximum of 10−5 was utilized. Priority was placed on predictions with smaller E-values when multiple KEGG or EC numbers were predicted. The results of the EC analyses are presented in Tables 1 and S4 and Figure 2. The KEGG results are presented in Table S5 and Figure 3.
Table 1. Unigene E.C. profile results.
Numbers assigned via enzyme PSSM-oriented Blast. Percentages <1% have been omitted in this table, but are shown in Figure 2.
| Represented Classes & Subclasses | Class Functions | Contribution to Total Profile |
|---|---|---|
|
| ||
| EC 1 | Oxidoreductases | 13.4% |
|
| ||
| EC 1.1.- | Acts on –OH groups | 2.7% |
| EC 1.5.- | Acts on CH-NH groups | 1.8% |
| EC 1.9.- | Acts on heme groups | 1.8% |
| EC 1.14.- | Acts on paired donors, incorporating/reducing O2 | 2.7% |
|
| ||
| EC 2 | Transferases | 30.4% |
|
| ||
| EC 2.1.- | Transfers 1C groups | 1.8% |
| EC 2.3.- | Acyltransferases | 1.8% |
| EC 2.4.- | Glycosyltransferases | 3.6% |
| EC 2.5.- | Alkyl- or aryltransferases, excluding CH3 transfer | 1.8% |
| EC 2.7.- | Phosphotransferases | 21.4% |
|
| ||
| EC 3 | Hydrolases | 43.7% |
|
| ||
| EC 3.1.- | Esterase | 8.0% |
| EC 3.3.- | Acts on ether bonds | 2.7% |
| EC 3.4.- | Peptidases | 8.9% |
| EC 3.5.- | Acts on non-peptide C-N bonds | 1.8% |
| EC 3.6.- | Acts on acid anhydrides | 21.4% |
|
| ||
| EC 4 | Lyases | 3.6% |
|
| ||
| EC 4.2.- | Carbon-oxygen lyases | 1.8% |
| EC 4.3.- | Carbon-nitrogen lyases | 1.8% |
|
| ||
| EC 5 | Isomerases | 4.5% |
|
| ||
| EC 5.2.- | Cis-trans isomerases | 2.7% |
| EC 5.3.- | Intramolecular isomerase | 1.8% |
|
| ||
| EC 6 | Ligases | 3.6% |
|
| ||
| EC 6.3.- | Forms C-N bonds | 3.6% |
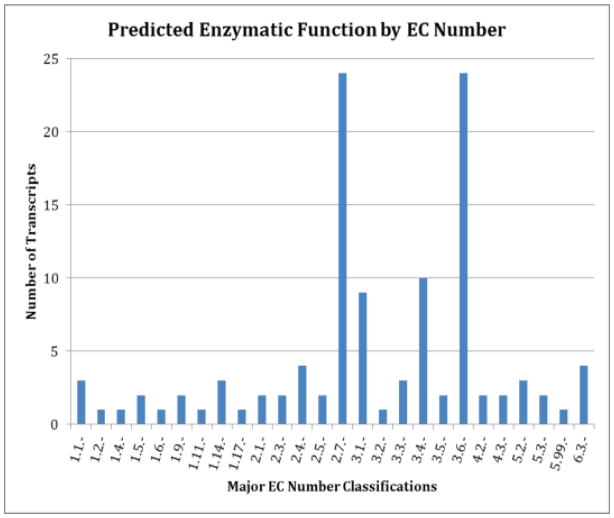
Figure 2. Enzymatic function profile.
Predicted functionality by Enzyme Commission (E.C.) number. Number descriptions given in Table 1.
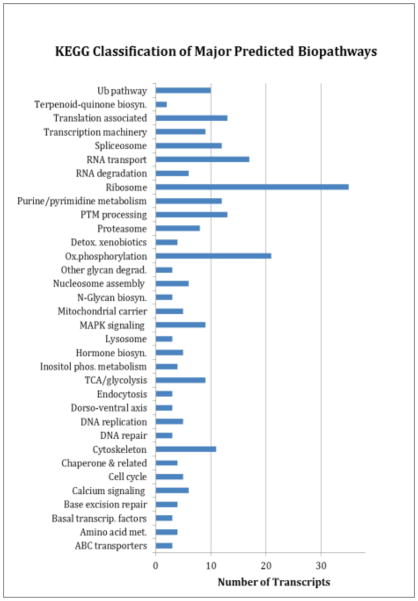
Figure 3. KEGG profile.
Predicted functionality by KEGG descriptions for the major pathways, systems, and functions. Only those groups with more than three transcripts are shown. (Ub: Ubiquitination; PTM: Post-Translational Modification; Ox: Oxidative).
3.0 RESULTS AND DISCUSSION
3.1 The transcripts
3.1.1 Overview
More than 950 original clone sequences from Lh venom gland expression were cleaned and assembled using pred/phrap methodology (Ewing, 1998b; Ewing, 1998a) to yield 827 preliminary unigenes. 153 (145 singlets and 8 contigs) of the 827 are novel, lacking reliable domain identifications and/or significant similarity to published sequences. An additional 42 sequences (37 singlets and 5 contigs) are similar to hypothetical proteins that lack annotation. Here, we present 281 unique putative identities within standard limits of similarity and homology searches (see Methods and Supplemental Tables S1 and S2). The unigene singlet sequences (characterized, novel, and hypothetical), have been deposited in the NCBI expressed sequence tag database, dbEST (LIBEST_028179, submitted 04/15/2013, http://www.ncbi.nlm.nih.gov/dbEST/ (Boquski, 1993), see Supplemental Table S6). These sequences have been submitted in their raw forms, with no base-calling, and have been trimmed of ligation sites, polyA tails, and base-call ambiguity of greater than 5%.
Of the 281 sequences presented here, we have classified 261 unigenes as part of venom gland cellular function, metabolism, and physiology (Supplementary Table S1) and also into more specific functional subclasses (e.g. cell cycle, energetics). At least some of these proteins may contribute to the venom gland physiology and may be important in producing or maintaining functional venom components. Noteworthy molecules include those similar to proteins in MAP kinase signaling (Figure 3) and to immunity proteins such as a NF-κB inhibitor-interacting Ras-like protein, and a Drac1 Ras-related protein (Table S1). Significant similarities to cytoskeletal regulators include a kalirin-like (Rho GEF) protein and rasputin CG9412-PB (Table S1). Proteins with pleiotropic effects ranging from apoptosis to developmental cascades were found among the Blast results, including Roadkill and an enhancer of sevenless 2B-like protein (Table S1).
The remaining unigenes are categorized as putative venom-effector proteins that may target host cells (Supplementary Table S2) and are divided into putative venomic bioactivities possibly affecting behavior, reproduction, or metabolism. Specific proteins are discussed in Sections 3.2, 3.3, and 3.4, including examples that affect the development and nutritional status of the host partner in other parasite-host systems.
3.1.2 Taxonomic relationships predicted via protein similarity
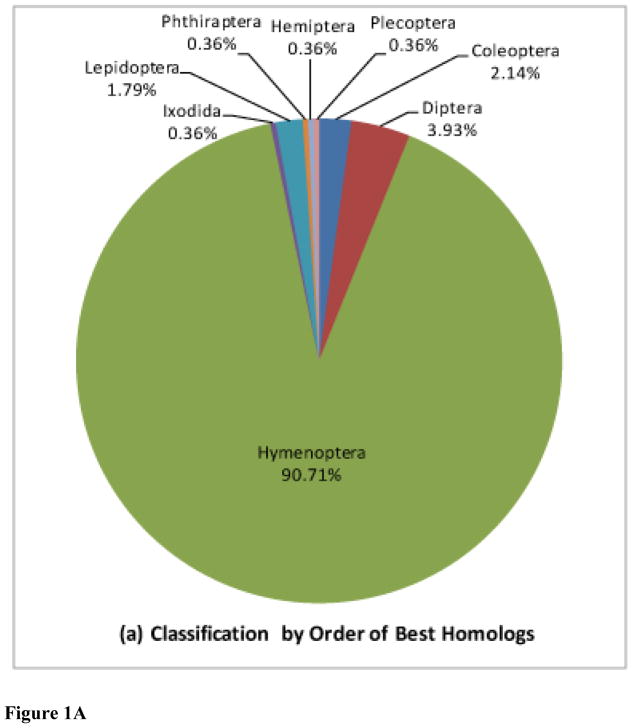
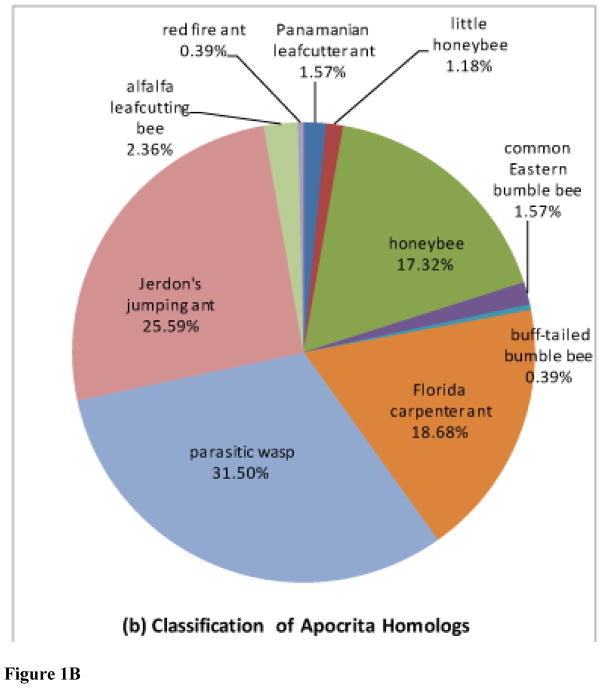
Taxonomic binning of 281 unigenes conducted according to the most similar sequences is presented in Figure 1 and Supplementary Table S3. 90% of the most closely related sequences originate in Apocrita species. Of this number, half have been sequenced from ants (e.g. Florida carpenter ant, Camponotus floridanus, and Jerdon’s jumping ant, Harpegnathos saltator), while the remaining are almost evenly split between bees (e.g. A. mellifera) and parasitic wasps (e.g. N. vitripennis). These numbers are likely biased because of limited sequence availability; as more Apocrita genomes become sequenced, closer relationships between the genes of these individual Hymenoptera will become more evident. We also found one sequence each with some similarity to viral and bacterial proteins. A domain (PF00740) from the Parvovirus VP2 coat protein, associated with viral assembly, was identified by Pfam (E = 1.7e-6) in one transcript with high identity to the Maverick capsid-like p31.10 protein from Cotesia congregata bracovirus [GenBank CBZ06032.1]. Maverick elements are integrated in the chromosomes of a number of related insects (Dupuy, 2011). Another transcript is similar [E = 1e-126, 82% identity] to a conserved outer membrane protein from Acetobacter pasteurianus and other acetic-acid bacteria. A bacterial intein domain [Pfam Hint_2 PF13403, at E = e-18, or better] is present in the same transcript suggesting that the encoded protein is self-splicing. Both these sequences merit verification and analysis and further details will be reported elsewhere.
Figure 1. Sequence classifications using taxonomic binning.
Sequences are classified (a) by order, and (b) by species among Apocrita based on highest similarity between proteins
3.1.3 Enzyme Profiling
The PRIAM webserver was used to predict the enzymatic character of the Lh venom gland transcriptome (See Figure 2, Supplementary Table S4). Table 1 lists the EC number classes found within the profile. The major classes include the EC 2.7.- transferases and the EC 3.1.-, 3.4.-, and 3.6.- hydrolases. Phosphorus group transferases are part of the dominant EC 2.7.- group (21%), which includes kinases, enzymes that are expected in high concentration given their prominent roles in cell signaling and energy metabolism. The EC 3.6.- subclass, the other major predicted group (also 21%), are enzymes that hydrolyze acid anhydrides, such as the DNA and RNA helicases (3.6.12.- and 3.6.13.-). The next largest groups are the esterases (EC 3.1.-, 8%) and peptidases (EC 3.4.-, 9%).
Within the EC 2.7.- group there is heavy representation of enzymes such as mitogen-activated (EC 2.7.11.24) and Ser/Thr (EC 2.7.11.1) kinases. The esterases (EC 3.1.-) are most highly represented by the phosphatases (EC 3.1.3.-), while the peptidases (EC 3.4.1.-) most frequently predicted are related to de-ubiquitination (EC 3.4.19.12) and the proteasome (EC 3.4.25.1). These profiles fall within normal cellular function, but are also suggestive of higher levels of protein trafficking and secretion.
3.1.4 Functional KEGG Profiling
Figure 3 presents the major functional groupings classified by KEGG numbers (Supplementary Table S5). The largest transcript group, accounting for 12% of the total, is associated with ribosome assembly and protein synthesis. Also related to protein production are the functional groups of translation factors (5%) and post-translation modifications (PTM) (5%). KEGG pathways associated with energy production, including the TCA cycle, glycolysis, and oxidative phosphorylation, accounted for 10% of the total. Also significant, were transcriptional functionalities (15%), cytoskeletal proteins (4%), and the ubiquitination pathway (4%).
3.2 Host hormone/pheromone and metabolism modulation
3.2.1 Host maturation
3.2.1.1 Pupation: Juvenile hormone
Pupation is controlled by juvenile hormone (JH) with high levels inhibiting metamorphosis (Nijhout, 1974; Beckage, 1982). JH titer increases in the Lepidoptera Pieris rapae upon parasitism by the endoparasitic wasp Pteromalus puparum (Zhu, 2009). An impressive increase in JH titer of 100 times has been detected in the Lepidoptera Lacanobia oleracea upon parasitism, leading to the arrest of its maturation (Bell, 2010). Most commonly, these effects are a result of JH esterase inhibition in parasitism by PDV wasps such as Glyptapanteles liparidis and Microplitis demolitor (Dover, 1995; Schafellner, 2007). The more recent venomic studies notably have not identified proteins that effect JH titers (Crawford, 2008; deGraaf, 2010; Vincent, 2010).
Methyltransf_FA, a domain closely associated with enzymes of the JH biosynthetic pathway, has been identified in the transcript 5A01 (Table S2) at high levels of significance [Pfam 12248, Methyltransf_FA; E = 3.3e-20]. Although the top scoring BlastX results (Altschul, 1997) are unannotated, they contain this domain and are encoded in closely related Hymenoptera. Also found within these hits, are Drosophila spp. sequences. The D. melanogaster homolog to 5A01 is CG10527 [GenBank NP_611544; E = 9e-55; 49% identity], a gene that is not necessary for JH production, but may be involved with JH pathways (Zhang, 2010). CG10527 mutants are resistant to the effects of JH (Zhang, 2010).
As an additional potential source of developmental control, Contig88 (Table S2), aligns with high significance and identity to a N. vitripennis sequence [GenBank: E = 7e-59; 38% identity] with putative methyltransferase 235L-like function. This Nasonia gene is associated with the JH biosynthetic pathway [KEGG ko00981]. However, Contig88 shows slightly higher sequence similarity to a putative malonyl-CoA O-methyltransferase BioC-like protein [GenBank XP_003708425.1; E = 3e-61; 40% identity]. Domain identification within this transcript cannot at present be narrowed to a specific methyltransferase due to multiple borderline CDD database hits.
3.2.1.2 Host molting and eclosion
Transcript 9C12 (Table S2) demonstrates strong similarity (E-value = e-82; 56% identity) to the N-terminus of a N. vitripennis [GenBank XP_001604327] protein containing an ecdysteroid kinase domain (CDD: E-value = e-11). Molting, which involves both cuticle loosening and peristaltic contractions, is under the control of a hormone and neuropeptide cascade: eclosion hormone ecdysis-triggering hormone and crustacean cardioactive peptide (Gammie, 1999). Phosphorylation of ecdysteroids inactivates these molecules, suppressing morphogenesis until it is appropriate (Makka, 2002). In silkworm Bombyx mori ovaries, ecdysteroids are sequestered and then reactivated, or synthesized de novo, often through the opposing actions of the specific kinase and phosphates (Sonobe, 1999). Venomic modulation of ecdysteroid levels, and repression of host metamorphosis, has been recorded in multiple wasp-host pairs (Beckage, 2004).
LARK RNA-binding protein mutants show a disruption in circadian clock-related events, in particular, eclosion (Newby, 1993). LARK is a RNA Recognition Motif (RRM) domain-containing protein with multiple circadian associated protein binding partners (Huang, 2007). RRM domains perform various RNA-binding events (Maris, 2005). In D. melanogaster, levels of Ecdysone-induced-protein 74EF (E74), a repressor of eclosion, positively correlate with LARK expression levels (Huang, 2007). These results suggest that LARK controls Drosophila metamorphosis via translational modulation of eclosion effectors (Huang, 2007) and that exogenously-supplied LARK could suppress pupation. A Lh venom gland transcript (6B05, Table S1) with very high identity (93%) to the New World ant Acromyrmex echinatior, GenBank EGI70876 ortholog suggests yet another mechanism by which host development is retarded.
3.2.2 Xenobiotic detoxification and hormone synthesis
Commonalities in the enzymes in xenobiotic detoxification and hormone synthesis has complicated the understanding of host-parasite interactions; it is difficult to tease out the evolutionary importance in favor of one pathway or the other. These oxidative enzymes (e.g. cytochrome P450s, various esterases, glutathione S-transferases) detoxify and catalyze hormone/pheromone biosynthesis (Scott, 2008); functions that are potentially advantageous within a parasite’s chemical strategy (Oakenshott, 2010).
Multiple transcripts (e.g. 2D05, 7E01, 3F11, Table S2) associated with detoxification and/or hormone/pheromone biosynthesis have been annotated in the Lh venom gland. This functional group includes sequences similar to Glu—Cys ligase [GenBank XP_001605407], cytochrome P450 [GenBank NP_001165992], and epoxide hydrolase 1 precursor [GenBank NP_001128399]. The presence of such enzymes within the venom gland of a parasitic wasp suggests either hormone biosynthetic or detoxification functions, both potentially contributing to the ultimate goal of parasite survival within its host.
3.2.3 Energy balance modulation
cGMP-dependent protein kinases (PKG) catalyze the addition of a phosphate group to serine or threonine in the presence of the secondary messenger molecule cGMP. Leptopilina heterotoma venom modulation of host energetics is suggested by a transcript (2H01, Table S2) with similarity to the kinase domain from the leafcutter bee, Megachile rotundata [GenBank XP_003704405]. Identity is at 86% within their predicted STKc_PKA domains. Interestingly, M. rotundata XP_003704405 is orthologous to the product of the D. melanogaster foraging gene, for (CG10033). In Drosophila, polymorphism in for creates two modes of food seeking behavior in larvae with “rovers” showing higher sucrose responsiveness (Osborne, 1997; Belay, 2007). These behavioral phenotypes are correlated to allele-specific PKG enzymes with higher catalytic activity (Osborne, 1997). Acceleration of carbohydrate and lipid catabolism is a well-known parasitic strategy (Vinson, 1980). An increase in PKG catalytic activity in the venom via the expression of a for ortholog could possibly raise nutrient levels in the host.
3.3 Modulation of host behavior and environmental interactions
3.3.1 Yellow protein
The major royal jelly proteins (MRJPs), or yellow proteins, have been investigated in the venoms of both the honey bee (Apis mellifera) (Peiren, 2005; Peiren, 2008) and the Chelonus inanitus wasp (Vincent, 2010). MRJP genes show extensive duplication and diversification (Albert, 2004; Drapeau, 2006; Ferguson, 2011). The largest currently-known MRJP gene family is in the Nasonia genomes (The Nasonia Working Group, 2010), suggesting that they are important to both caste-dependent and -independent insects (Drapeau, 2006; Ferguson, 2011).
Yellow proteins function both in Drosophila male courtship behaviors, starting in the third instar (Drapeau, 2003), and in melanization (Brehme, 1941; Biessmann, 1985), although their exact roles in either process are not clear (Han, 2002; Drapeau, 2003; Ferguson, 2011). Melanin is used in wound healing and encapsulation and its expression is up-regulated upon immune challenge (De Gregori, 2001).
Sequence 3C06 (Table S2) shows similarity (percent identity = 26% and similarity = 45%) to yellow-like proteins from at least 100 other Drosophila species [e.g. D. subobscura GenBank CAC16206] and may indicate specific host targeting. Although 3C06 is certainly related to many Apocrita yellow proteins (approximately 50% identity), the well-studied MRJP 8- and 9-related sequences from the honeybee (Peiren, 2005; Peiren, 2008) and Chelonus (Vincent, 2010) venoms were notably absent from the top 100 Blast hits. Experimental data is needed to test if 3C06 can disrupt melanization, delay egg encapsulation, or modulate sexual maturation in their larval hosts.
3.3.2 Chemosensory and hormone/pheromone-binding proteins
Odorant-binding and other chemosensory-binding proteins (OBPs and CBPs, respectively) are significant to communication in insects. These small (14 to 20 kD) extracellular proteins possibly aid in the solubilization and transport of small hydrophobic odorant molecules and pheromones (Pelosi, 1994; Pelosi, 1996; Pelosi, 2005). The functions of OBPs in insect olfaction are crucial to the environmental, reproductive, and social success of insects. The largest class of OBPs, to date, has been found in N. vitripennis (Vieira, 2012).
One transcript and two predicted contigs show putative homology to proteins within this hydrophobic sequence binding class. Notable identity exists between Contig46 (Table S2) and a predicted N. vitripennis sequence, a B1-like protein [GenBank XP_001601068.1; 5e-43, 57% identity]. Contig46 is characterized by a pheromone-binding protein/general odorant-binding protein (PBP_GOBP) six cysteine-containing domain [Pfam 01395: E = 1.2e-23]. Additionally, significant similarity has been found between Contig84 (Table S2) and the predicted ant Harpegnathos saltator sequence GenBank EFN85227.1: Ejaculatory bulb-specific protein 3 [GenBank: E = 2e-23, 62% identity]. A slightly different insect-specific pheromone-binding A10/OS_D domain [Pfam 03392, E = 2.2e-25], is found in this contig. Transcript 9F05 (Table S2) shows enough sequence similarity with the predicted N. vitripennis PBP_GOBP domain-containing general odorant-binding 56d-like protein (OBP08) to suggest homology, but at a distant level [GenBank XP_001600573; E = 1e-09, 33% identity]. The presence of multiple transcripts and multiple pheromone/odorant-binding domains in the Lh venom proteins suggests that they may be associated with host selection (e.g., superparasitism) or oviposition behavior.
3.4 Venom Proteins with Enzymatic Activity: Proteases, Phosphatases, and Lipases
3.4.1 Evidence of protease activity in parasites
Cysteine proteases are well-established as components of parasitic wasp venoms (Parkinson, 2002a; Parkinson, 2002b; Crawford, 2008; deGraaf, 2010; Vincent, 2010), but are also utilized by other parasites, including helminthes and protozoa such as Anisakis and Leishmania (McKerrow, 2006a). Lysosomal-type proteases, which include cathepsin and aspartic proteases, facilitate parasite entry through tissue degradation, immune activation and/or repression, and nutrient release from host proteins (McKerrow, 2006b).
3.4.1.1 Cathepsin D-Like Aspartic Protease
Cathepsin-D is a lysosomal protease active at acidic pH (Lee, 1998; Fusek, 2005). It is an aspartic endopeptidase in the pepsin family (EC.3.4.23). The active site is characterized by two catalytic aspartate residues in a conserved triad of Asp-Tyr-Asp, separated by approximately 200 residues (Baldwin, 1993; Fusek, 2005).
The transcript 10A02 (Table S2) is most similar to, at no less than 65% identity, (1) a N. vitripennis protein, tentatively annotated as a lysosomal aspartic protease-like protein [GeneBank XP_001600543; E = 3e-77], and (2) a beetle Tribolium castaneum protein similar to cathepsin D isoform 1 [GenBank XP_966517; E = 9e-76]. Additionally, a cathepsin_D_like domain [CDD domain cd05485] is identified between nucleotides 107 and 260 of 10A02 at E = 7e-63.
The presence of cathepsin D in the midgut of Hymenoptera has long been established (Houseman, 1983) and an increase in its expression has been correlated to breakdown of cysteine protease inhibitors such as the cystatins, in particular phytocystatins (Ahn, 2009). Cathepsin D has also been found to cleave antimicrobial peptide precursors such as prohemocidins in ticks (Rhipicephalus (Boophilus) microplus) (Cruz, 2010) and pro-antimicrobial peptides in social insects (Camponotus pennsylvanicus) (Hamilton, 2010). Ecdysone-induced expression of cathepsin D is necessary for tissue remodeling during metamorphosis in the silkworm, Bombyx mori (Gui, 2006).
Degradation of the vitellogenin production cellular machinery in the fat body of the mosquito (Aedes aegypti) has been linked to cathepsin D, E, and similar proteins (Cho, 1991; Cho, 1992). Permeabilization of the lysosomal membrane and the subsequent release of various proteases, particularly cathepsin D, activate intrinsic apoptotic pathways in multiple cell types (Roberg, 1999; Stoka, 2007). Although the role of cathepsin D in parasitic Hymenoptera remains elusive, Lh 10A02 may play a role in venom production or in blocking host immunity and supporting wasp egg development.
3.4.2 Phosphatases
Acid phosphatases are commonly known components of the Hymenoptera venoms of Apis mellifera (Grunwald, 2006), N. vitripennis (deGraaf, 2010), Pimpla hypochondriaca (Dani, 2005), and Pteromalus puparum (Zhu, 2008). These enzymes cleave phosphoric acid monoester bonds to yield free protein and phosphate ions. Potential functions of phosphatases as components of venom include nutrient release and modulation of immune signaling (Xia, 2000; Xia, 2001; Dani, 2005).
Transcript 9B06 (Table S2) shows similarity to multiple histidine phosphatases and the highest levels of identity (34–35%) to acid phosphatase sequences from (1) N. vitripennis [GenBank XP_001605452; PREDICTED: venom acid phosphatase Acph-1-like isoform 1], (2) Harpegnathos saltator [GenBank EFN76082.1; Testicular acid phosphatase-like protein], and (3) the well-known Apis mellifera Api m 3 protein [GenBank ACPH1_APIME]. The significance levels (E-values) are comparable for all and are no greater than 2e-21. In the honeybee, the presumably homologous phosphatases Api m 3 and Api m 5, are known to be important antigens (Hoffman, 1977; Grunwald, 2006). Api m 3 is significant to honey bee stings as the major antigen with multiple epitopes that interact with human IgE and induce histamine release (Barboni, 1987; Grunwald, 2006; Georgieva, 2009). In the endoparasitic wasp Pteromalus puparum, expression of phosphate hydrolases have been localized to the long gland nuclei and secretory cells, but show activity in a range centered around pH 4.8 (Zhu, 2008), well below the alkaline to neutral pH of their host hemolymphs. In Pimpla hypochrondriaca, specific phosphatase inhibitors failed to show a reduction of antihemocytic activities (Dani, 2005).
3.4.3 Lipases
Transcript 3H06 (Table S2) shows similarity, and perhaps homology, to the C-termini of phospholipase B (PLB) orthologs from ants and bees: Megachile rotundata (alfalfa leafcutting bee) [GenBank XP_003704073; 1e-40, 41% identity], Solenopsis invicta (red fire ant) [GenBank EFZ13332; 6e-37, 41% identity], and Acromyrmex echinatior (Panamanian leafcutter ant) [GenBank EGI65669; 7e-37, 42% identity]. PLB is a novel enzyme with both Phospholipase A1- and A2-like activities. It is widely encoded, except in yeast (Morgan, 2004). PLB is established as an important component of many venoms and was reported as early as 1964 for bee and various snake venoms (Doery, 1964).
PLB is thought to be the second most concentrated component in the ichneumonid endoparasitoid wasp Pimpla turionelle venom (Uckan, 2006). A lipase-like protein has been detected both by ESTs and mass spectrometry in the braconid endoparasitoid Chelonus inanitus (Vincent, 2010). Lipases have also been found in the venoms of Pimpla hypochondriaca (Dani, 2005) and N. vitripennis (deGraaf, 2010). The exact role of these lipases is unknown, but positive correlation between parasite success and opportunistic modulation of host metabolism is available (Rivers, 1995). N. vitripennis venom alters lipid content in host hemolymph and fat bodies upon envenomation in its host, Sarcophaga bullata (Rivers, 1995). Ectoparasitoid Euplectrus separatae (previously Euplectrus sp near plathypenae) envenomation of its host oriental armyworm Pseudaletia separata also causes an increase in lipid content in the hemolymph which is possibly related to concurrent lysis of fat body cells (Nakamatsu, 2003a; Nakamatsu, 2003b; Nakamatsu, 2004).
4.0 CONCLUDING REMARKS
Parasitism requires bioactive venom proteins and peptides for immune evasion or immune suppression, to facilitate nutrient acquisition, and to cause some level of host subdual (Rivers, 2002). The most critical determinants of venom protein profiles in relation to host strategy and host range have remained intractable until recently. Powerful transcriptomic and venom proteomic approaches (deGraaf, 2009) are now providing thorough characterizations to understand the roles of individual venom components in wasp parasitism.
The goal of this study was to pilot an analysis of venom gland components of a natural parasite of the most-highly studied insect host. Enzymatic and KEGG profiles of a limited number of molecules has revealed that the transcriptome contains a significant number of novel proteins whose functions may be unique to the parasitoid life history or to the function of the venom gland organ, including VLP biogenesis. The novel sequences found in this study must be addressed by future works in other Leptopilina species. Transcripts with similar sequence expressed in the same tissues will establish sequence and promote functional studies. The transcriptome contains numerous sequences for augmented protein production and robust secretion, which support the largely secretory function of the venom gland and its contribution to active venom production.
The sequence similarities reveal a set of putative effectors with predicted enzymatic activities (protease Cathepsin-D, acid and histidine phosphatases, and phospholipase B) conserved among other parasitoids and eusocial Hymenoptera. We have identified specific candidate molecules that might perturb host development (e.g., JH biosynthesis), host energetics, behavior, and nutrient availability (e.g, Drosophila foraging homolog, odorant-binding proteins), or host immune physiology (e.g., NF-κB inhibitor-interacting Ras-like protein, yellow family proteins, cytochrome P450s, various esterases, glutathione S-transferases) to support parasite progeny. The roles of these predicted proteins in the Lh venom remain to be tested. Prokaryotic and viral sequences are present in this dataset; their quantities are however too low to reveal the nature of this species’ VLPs. We have undertaken proteomic analysis of purified VLPs to address this question more directly.
Parasitoid wasps are known agents for biological control of insect pests. The cDNA clones and sequences reported here can be used to examine specific gene expression patterns, to develop physical maps of the wasp genome (Gokhman, 2011), and to confirm DNA assemblies derived from deep sequencing methods. Drosophila genetics will facilitate the analysis of specific Lh venom proteins with potential effects on host physiology in vivo. These studies will have a bearing on understanding similar host-parasite interactions. The characterization of inhibitory factors in the Lh venom has the potential to improve agriculture and human health as some proteins of this Drosophila parasite may also modulate physiologies of economically significant insect pests.
Supplementary Material
Supplementary Table S1: Cellular homeostasis pathways: L. heterotoma venom gland transcripts and the most significantly similar database protein or putative domain identities. Slight variations may exist between E-values listed here and Blasts with dbEST clone sequences due to phred base-calling and sequence length. * E-values are subject to change as the size of the nr NCBI database increases with time. Typically these values have been found to decrease, suggesting that these values are high-end estimates.
Supplementary Table S2: Putative venom-related proteins: L. heterotoma venom gland transcripts and the most significantly similar database protein or putative domain identities. Slight variations may exist between E-values listed here and Blasts with dbEST clone sequences due to phred base-calling and sequence length. * E-values are subject to change as the size of the nr NCBI database increases with time. Typically these values have been found to decrease, suggesting that these values are high-end estimates.
Supplementary Table S3: Taxonomic binning based on presence of highest similarity scoring protein: Data compiled from Tables S1 and S2.
Supplementary Table S4: L. heterotoma unigene hits within the PRIAM database.
Supplementary Table S5: L. heterotoma unigene hits within the KEGG database.
Supplementary Table S6: L. heterotoma clones and their associated National Center for Biotechnology Information (NCBI) Accession Numbers assigned upon acceptance into the database of Expressed Sequence Tags (dbEST).
HIGHLIGHTS.
A pilot transcriptome of the L. heterotoma venom gland complex yields 827 unigenes.
More than 150 novel transcripts revealed, lacking significant known similarities.
The remaining unigenes support conservation with venomous and stinging Hymenoptera.
A subset of these reported unigenes likely contribute to venom and host control.
A leading report of a figitid venom transcriptome targeting Drosophila hosts.
Acknowledgments
We thank undergraduate students of spring 2010 Bioinformatics class (Bio31312) and C. Chand for initial analyses of the sequence data. Thanks to W. Qiu (Hunter College) and R. Walsh (CUNY High Performance Computing Center) for help with bioinformatics and computing; to I. Paddibhatla and Z. Papadopol for help with experiments; and to A. Berkov and V. Gokhman for feedback on the manuscript. This publication was made possible by grants from NSF (1121817), USDA (NRI/USDA CSREES 2006-03817 and 2009-35302-05277), and NIH (8G12MD007603-27 and RISE 41399-009). Funding for cDNA library preparation came from P50-GM68762 to Peter Sorger (Harvard Medical School).
List of abbreviation GENE 38615
- A
adenosine
- Acph/ACPH–
acid phosphatase
- APIME
Apis mellifera
- Asp-Tyr-Asp
aspartate-tyrosine-aspartate
- bp
base pair
- C
cytidine
- CBP
chemosensory-binding protein
- CDD
Conserved Domain Database
- cGMP
cyclic guanosine monophosphate
- CO2
carbon dioxide
- cDNA
complementary deoxyribonucleic acid
- CUNY
City University of New York
- dbEST
Expressed Sequence Tags database
- dir
direct
- DNA
deoxyribonucleic acid
- EBI
European Bioinformatics Institute
- EC
Enzyme Commission
- EST
expressed sequence tag
- FA
farnesoic acid
- for
foraging gene
- G
guanosine
- GEF
guanine nucleotide exchange factor
- Glu-Cys
glutamate-cysteine
- hrs
hours
- IgE
immunoglobulin E
- JAK-STAT
Janus kinase-signal transducer and activator of transcription
- JH
juvenile hormone
- KAAS
KEGG Automatic Annotation Server
- kD
kiloDalton
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- Lh
Leptopilina heterotoma
- MAP
mitogen-activated protein
- mer
repeating unit
- ml
milliliter
- MUSCLE
Multiple Sequence Comparison by Log-Expectation
- MRJP
major royal jelly protein
- N
any nucleotide
- NCBI
National Center for Biotechnology Information
- NF-kappa B
nuclear factor kappa-light-chain-enhancer of activated B cells
- ng
nanogram
- NIH
National Institutes of Health
- NSF
National Science Foundation
- nr
non-redundant
- NRI
National Research Initiative
- OBP
odorant-binding protein
- ORF
open reading frame
- PBP_BOBP
pheromone-binding protein/general odorant-binding protein
- PCR
polymerase chain reaction
- PDV
polyDNA virus
- PF
Pfam accession number
- PKG
cyclic guanosine monophosphate-dependent protein kinase
- PLB
phospholipase B
- polyA
poly adenosine monophosphate
- PRIAM
Profils pour l’Identification Automatique du Métabolisme
- PSI
Position-Specific Iterated
- PTM
post-translation modification
- RISE
Research Initiative for Scientific Enhancement
- RNA
ribonucleic acid
- RRM
RNA Recognition Motif
- Ser/Thr
serine/threonine
- SMART
Simple Modular Architecture Research Tool
- spp
species pluralis
- STKc-PKA
Serine/Threonine Kinase, cAMP-dependent Protein Kinase
- T
thymidine
- TCA
tricarboxylic acid
- μg
microgram
- USDA
United States Department of Agriculture
- rev
reverse
- VLP
virus-like particle
- y w
yellow white
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn JE, Zhu-Salzman K. CmCatD, a cathepsin D-like protease has apotential role in insect defense against a phytocystatin. J Insect Physiol. 2009;55:678–685. doi: 10.1016/j.jinsphys.2009.04.016. [DOI] [PubMed] [Google Scholar]
- Albert S, Klaudiny J. The MRJP/YELLOW protein family of Apis mellifera: Identification of new members in the EST library. J of Insect Physiol. 2004;50:51–59. doi: 10.1016/j.jinsphys.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Allemand R, Lemaitre C, Frey F, Bouletreau M, Vavre F, Norlander G, van Alphen J, Carton Y. Phylogeny of six African Leptopilina species (Hymenoptera: Cynipoidea, Figitidae), parasitoids of Drosophila, with description of three new species. Ann Soc entomol Fr (ns) 2002;38:319–332. [Google Scholar]
- Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin ETB, Gulnik TN, Hosur S, Sowder MV, Cachau RC, Collins RE, Silva J, AM, Erickson JW. Crystal structures of native and inhibited forms of human cathepsin D: Implications for lysosomal targeting and drug design. PNAS. 1993;90:6796–6800. doi: 10.1073/pnas.90.14.6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboni E, Kemeny DM, Campos S, Vernon CA. The purification of acid phosphatase from honey bee venom (Apis mellifica) Toxicon. 1987;25:1097–1103. doi: 10.1016/0041-0101(87)90266-2. [DOI] [PubMed] [Google Scholar]
- Beckage N, Gelman D. Wasp parasitoid distruption of host development: Implications for new biologically based strategies for insect control. Annu Rev Entomol. 2004;49:299–330. doi: 10.1146/annurev.ento.49.061802.123324. [DOI] [PubMed] [Google Scholar]
- Beckage N, Riddiford L. Effects of parasitism by Apanteles congregatus on the endocrine physiology of the tobacco hornworm Manduca sexta. Gen Comp Endocrinol. 1982;47:308–322. doi: 10.1016/0016-6480(82)90238-6. [DOI] [PubMed] [Google Scholar]
- Belay A, Scheiner R, So AKC, Douglas S, Chakaborty-Chatterjee M, Levine J, Sokolowski M. The foraging gene of Drosophila melanogaster: spatial-expression analysis and sucrose responsiveness. J of Com Neurol. 2007;504:570–582. doi: 10.1002/cne.21466. [DOI] [PubMed] [Google Scholar]
- Bell HA, Powell ME, Price DRG, Weaver RJ. The biological effects of venom derived from the Ectoparasitic wasp Eulophus pennicornis (Nees) (Hymenoptera: Eulophidae): Evidence for dual endocrine regulation. The Open Entomology Journal. 2010;4:8–17. [Google Scholar]
- Biessmann H. Molecular analysis of the yellow gene (y) region of Drosophila melanogaster. PNAS. 1985;82:7369–7373. doi: 10.1073/pnas.82.21.7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitra K, Suderman R, Strand M. Polydnavirus Ank proteins bind NF-κB homodimers and inhibit processing of relish. PLoS Pathog. 2012;8:e1002722. doi: 10.1371/journal.ppat.1002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boquski M, Lowe T, Tolstoshev C. dbEST--database for “espressed sequence tags”. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Brehme KS. The effect of adult body color mutations upon the larva of Drosophila melanogaster. PNAS. 1941;27:254–261. doi: 10.1073/pnas.27.6.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carton Y, Bouletreau M, van Alphen JJM, van Lenteren JC. The Drosophila parasitic wasps. In: Ashburner MCL, Thompson JN, editors. The genetics and biology of Drosophila. Academic Press; London: 1986. pp. 347–394. [Google Scholar]
- Cherry S, Silverman N. Host-pathogen interactions in drosophila: new tricks from an old friend. Nat Immunol. 2006;7:911–917. doi: 10.1038/ni1388. [DOI] [PubMed] [Google Scholar]
- Chiu H, Govind S. Natural infection of D. melanogaster by virulent parasitic wasps induces apoptotic depletion of hematopoietic precursors. Cell Death Differ. 2002;9:1379–1381. doi: 10.1038/sj.cdd.4401134. [DOI] [PubMed] [Google Scholar]
- Chiu H, Morales J, Govind S. Identification and immuno-electron microscopy localization of p40, a protein component of immunosuppressive virus-like particles from Leptopilina heterotoma, a virulent parasitoid wasp of Drosophila. J Gen Virol. 2006;87:461–470. doi: 10.1099/vir.0.81474-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WL, Dhadialla TS, Raikhel AS. Purification and characterization of a lysosomal aspartic protease with cathepsin D activity from the mosquito. Insect Biochem. 1991;21:165–176. [Google Scholar]
- Cho WL, Raikhel AS. Cloning of cDNA for mosquito lysosomal aspartic protease. J of Biol Chem. 1992;267:21823–21829. [PubMed] [Google Scholar]
- Claudel-Renard C, Chevalet C, Faraut T, Kahn D. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2003;31:6633–6639. doi: 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford AM, Brauning R, Smolenski G, Ferguson C, Barton D, Wheeler T, McCulloch A. The consistuents of Microctonus sp. parasitoid venoms. Insect Mol Biol. 2008;17:313–324. doi: 10.1111/j.1365-2583.2008.00802.x. [DOI] [PubMed] [Google Scholar]
- Cruz CE, Fogaca AC, Nakayasu ES, Angeli CB, Belmonte R, Almeida IC, Mianda A, Miranda MTM, Tanaka AS, Braz GR, Craik CS, Schneider E, Caffrey CR, Daffre S. Characterization of proteinases from the midgut of Rhipicephalus (Boophilus) microplus involved in the generation of antimicrobial peptides. Parasites Vectors. 2010;3:63. doi: 10.1186/1756-3305-3-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani MP, Edwards JP, Richards EH. Hydrolase activity in the venom of the pupal endoparasitic wasp, Pimpla hypochondriaca. Comp Biochem Phys B. 2005;141:373–381. doi: 10.1016/j.cbpc.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Danneels EL, Rivers DB, de Graaf DC. Venom proteins of the parasitoid wasp Nasonia vitripennis: Recent discovery of an untapped pharamacopee. Toxins. 2010;2:494–516. doi: 10.3390/toxins2040494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregori E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. PNAS. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deGraaf DC, Aerts M, Brunain M, Desfardins CA, Jacobs FJ, Werren JH, Devreese B. Insights into the venom composition of the ectoparsitoid wasp Nasonia vitripennis form bioinformatic and proteomic studies. Insect Molecular Biology. 2010;19:11–26. doi: 10.1111/j.1365-2583.2009.00914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deGraaf DC, Aerts M, Dannells E, Devreese B. Bee, wasp and ant venomics pave the way for a component-resolved diagnosis of sting allergy. J Proteomics. 2009;72:145–154. doi: 10.1016/j.jprot.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Doery HM, Pearson JE. Phospholipase B in snake venoms and bee venom. Biochem J. 1964;92:599–602. doi: 10.1042/bj0920599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover BA, Menon A, Brown RC, Strand MR. Suppression of jevenile hormone esterase in Heliothis virescens by Microplitis demolitor calyx fluid. J Insect Physiol. 1995;41:809–817. [Google Scholar]
- Drapeau M, Albert S, Kucharski R, Prusko C, Maleszka R. Evolution of the Yellow/MRJP family and the emergence of social behavior in honey bees. Genome Res. 2006;16:1385–1394. doi: 10.1101/gr.5012006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapeau MD, Radovic A, Wittkopp PJ, Long AD. A gene necessary for normal male courtship, yellow, acts downstream of fruitless in the Drosophila melanogaster larval brain. J Neurobiol. 2003;55:53–72. doi: 10.1002/neu.10196. [DOI] [PubMed] [Google Scholar]
- Dubuffet A, Colinet D, Anselme C, Dupas S, Carton Y, Poirie M. Chapter 6 Variation of Leptopilina boulardi success in Drosophila hosts: What is inside the black box? In: Prevost G, editor. Advances in Parasitology. Academic Press; 2009. pp. 147–188. [DOI] [PubMed] [Google Scholar]
- Dupuy C, Periquet G, Serbielle C, Bezier A, Louis F, Drezen JM. Transfer of a chromosomal Maverick to endogenous bracovirus in a parasitoid wasp. Genetica. 2011;139:489–496. doi: 10.1007/s10709-011-9569-x. [DOI] [PubMed] [Google Scholar]
- Edgar R. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004a;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004b;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998a;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl M, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998b;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Ferguson LC, Green J, Surridge A, Jiggins CD. Evolution of the Insect Yellow Gene Family. Mol Biol Evol. 2011;28:257–272. doi: 10.1093/molbev/msq192. [DOI] [PubMed] [Google Scholar]
- Ferrarese R, Morales J, Fimiarz D, Webb B, Govind S. A supracellular system of actin-lined canals controls biogenesis and release of virulence factors in parasitoid venom glands. J Exp Biol. 2009;212:2261–2268. doi: 10.1242/jeb.025718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate P, Coggill P, Heger A, Polington JE, Gavin OL, Gunesekaran G, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Batemen A. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusek M, Vetvicka V. Dual Role of Cathepsin D: Ligand and Protease. Biomed Papers. 2005;149:43–50. doi: 10.5507/bp.2005.003. [DOI] [PubMed] [Google Scholar]
- Gammie S, Truman J. Ecolsion hormone provides a link between ecdysis-triggering hormone and crustacean cardioactive peptide in the neuroendocrine cascade that controls ecdysis behavior. J Exp Biol. 1999;202:343–352. doi: 10.1242/jeb.202.4.343. [DOI] [PubMed] [Google Scholar]
- Georgieva D, Greunke K, Genov N, Betzel C. 3-D Model of the bee venom acid phosphatase: Insights into allergenicity. Biochem Bioph Res Co. 2009;378:711–715. doi: 10.1016/j.bbrc.2008.11.101. [DOI] [PubMed] [Google Scholar]
- Gokhman V, Johnston J, Small C, Rajwani R, Hanrahan S, Govind S. Genomic and karyotypic variation in Drosophilia parasitoids (Hymenoptera, Cynipoidea, Figitidae) Comp Cytogen. 2011;5:211–221. doi: 10.3897/CompCytogen.v5i3.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind S. Innate immunity in Drosophila: Pathogens and pathways. Insect Sci. 2008;15:29–43. doi: 10.1111/j.1744-7917.2008.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald T, Bockisch B, Spillner E, Ring J, Bredehorst R, Ollert MW. Molecular clonign and expression in insce cells of honeybee venom allergen acid phosphatase (Api m 3) J Allergy Clin Immun. 2006;117:8948–854. doi: 10.1016/j.jaci.2005.12.1331. [DOI] [PubMed] [Google Scholar]
- Gui ZZ, Lee KS, Kim BY, Choi YS, Wei YD, Choo YM, Kang PD, Yoon HY, Kim I, Je YH, Seo SJ, Lee SM, Guo X, Shon HD, Jin BR. Functional role of aspartic proteinase cathepsin D in insect metamorphosis. BMC Developmental Biology. 2006;6:1–11. doi: 10.1186/1471-213X-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton C, Lejeune BT, Rosengaus RB. Trophallaxis and prophylaxis: social immunity in the carpenter ant Camponotus pennsylvanicus. Biol Lett. 2010;7:89–92. doi: 10.1098/rsbl.2010.0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q, Fang J, Ding H, Johnson JK, Christensen BM, Li J. Identification of Drosophila melanogaster yellow-f and yellow-f2 proteins as dopachrome-conversion enzymes. Biochem J. 2002;368:333–340. doi: 10.1042/BJ20020272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DR. Allergens in bee venom: III. Identification of allergen B of bee venom as an acid phosphatase. J Allergy Clin Immun. 1977;59:364–366. doi: 10.1016/0091-6749(77)90019-7. [DOI] [PubMed] [Google Scholar]
- Houseman JG, Downe AER. Cathepsin D-like activity in the posterior midgut of hemipteran insects. Comparative Biochemistry and Physiology B-Biochemistry & Molecular Biology. 1983;75:509–512. [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Genova G, Roberts M, Jackson FR. The LARK RNA-binding protein selectely regulates the cricadian ecolsion rhythm by controling E74 protein expression. PLos ONE. 2007:e1107. doi: 10.1371/journal.pone.0001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AY, Gulnik SV, Erickson JW. Conformational switching in an aspartic proteinase. Nat Struct Mol Biol. 1998;5:866–871. doi: 10.1038/2306. [DOI] [PubMed] [Google Scholar]
- Lee M, Kalamarz M, Paddibhatla I, Small C, Rajwani R, Govind S. Chapter 5 Virulence factors and strategies of Leptopilina spp.: Selective responses in Drosophila hosts. In: Prevost G, editor. Advances in Parasitology. Academic Press; 2009. pp. 123–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M Uni Prot Consortium. UniProt Knowledgebase: a hub of integrated protein data. Database. 2011 doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makka T, Seino A, Tomita S, Fujiwara H, Sonobe H. A possible role of 20-hydroxyecdysone in embryonic development of the silworm Bombyx mori. Insect Biochem and Phys. 2002;51:111–120. doi: 10.1002/arch.10055. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–31. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010;39:D225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FHT. The RNA recognition motif, a plastic RNA-binding platform to regualate post-transcriptional gene expression. FEBS Jol. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- McKerrow J, Caffrey C, Kelly B, Like P, Sajid M. Proteases in Parasitic Diseases. Annu Rev Pathol Mech Dis. 2006a;1:497–536. doi: 10.1146/annurev.pathol.1.110304.100151. [DOI] [PubMed] [Google Scholar]
- McKerrow JH, Caffrey C, Kelly B, Loke P, Sajid M. Proteases in Parasitic Diseases. Annu Rev Pathol Mech Dis. 2006b;1:497–536. doi: 10.1146/annurev.pathol.1.110304.100151. [DOI] [PubMed] [Google Scholar]
- Morales J, Chiu H, Oo T, Plaza R, Hoskins S, Govind S. Biogenesis, structure, and immune-suppressive effects of virus-like particles of a Drosophila parasitoid, Leptopilina victoriae. J Insect Physiol. 2005;51:181–195. doi: 10.1016/j.jinsphys.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Morgan CP, Insall R, Hynes L, Cockcroft S. Identification of phospholipase B from Dictyostelium discoideum reveals a new lipase family present in mammals, flies, and nemaodes, but not yeast. Biochem J. 2004;382:441–449. doi: 10.1042/BJ20040110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa A, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamatsu Y, Tanaka T. Development of a gregarious ectoparasitoid, Euplectrus separatae (Hymenoptera; Hymenoptera: Eulophidae), that parasitizes Pseudaletia separata (Lepidoptera: Noctuidae) Arthropod Struct Dev. 2003a;32:329–336. doi: 10.1016/j.asd.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Nakamatsu Y, Tanaka T. Venom of ectoparasitoid, Euplectrus sp. near plathypenae (Hymenoptera: Eulophidae) regulates the physiological state of Pseudaletia separata (Lepidoptera: Noctuidae) host as a food resource. J Insect Physiol. 2003b;49:149–159. doi: 10.1016/s0022-1910(02)00261-5. [DOI] [PubMed] [Google Scholar]
- Nakamatsu Y, Tanaka T. Venom of Euplectrus separatae causes hyperlipidemia by lysis of host fat body cells. J Insect Physiol. 2004;50:267–275. doi: 10.1016/j.jinsphys.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Nappi A, Poirie M, Carton Y. Chapter 4 The role of melanization and cytotoxic by-products in the cellular immune responses of Drosophila against parasitic wasps. In: Prevost G, editor. Advances in Parasitology. Academic Press; 2009. pp. 99–121. [DOI] [PubMed] [Google Scholar]
- Needleman S, Wunsch C. A general method applicable to the search for similarities in th eamino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Newby L, Jackson FR. A new biological rhythm mutant of Drosophila melanogaster that identifies a gene with an essential embryonic function. Genetics. 1993;135:1077–1090. doi: 10.1093/genetics/135.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhout H, Williams C. Control of moulting and metamorphosis in the tobacco hornworm, Manduca sexta (L.): cessation of juvenile hormone secretion as a trigger for pupation. J Exp Biol. 1974;61:493–501. doi: 10.1242/jeb.61.2.493. [DOI] [PubMed] [Google Scholar]
- Noguchi H. Snake venoms; an investigation of venomous snakes: with special reference to the phenomena of their venoms. The Carnegie Institute of Washington; 1909. [Google Scholar]
- Oakenshott J, Johnson R, Berenbaum M, Ranson H, Cristino A, Claudianos C. Metabolic enzymes associated with xenobiotic and chemosensory responses in Nasonia vitripennis. Insect Mol Biol. 2010;19:147–163. doi: 10.1111/j.1365-2583.2009.00961.x. [DOI] [PubMed] [Google Scholar]
- Osborne K, Robichon A, Burgess E, Butland S, Shaw R, Coulthard A, Pereira H, Greenspan R, Sokolowski M. Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science. 1997;277:834–836. doi: 10.1126/science.277.5327.834. [DOI] [PubMed] [Google Scholar]
- Parkinson N, Conyers C, Smith I. A venom protein from the endoparasitoid wasp Pimpla hypochondriaca is similar to snake venom reprolysin-type metalloproteases. J of Invertebr Pathol. 2002a;79:129–131. doi: 10.1016/s0022-2011(02)00033-2. [DOI] [PubMed] [Google Scholar]
- Parkinson N, Richards EH, Conyers E, Smith I, Edwards JP. Analysis of venom consistuents from the parasoitoid wasp Pimpla hypocondriaca ad clonign of a cDNA eoncoding a venom protein. Insect Biochem Molec. 2002b;32 doi: 10.1016/s0965-1748(01)00155-2. [DOI] [PubMed] [Google Scholar]
- Peiren N, de Graaf D, Vanrobaeys F, Danneels E, Devreese B, Van Beeumen F, Jacobs F. Proteomic analysis of the honey bee worker veom gland focusing on the mechanisms of protection against tissue damage. Toxicon. 2008;52 doi: 10.1016/j.toxicon.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Peiren N, Vanrobaeys F, de Graaf D, Devreese B, Van Beeumen J, Jacobs F. The protein composition of honeybee venom reconsidered by a proteomic approach. BBA. 2005;1752:1–5. doi: 10.1016/j.bbapap.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Pelosi P. Odorant-binding proteins. Crit Rev Biochem Mol Biol. 1994;29:199–228. doi: 10.3109/10409239409086801. [DOI] [PubMed] [Google Scholar]
- Pelosi P. Perireceptor events in olfaction. J Neurobiol. 1996;30:3–19. doi: 10.1002/(SICI)1097-4695(199605)30:1<3::AID-NEU2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Pelosi P, Calvello M, Ban L. Diversity of odorant-binding proteins and chemosensory proteins in insects. Chem Senses. 2005;30:i291–i292. doi: 10.1093/chemse/bjh229. [DOI] [PubMed] [Google Scholar]
- Pennacchio F, Strand MR. Evolution of developmental strategies in parasitic Hymenoptera. Annu Rev Entomol. 2006;51:233–258. doi: 10.1146/annurev.ento.51.110104.151029. [DOI] [PubMed] [Google Scholar]
- Prevost G, Doury G, Mabiala-Moundoungou A, Cherqui A, Eslin P. Chapter 9 Strategies of avoidance of host immune defenses in Asobara species. In: Prevost G, editor. Advances in Parasitology. Academic Press; 2009. pp. 235–255. [DOI] [PubMed] [Google Scholar]
- Pruitt K, Tatusova T, Maglott D. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence databae of genomes, transcripts and proteins. Nucleic Acids Res. 2004;33:D5010–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers D, Denlinger D. Redirection of metabolism in the flesh fly, Sarcophaga bullata, following envenomation by the ectoparasitoid Nasonia vitripennis and correlation of metabolic effects with the diapause status of the host. J of Insect Physiol. 1994;40:207–215. [Google Scholar]
- Rivers DB, Denlinger DL. Venom-induced alternations in fly lipid metabolism and its impact on larval development of the ectoparasitoid Nasonia vitripennis (Walker) (Hymenoptera: Pteromalidae) J Invertebr Pathol. 1995;66:104–110. [Google Scholar]
- Rivers DB, Ruggiero L, Hayes M. The ectoparasitic wasp Nasonia vitripennis (Walker) (Hymenoptera: Pteromalidae) differentially affects cells mediating the immune responseof its flesh fly host, Sarcophaga bullata Parker (Diptera: Sarcophagidae) J Insect Physiol. 2002;48:1053–1064. doi: 10.1016/s0022-1910(02)00193-2. [DOI] [PubMed] [Google Scholar]
- Rizki T, Rizki R. Lamellocyte differentiation in Drosophila larvae parasitized by Leptopilina. Dev Comp Immunol. 1992;16:103–110. doi: 10.1016/0145-305x(92)90011-z. [DOI] [PubMed] [Google Scholar]
- Roberg K, Johansson U, Ollinger K. Lysosomal release of Cathepsin D precedes relocation of Cytochrom C and loss of mitochondrial transmembrane potential during apoptosis induced by oxiative stress. Free Radical Mio Med. 1999;27:1228–1237. doi: 10.1016/s0891-5849(99)00146-x. [DOI] [PubMed] [Google Scholar]
- Altschul STM, Schäffer A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafellner C, Marktl RC, Schopf A. Inhibition of juvenile hormone esterase activiy in Lymantria dispar (Lepidoptera, Lymantriidae) larvae parasitized by Glyptanpanteles liparidis (Hymenoptera, Braconidae) J Insect Physiol. 2007;53:858–868. doi: 10.1016/j.jinsphys.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Schilthuizen M, Nordlander G, Stouthamer R, van Alphen J. Morphological and molecular phylogenetics in the genus Leptopilina (Hymenoptera: Cynipoidea: Eucoilidae) Syst Entomol. 1998;23:253–264. [Google Scholar]
- Schlenke T, Morales J, Govind S, Clark A. Contrasting infection strategies in generalist and specialist wasp parasitoids of Drosophila melanogaster. PLoS Pathog. 2007;26:1486–1501. doi: 10.1371/journal.ppat.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Hempel P. Natural insect host-parasite system show immune priming and specificity: puzzles to be solved. Bioessays. 2005;27:1026–1034. doi: 10.1002/bies.20282. [DOI] [PubMed] [Google Scholar]
- Scott J. Insect cytochrome P450s: Thinking beyond detoxification. In: Liu N, editor. Recent Advances in Insect Physiology, Toxicology, and Molecular Biology. Signpost; Kerala, India: 2008. pp. 117–124. [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson T, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson J, Higgins D. Fast, scalable generation of high-quality protien multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonobe H, Tokushige H, Makka T, Hara N, Fujimoto Y. Comparative studies of ecdysteroid metabolism between diapause eggs and non-diapause eggs of the silkworm, Bombyx mori. Zool Sci. 1999;16:935–943. [Google Scholar]
- Stoka V, Turk V, Turk B. Lysosomal cysteine cathepsins: signaling pathways in apoptosis. Biol Chem. 2007;388:555–560. doi: 10.1515/BC.2007.064. [DOI] [PubMed] [Google Scholar]
- Strand M, Burke G. Polydnaviruses as symbionts and gene delivery systems. PLoS Pathog. 2012;8:e1002757. doi: 10.1371/journal.ppat.1002757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji T, Ip Y. Regulators of the Toll and Imd pathways in the Drosophila innate immune response. Trends Immunol. 2005;26:193–198. doi: 10.1016/j.it.2005.02.006. [DOI] [PubMed] [Google Scholar]
- The Nasonia Working Group etal. Functional and Evolutionary Insights from the Genomes of Three Parasitoid Nasonia Species. Science. 2010;237:343–347. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd C. An anti-serum for scorpion venom. J Hyg (London) 1909;9:69–85. doi: 10.1017/s0022172400016144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uckan F, Ergin E, Rivers D, Gencer N. Age and diet influence the composition of venom from the endoparasitic wasp Pimpla turionellae L. (Hymenoptera: Ichneumonidae) Arch Insect Biochem. 2006;63:177–187. doi: 10.1002/arch.20154. [DOI] [PubMed] [Google Scholar]
- Vieira F, Foret S, He X, Rozas J, Field LM, Zhou JJ. Unique features of odorant-binding proteins of the paraisoid waps Nasonia vitripennis revealed by genome annotation and comparative analyses. PLos ONE. 2012;7:e43034. doi: 10.1371/journal.pone.0043034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent B, Kaeslin M, Roth T, Heller M, Poulain J, Cousserans F, Schaller J, Poirie M, Lanzrein B, Drezen JM, Moreau SJM. The venom composition of the parasitic wasp Chelonus inanitus resolved by combined expressed sequence tags analysis and proteomic approach. BMC Genomics. 2010;11:693–707. doi: 10.1186/1471-2164-11-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson S, Iwantsch G. Host Regulation by Insect Parasitoids. Quart Rev Biol. 1980;55:143–165. [Google Scholar]
- Walker I. Die Abwehrreaktion des Wirtes Drosophila melanogaster gegen die zoophage Cynipide Pseudeucoila bochei Weld. Rev Suisse Zool. 1959;66:569–632. [Google Scholar]
- Wu S, Zhu Z, Fu L, Niu B, Li W. WebMGA: a customizable web server for fast metagonoic sequence analysis. BMC Genomics. 2011;12:244. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Clarkson JM, Charnely AK. Acid phosphatases of Metarhizium anisopliae during infection of the tobacco hormworm Manduca sexta. Arch Microbiol. 2001;176:427–434. doi: 10.1007/s002030100342. [DOI] [PubMed] [Google Scholar]
- Xia Y, Dean P, Judge AJ, Gillespie JP, Clarkson JM, Charnely AK. Acid phosphatases in the haemolymph of the desert locust, Schistocerca gregaria, infected with the entomopathogenic fungus Metarhizium anisopliae. J Insect Physiol. 2000;46:1249–1257. doi: 10.1016/S0022-1910(00)00045-7. [DOI] [PubMed] [Google Scholar]
- Zhang H, Tian L, Tobe S, Xiong Y, Wang S, Lin X, Liu Y, Bendena W, Li S, Zhang YQ. Drosophila CG10527 mutants are resistant to juvenile hormone and its analog methoprene. Biochem Bioph Res Co. 2010;401:182–187. doi: 10.1016/j.bbrc.2010.09.019. [DOI] [PubMed] [Google Scholar]
- Zhu J-y, Ye G-y, Dong S-Z, Fang Q, Hu C. Venom of Pteromalus puparum (Hymenoptera: Pteromalidae) induced endocrine changes in the hemolymph of its host, Pieris rapae (Lepidoptera: Pieridae) Insect Biochem and Phys. 2009;71:45–53. doi: 10.1002/arch.20304. [DOI] [PubMed] [Google Scholar]
- Zhu J-y, Ye G-y, Hu C. Molecular cloning and characterization of acid phosphatase in venom of the endoparasitoid wasp Pteromalus puparum (Hymenoptera: Pteromalidae) Toxicon. 2008;51:1391–1399. doi: 10.1016/j.toxicon.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Machleder E, Chenchik A, Li R, Siebert P. Reserve transcriptase template switching: A SMART approach for full-length cDNA library construction. BioTechniques. 2001;30:892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: Cellular homeostasis pathways: L. heterotoma venom gland transcripts and the most significantly similar database protein or putative domain identities. Slight variations may exist between E-values listed here and Blasts with dbEST clone sequences due to phred base-calling and sequence length. * E-values are subject to change as the size of the nr NCBI database increases with time. Typically these values have been found to decrease, suggesting that these values are high-end estimates.
Supplementary Table S2: Putative venom-related proteins: L. heterotoma venom gland transcripts and the most significantly similar database protein or putative domain identities. Slight variations may exist between E-values listed here and Blasts with dbEST clone sequences due to phred base-calling and sequence length. * E-values are subject to change as the size of the nr NCBI database increases with time. Typically these values have been found to decrease, suggesting that these values are high-end estimates.
Supplementary Table S3: Taxonomic binning based on presence of highest similarity scoring protein: Data compiled from Tables S1 and S2.
Supplementary Table S4: L. heterotoma unigene hits within the PRIAM database.
Supplementary Table S5: L. heterotoma unigene hits within the KEGG database.
Supplementary Table S6: L. heterotoma clones and their associated National Center for Biotechnology Information (NCBI) Accession Numbers assigned upon acceptance into the database of Expressed Sequence Tags (dbEST).