Abstract
Objective
To investigate whether mRNA expression of TMPRSS2-ERG fusion gene, a suggested prostate cancer (PCa) biomarker, was specific to cancerous lesions alone and to study the expression of SPINK1 and PCA3 mRNAs in the same cohort to also explore the proposed mutual exclusivity of TMPRSS2-ERG and SPINK1 expression.
Methods
Levels of two TMPRSS2-ERG transcripts, PCA3, and SPINK1 mRNAs were measured with highly standardized RT-qPCR assays in cystoprostatectomy specimens from 19 invasive bladder cancer patients and in 174 radical prostatectomy (RP) samples [88 histologically benign prostate (HBP) tissues and 86 from cancerous lesions] from 87 patients with clinically localized PCa.
Results
Expression of TMPRSS2-ERG transcripts was detected in 45/88 (51%) HBP tissues from RP specimens, and more frequently (57/86, 66%) found in cancerous lesions. By contrast, TMPRSS2-ERG expression was detected in only 2/19 (11%) cystoprostatectomy specimens, both with incidental PCa foci elsewhere in the gland. Similar trends of changes in the expression of PCA3 and SPINK1 were present in HBP tissue from RP compared to cystoprostatectomy specimens.
Conclusions
Although expression of TMPRSS2-ERG, SPINK1, and PCA3 mRNA is higher or more frequently found in cancerous lesions, HBP tissues from patients with clinically localized PCa manifest molecular, mRNA level changes that are absent in cystoprostatectomy specimens lacking incidental PCa foci, or infrequent in cystoprostatectomy specimens containing incidental PCa. If this finding is replicated, these molecular assays could be used to inform men with negative biopsies about the likelihood of cancerous lesions in unsampled regions and hence the need for repeat biopsy.
Keywords: Prostatic Neoplasms, Gene Expression, Reverse Transcriptase Polymerase Chain Reaction, TMPRSS2-ERG, PCA3, SPINK1
INTRODUCTION
Fusion between genes TMPRSS2 (transmembrane protease serine 2) and ERG (ETV related gene) has been suggested as a novel, cancer-specific marker for prostate cancer (PCa)1. With the high prevalence of PCa, it may be the most common cancer-associated genetic rearrangement as studies have shown it to be present in about 40–70% of PCa cases. This gene fusion has been identified in a subset of prostatic tumors with potentially distinct disease characteristics. The details of those characteristics are still debated as contradictory results on different transcript isoforms and their effects on prognosis have been reported2,3. The cancer specificity of this gene fusion has been questioned as TMPRSS2-ERG expression has been detected also in histologically benign tissue of cancerous prostates4,5,6, but cancer-related changes of gene expression in non-neoplastic tissue of cancer-affected prostates are not unheard of and have been reported for other biomarkers as well7,8,9.
Another subgroup of PCa was suggested when SPINK1 mRNA, often studied in conjunction with pancreatic diseases but also overexpressed in PCa10, was found to be overexpressed preferentially in cancers lacking TMPRSS2-ERG fusion11,12. However, a recent study by Leinonen et al contradicts this by showing no signs of their mutually exclusive expression13.
As most of the previous TMPRSS2-ERG studies have been DNA- and FISH-based, we set out to study the mRNA transcript levels of the fusion gene, targeting two of the over 20 transcript isoforms that have been reported14: transcripts III and VI. TMPRSS2-ERG III is the most commonly expressed isoform1 and TMPRSS2-ERG VI was chosen for its claimed association with aggressive disease15. For this purpose, we developed highly sensitive, truly quantitative and internally standardized reverse-transcription PCR (RT-qPCR) assays for both TMPRSS2-ERG transcript isoforms as well as for SPINK1 mRNA, employing a previously described assay concept16. The mRNA levels of these three markers were measured in 86 cancerous prostate tissues and 88 histologically benign prostate tissues (HBP) from 87 PCa patients, and in prostatic tissue specimens obtained by cystoprostatectomy from 19 patients with invasive cancer of the urinary bladder and with no history of PCa. In addition to measuring the absolute levels of expression with a truly quantitative method, our aim was to find out whether the previously reported4,5,6, occasional presence of the gene fusion in benign tissue in men with PCa was evidence of a field effect or due to the gene fusion not being truly cancer specific. We also sought to explore the claimed mutual exclusivity between SPINK1 and TMPRSS2-ERG fusion gene and whether there were alterations in their expression levels correlating to the neoplastic characteristics of the tissue. Association with PCa stage and tumor percentage was also assessed for all transcripts.
Our previously reported data on higher-than-expected levels of PCA3 in HBP tissue of cancer-affected prostates compared to cancerous tissues17 raised questions on the field effect hypothesis regarding that gene. Those issues were also explored further in this study by measuring the mRNA levels of PCA3 in the 19 cystoprostatectomy specimens and by comparing those levels to the previously obtained values of PCA3 expression in HBP and cancerous tissue of these 87 PCa patients.
MATERIALS AND METHODS
Tissue samples
We obtained 174 prostate tissue samples from 87 PCa patients immediately after radical prostatectomy (RP) at Turku University Hospital, Turku, Finland. Two small sample wedges were taken from each prostate, one intending to sample the suspected cancer area and the other the control area. A small tissue sample size ensured the best possible homogeneity of material. Based on the histological examination of the immediately adjacent tissue surrounding each sampling site, an experienced senior genitourinary pathologist classified 76 samples as histologically benign tissue, 12 as prostatic intraepithelial neoplasia (PIN), and 86 as cancerous tissue ranging from Gleason grade 2 to 5. The PIN samples were considered as histologically benign tissue in all subsequent analyses unless stated otherwise. The estimated median proportion of cancerous tissue in cancerous samples was 30% (interquartile range: 10, 55). Patient characteristics are shown in Supplemental Table 1 and described previously17. None of the patients received prior radiation therapy. Samples were stored as previously described18.
We also obtained prostate tissue samples from 19 patients undergoing cystoprostatectomy for treatment of invasive bladder cancer at Skåne University Hospital, Malmö, Sweden. Samples were obtained from the peripheral apical or dorsolateral portion of the prostate within 15 minutes after surgical removal and stored deep-frozen. Incidental, small foci of prostate adenocarcinomas were found upon further histopathological assessment in 12/19 of the cystoprostatectomy specimens but not anywhere close to the areas sampled for RNA isolation. Seven cystoprostatectomy specimens contained no evidence of incidental PCa in the prostate.
The study protocol was approved by local Ethics committees and it was in accordance with the Helsinki Declaration of 1975, as revised in 1996, with written informed consent obtained from each participant.
RNA extraction and reverse transcription
All tissue samples were processed as previously described18. RNA extraction protocol included the addition of a known amount of artificial RNA19 to the samples to act as an internal control, tracking the inherent loss of RNA material throughout the process.
Real-time qPCR
Expression levels of SPINK1 and PCA3 mRNAs and TMPRSS2-ERG fusion transcripts types III and VI were measured with optimized RT-qPCR assays using target-specific oligonucleotide probes and time-resolved fluorometry in detection as described previously18 and in Supplemental Tables 2, 3, and 4. KLK3 mRNA levels were also determined with an RT-qPCR assay with the same concept16,21,22 to ensure the presence of prostate cells. Data analysis is described in detail in Supplemental Data 1.
RESULTS
KLK3
There were no significant differences in KLK3 mRNA levels when we compared the expression in cancerous versus HBP tissue from RP specimens (p=0.2) from men with clinically localized PCa as previously reported17. However, KLK3 expression tended to be higher in HBP tissue from RP compared to cystoprostatectomy specimens (p=0.032).
TMPRSS2-ERG
Overall, there was evidence of TMPRSS2-ERG III expression in 55/87 (63%) and TMPRSS2-ERG VI expression in 51/87 (59%) men with clinically localized PCa. Individual tissue samples contained either both of the fusion transcript types, only one of them or neither. Both transcripts were detected in 43/87 (49%) patients, with at least one of the fusion transcripts detected in 63/87 (72%) of these patients.
Of the 88 HBP tissue samples from RP specimens from men with clinically localized PCa, more than half (45/88 or 51%) showed expression of at least one of the TMPRSS2-ERG transcripts (Table 1). Expression of either or both of the TMPRSS2-ERG transcripts was detected in 57/86 (66%) cancerous tissue samples from the RP specimens. By contrast, TMPRSS2-ERG expression was not detected in prostatic tissue from 17 patients with invasive bladder cancer treated with cystoprostatectomy, whereas it was detectable in prostate tissue from two cystoprostatectomy specimens which contained foci of incidental PCa elsewhere in the gland.
Table 1.
Frequencies of TMPRSS2-ERG III and VI mRNA expression detected in histologically benign tissues of prostates with clinical PCa, PIN tissue, cancerous prostate tissues and in tissues of clinical PCa-free prostates. PIN, prostatic intraepithelial neoplasia; RP, radical prostatectomy.
| Number of samples | |||||
|---|---|---|---|---|---|
| Only TMPRSS2- ERG III mRNA was detected |
Only TMPRSS2- ERG VI mRNA was detected |
Both TMPRSS2- ERG III and VI mRNAs were detected |
Neither TMPRSS2- ERG III nor VI mRNA were detected |
Total | |
| Histologically benign tissue from RP specimens (including the 12 PIN samples) |
11 (13%) | 10 (11%) | 24 (27%) | 43 (49%) | 88 |
| PIN tissue from RP specimens |
0 (0%) | 4 (33%) | 3 (25%) | 5 (42%) | 12 |
| Cancerous prostate tissue from RP specimens |
10 (12%) | 8 (9%) | 39 (45%) | 29 (34%) | 86 |
| Prostate tissue from cystoprostatectomy specimens |
0 (0%) | 1 (5%) | 1 (5%) | 17 (89%) | 19 |
Trends of field effect were observed particularly in the paired samples. Of the 48 men from whom both a cancerous and benign sample were obtained, 30 manifested either or both of the TMPRSS2-ERG transcripts in the cancerous sample and 21/30 (70%) of them had detectable TMPRSS2-ERG transcripts also in the matched HBP tissue. In addition, evidence of multi-clonality was implicated in glands presenting with two cancerous samples. Of the 19 men from whom two cancerous RP samples were obtained, 2 manifested only TMPRSS2-ERG III mRNA in one of the samples and only TMPRSS2-ERG VI mRNA in the other. Concordance of expression of the two TMPRSS2-ERG transcripts between the paired samples is described in full detail in Supplemental Tables 5 and 6.
Both TMPRSS2-ERG fusion transcripts were statistically significantly more frequently detected in cancerous than in HBP tissue of the RP specimens from men with clinically localized PCa (for TMPRSS2-ERG III, p=0.022 and for TMPRSS2-ERG VI, p=0.026). Also the differences in the presence of fusion transcripts between HBP tissue from prostates with clinically localized PCa and cystoprostatectomy specimens were statistically significant for both fusion transcripts (p=0.020 for TMPRSS2-ERG III and p=0.033 for TMPRSS2-ERG VI). There was no statistically significant difference in TMPRSS2-ERG III and VI levels between PIN samples and HBP tissue from RP specimens (p=0.2 and 0.11 respectively).
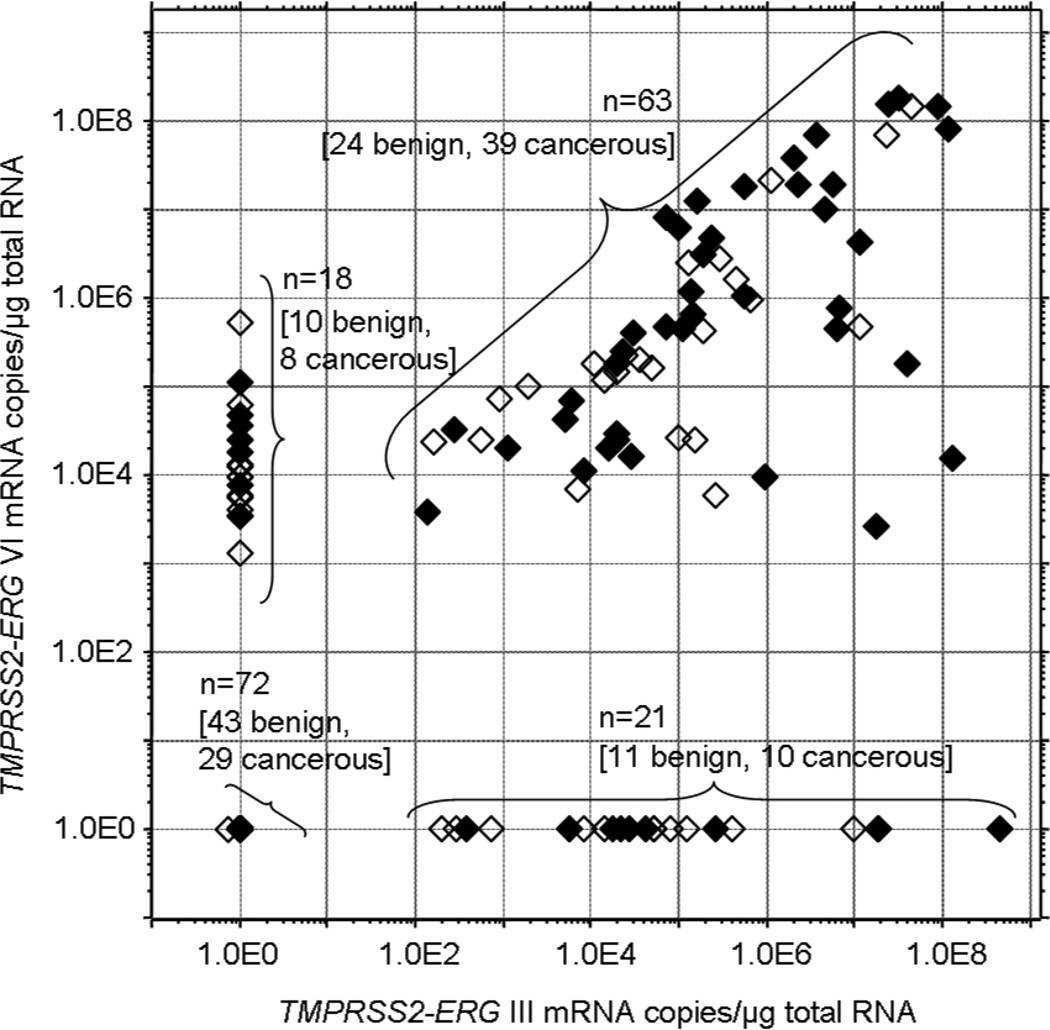
Among the fusion transcript positive samples of the RP specimens from clinically localized PCa, the expression levels of each isoform varied over 5–6 orders of magnitude with no statistically significant difference of mRNA levels between the cancerous and histologically benign groups (Figure 1). The TMPRSS2-ERG VI expression levels of the two cystoprostatectomy samples in which the expression of this mRNA was detectable, were 3.5×104 and 4.6×105 mRNA copies per µg of total RNA. The latter sample also presented TMPRSS2-ERG III mRNA expression on the level of 3.2×103 mRNA copies per µg of total RNA.
Figure 1.
TMPRSS2-ERG III and VI mRNA levels in 174 radical prostatectomy samples. The open diamonds denote histologically benign tissue samples and the closed diamonds represent cancerous sample. When mRNA was not detected, the sample was given the value 1.
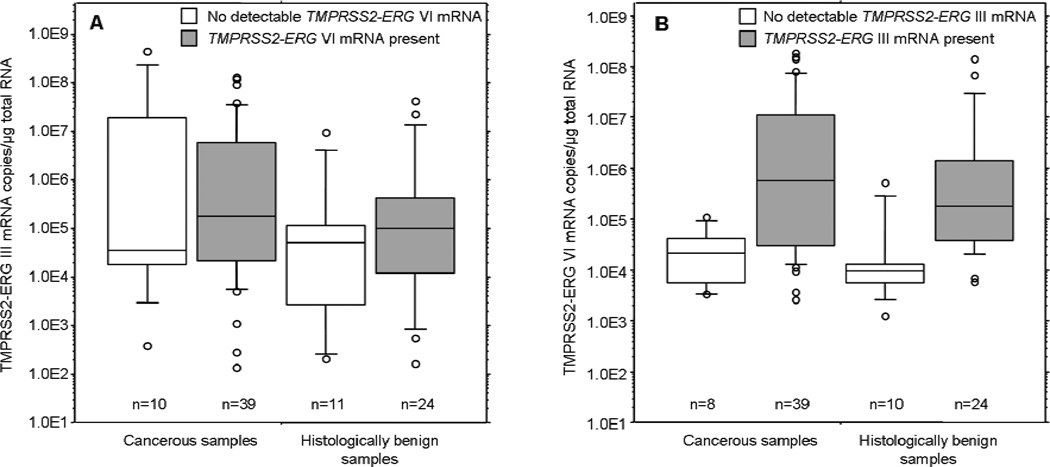
Within the samples where TMPRSS2-ERG VI mRNA was detectable, isoform VI levels were statistically significantly higher if the same sample also showed detectable III isoform expression (Figure 2). This phenomenon was seen both in cancerous (p=0.004 by Mann-Whitney) and HBP samples (p=0.001). However, no statistically significant difference (p=0.18) was observed in TMPRSS2-ERG III levels regarding whether the samples also expressed TMPRSS2-ERG VI or not (Figure 2).
Figure 2.
A. TMPRSS2-ERG III mRNA levels in cancerous and histologically benign samples with (grey) or without (white) detectable TMPRSS2-ERG VI mRNA expression.
B. TMPRSS2-ERG VI mRNA levels in cancerous and histologically benign samples with (grey) or without (white) detectable TMPRSS2-ERG III mRNA expression.
The 10/25/50/75/90th percentiles are marked in the figures and open circles denote the outlier values.
TMPRSS2-ERG expression did not associate with Gleason grade of the tissue sample (p=0.5 for both III and VI) but advanced pathologic stage was positively associated with the presence of TMPRSS2-ERG III (p=0.003), and TMPRSS2-ERG VI mRNAs (p=0.020) (Supplemental Table 7). There was no significant association between TMPRSS2-ERG III and VI mRNA levels and the cancer cell percentage of the tissue samples (p=0.8 and p=0.7, respectively).
SPINK1
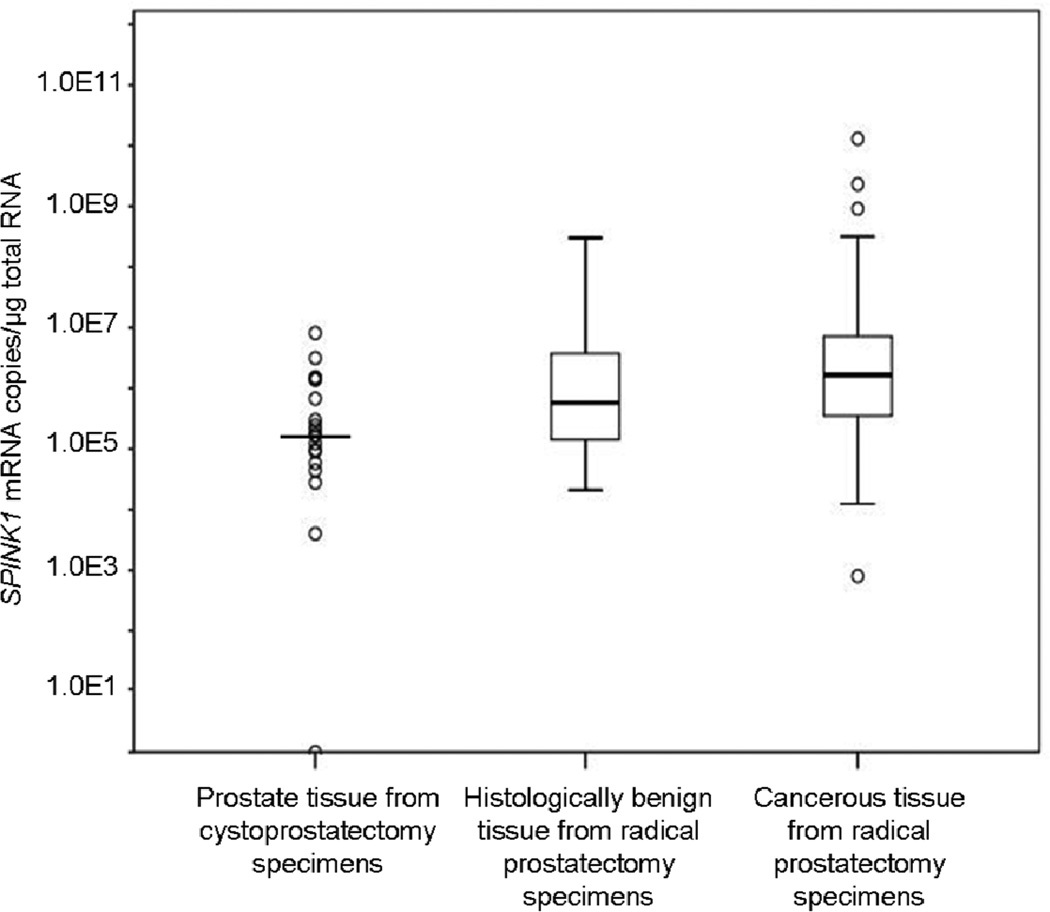
All samples except one cystoprostatectomy sample contained measurable SPINK1 mRNA (Figure 3). The cancerous tissue from RP specimens had significantly higher mRNA levels (2.9-fold difference in medians, p=0.047) than the HBP tissue of RP specimens from men with clinically localized PCa. Furthermore, SPINK1 mRNA levels were statistically significantly lower in the cystoprostatectomy samples than in the HBP samples from RP specimens (3.4-fold difference in medians, p=0.010 after adjusting for clustering). The difference in median expression between cancerous tissue of RP specimens and cystoprostatectomy samples was 10-fold (p<0.001).
Figure 3.
SPINK1 mRNA levels in cystoprostatectomy specimens and histologically benign or cancerous areas from radical prostatectomy specimens. For the cystoprostatectomy group, the horizontal line denotes the median value and the open circles denote SPINK1 mRNA values in individual samples. For the other two groups the 10/25/50/75/90th percentiles are marked in the figure and open circles denote the outlier values.
Neither Gleason grade of the tissue sample (p=0.4) nor advanced pathologic stage (p=0.3) was significantly associated with SPINK1 mRNA expression levels (Supplemental Table 7). There was no significant association between SPINK1 mRNA levels and the cancer cell percentage of the tissue samples (p=0.3).
The presence of TMPRSS2-ERG transcripts did not correlate with SPINK1 levels (p=0.8 for TMPRSS2-ERG III and p=0.6 for TMPRSS2-ERG VI), but there was a statistically significant association between SPINK1 mRNA levels and TMPRSS2-ERG III and VI levels (p=0.001 and p=0.041, respectively) in those samples where TMPRSS2-ERG fusion transcripts were detected.
PCA3
PCA3 mRNA was detectable in all 19 cystoprostatectomy samples. The median PCA3 mRNA level in those samples was 7.9×104 mRNA copies per µg of total RNA, which is 613-fold lower than the previously reported median PCA3 expression level in cancerous tissues17 and 107-fold lower than the previously reported median17 in HBP tissue of cancer-affected prostates. These differences were statistically significant (p<0.0001), but there was no statistically significant difference between PCA3 mRNA levels in cystoprostatectomy samples from cancer-free prostates (n=7) and levels in samples from prostates with incidental cancer foci elsewhere in the prostate (n=12) (p=0.18).
DISCUSSION
Although a significantly higher frequency of TMPRSS2-ERG transcripts was found in the cancerous tissue compared to the HBP tissue from the prostates harboring clinically localized cancer, the frequency of TMPRSS2-ERG expression remained high in samples from histologically benign areas. In contrast, fusion gene transcripts were detectable only rarely in cystoprostatectomy samples from bladder cancer patients and never without concomitant incidental tumor foci present in the same prostate. Thus TMPRSS2-ERG expression can be detected in histologically normal prostate tissue obtained from a prostate in which tumors were detected. A similar trend was seen in PCA3 and SPINK1 mRNA levels.
The presence of TMPRSS2-ERG fusions has been widely considered to be a cancer-specific event1, but we detected TMPRSS2-ERG III and/or VI mRNAs also in 51% of HBP tissues of RP specimens from prostates with clinically localized PCa (compared to 66% in the cancerous RP samples). The phenomenon of TMPRSS2-ERG positive, benign-appearing prostate tissue has been reported before on a few occasions4,5,6. In previous studies, it has mainly been considered an anomaly, but a conceivable explanation could be a carcinogenic field effect23, which suggests that a larger area than just the tumor focus is originally changed in terms of neoplastic events due to a carcinogenic signal that has affected the entire area24. This type of area can appear histologically normal, albeit molecular changes may still be detectable. Field effect-related observations in PCa8,9,25,26 led us to the hypothesis that the HBP tissue of PCa patients in this study may have undergone molecular changes with regard to PCA3 and SPINK1 genes and TMPRSS2-ERG fusion events even though the histological examination defined the samples as non-neoplastic. This is further supported not only by the fact that the two cystoprostatectomy samples that presented TMPRSS2-ERG expression, were taken from prostates that in pathologic sectioning studies were found to contain incidental prostate tumor foci, but also by the higher likelihood of the HBP samples to contain detectable TMPRSS2-ERG mRNA if the matched cancerous sample was also TMPRSS2-ERG fusion-positive.
The previously reported ranges of 10–100-fold overexpression of PCA3 in cancerous tissue27,28 did not seem to be in line with our recent results where the control tissue was HBP tissue from prostates with clinical PCa and only a 5.7-fold overexpression was observed17. However, in light of this study where we determined the PCA3 expression in benign prostate tissue from cystoprostatectomy specimens and compared it with the previously measured levels of PCA3 mRNA in tissue of cancer-affected prostates, the overexpression of PCA3 is more pronounced when the cancerous tissue is, in fact, compared to tissue from prostates without clinical PCa. The cohort of prostates without clinical PCa is admittedly limited in this study, but nevertheless, this suggests that the PCA3 levels may also be elevated in histologically benign areas of the cancerous prostate when compared to cancer-free prostates – as was seen here as a 107-fold difference in mRNA levels.
SPINK1 has been claimed to be overexpressed in PCa9,29 and also to exclusively identify a subgroup of PCas lacking TMPRSS2-ERG fusion, although contradictory results have been reported as well13. In this study, we did not find them to be mutually exclusive, but did observe a positive correlation between SPINK1 and TMPRSS2-ERG mRNA levels in the samples where TMPRSS2-ERG fusion transcripts were detected. However, measuring transcripts in tissue extracts does not allow a specific detection of different transcripts in the individual tumor cells which is a limitation with our and previous studies. A possible co-expression of gene fusions and SPINK1 mRNA in the same tumor cells still needs to be clarified.
Despite the small size of the studied cohort, these findings reported here suggest that detection of TMPRSS2-ERG fusion transcripts is indicative of presence of prostatic tumor in the corresponding prostate, whether or not the sampled cells appear neoplastic in histological examinations. As it was possible to examine histologically only the immediately adjacent tissue, and not the actual tissue used for the RT-qPCR analyses, we cannot exclude the possibility that cancer cells may have resided in the tissue deemed benign using established state-of-art histopathological analyses. It however seems unlikely that this would be the case for such a large number of samples used in our study. However, this study does not reveal whether the finding of TMPRSS2-ERG transcripts can be made even before the emergence of the tumor. If the expression of fusion transcripts is considered an early event, it may, as an inducing carcinogenic signal, be present also before the formation of histologically evident tumor foci. This could therefore give very early information not only about the likelihood of already established disease but also about increased future risk of tumor formation in the prostate even in cases where no tumor tissue was found by the biopsy procedure. Using RT-qPCR assays for determining the molecular composition of biopsy material could conceivably constitute an informative adjunct to the traditional histological examination, and this creates an interesting option calling for future studies. Extraction of nucleic acids from prostate biopsy material is technologically feasible30 although the amount of tissue used to extract nucleic acids would compete with the amount used for histopathological evaluation.
CONCLUSIONS
To conclude, histologically benign tissue from a prostate affected by PCa can present molecular level changes in mRNA expression regarding the fusion gene TMPRSS2-ERG and PCA3 and SPINK1 genes. Thus, molecular tools such as the markers studied here could provide a means to identify an enhanced risk of PCa or presence of already established PCa missed by the biopsy procedure.
Supplementary Material
Acknowledgments
Research support: Academy of Finland (Project 206690), European Union 6th Framework contract LSHC-CT-2004-503011 (P-Mark), National Cancer Institute (R33 CA 127768-03, R01 CA160816-02, P50-CA92629), Swedish Cancer Society (11-0624), the Sidney Kimmel Center for Prostate and Urologic Cancers, David H Koch through the Prostate Cancer Foundation, the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre based at Oxford University Hospitals NHS Trust and University of Oxford, and Fundación Federico SA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest:
Riina-Minna Väänänen: none
Hans Lilja: Dr Lilja holds patents for free PSA, intact PSA and hK2 assays.
Leni Kauko: none
Pauliina Helo: none
Henna Kekki: none
Angel M Cronin: none
Andrew J Vickers: none
Martti Nurmi: none
Kalle Alanen: none
Anders Bjartell: none
Kim Pettersson: none
REFERENCES
- 1.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 2.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 3.Saramäki OR, Harjula AE, Martikainen PM, et al. TMPRSS2:ERG fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin Cancer Res. 2008;14:3395–3400. doi: 10.1158/1078-0432.CCR-07-2051. [DOI] [PubMed] [Google Scholar]
- 4.Clark J, Merson S, Jhavar S, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2007;26:2667–2673. doi: 10.1038/sj.onc.1210070. [DOI] [PubMed] [Google Scholar]
- 5.Furusato B, Gao CL, Ravindranath L, et al. Mapping of TMPRSS2-ERG fusions in the context of multi-focal prostate cancer. Mod Pathol. 2008;21:67–75. doi: 10.1038/modpathol.3800981. [DOI] [PubMed] [Google Scholar]
- 6.Watson SK, Woolcock BW, Fee JN, et al. Minimum altered regions in early prostate cancer progression identified by high resolution whole genome tiling path BAC array comparative hybridization. Prostate. 2009;69:961–975. doi: 10.1002/pros.20949. [DOI] [PubMed] [Google Scholar]
- 7.Luo JH. Gene expression alterations in human prostate cancer. Drugs Today (Barc) 2002;38:713–719. doi: 10.1358/dot.2002.38.10.704653. [DOI] [PubMed] [Google Scholar]
- 8.Yu YP, Landsittel D, Jing L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 9.Kosari F, Cheville JC, Ida CM, et al. Shared gene expression alterations in prostate cancer and histologically benign prostate from patients with prostate cancer. Am J Pathol. 2012;181:34–42. doi: 10.1016/j.ajpath.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paju A, Hotakainen K, Cao Y, et al. Increased expression of tumor-associated trypsin inhibitor, TATI, in prostate cancer and in androgen-independent 22Rv1 cells. Eur Urol. 2007;52:1670–1679. doi: 10.1016/j.eururo.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 11.Tomlins SA, Rhodes DR, Yu J, et al. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell. 2008;13:519–528. doi: 10.1016/j.ccr.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jhavar S, Brewer D, Edwards S, et al. Integration of ERG gene mapping and gene-expression profiling identifies distinct categories of human prostate cancer. BJU Int. 2009;103:1256–1269. doi: 10.1111/j.1464-410X.2008.08200.x. [DOI] [PubMed] [Google Scholar]
- 13.Leinonen KA, Tolonen TT, Bracken H, et al. Association of SPINK1 Expression and TMPRSS2:ERG Fusion with Prognosis in Endocrine-Treated Prostate Cancer. Clin Cancer Res. 2010;16:2845–2851. doi: 10.1158/1078-0432.CCR-09-2505. [DOI] [PubMed] [Google Scholar]
- 14.Tomlins SA, Bjartell A, Chinnaiyan AM, et al. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. Eur Urol. 2009;56:275–286. doi: 10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Cai Y, Ren C, et al. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 16.Nurmi J, Wikman T, Karp M, et al. High-performance real-time quantitative RT-PCR using lanthanide probes and a dual-temperature hybridization assay. Anal Chem. 2002;74:3525–3532. doi: 10.1021/ac020093y. [DOI] [PubMed] [Google Scholar]
- 17.Väänänen RM, Lilja H, Cronin A, et al. Association of transcript levels of 10 established or candidate-biomarker gene targets with cancerous versus non-cancerous prostate tissue from radical prostatectomy specimens. Clin Biochem. 2013;46:670–674. doi: 10.1016/j.clinbiochem.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Väänänen RM, Rissanen M, Kauko O, et al. Quantitative real-time RT-PCR assay for PCA3. Clin Biochem. 2008;41:103–108. doi: 10.1016/j.clinbiochem.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Nurmi J, Lilja H, Ylikoski A. Time-resolved fluorometry in end-point and real-time PCR quantification of nucleic acids. Luminescence. 2000;15:381–388. doi: 10.1002/1522-7243(200011/12)15:6<381::AID-BIO623>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 20.Cerveira N, Ribeiro FR, Peixoto A, et al. TMPRSS2-ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia. 2006;8:826–832. doi: 10.1593/neo.06427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rissanen M, Helo P, Väänänen RM, et al. Novel homogenous time-resolved fluorometric RT-PCR assays for quantification of PSA and hK2 mRNAs in blood. Clin Biochem. 2007;40:111–118. doi: 10.1016/j.clinbiochem.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Nurmi J, Ylikoski A, Soukka T, et al. A new label technology for the detection of specific polymerase chain reaction products in a closed tube. Nucleic Acids Res. 2000;28:E28. doi: 10.1093/nar/28.8.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nonn L, Ananthanarayanan V, Gann PH. Evidence for field cancerization of the prostate. Prostate. 2009;69:1470–1479. doi: 10.1002/pros.20983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 25.Mazzucchelli R, Barbisan F, Santinelli A, et al. Immunohistochemical expression of prostate tumor overexpressed 1 in cystoprostatectomies with incidental and insignificant prostate cancer. Further evidence for field effect in prostatic carcinogenesis. Hum Pathol. 2011;42:1931–1936. doi: 10.1016/j.humpath.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 26.Popa I, Fradet Y, Beaudry G, et al. Identification of PCA3 (DD3) in prostatic carcinoma by in situ hybridization. Mod Pathol. 2007;20:1121–1127. doi: 10.1038/modpathol.3800963. [DOI] [PubMed] [Google Scholar]
- 27.Bussemakers MJ, van Bokhoven A, Verhaegh GW, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–5979. [PubMed] [Google Scholar]
- 28.Hessels D, Klein Gunnewiek JM, van Oort I, et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44:8–15. doi: 10.1016/s0302-2838(03)00201-x. [DOI] [PubMed] [Google Scholar]
- 29.Laxman B, Morris DS, Yu J, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68:645–649. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukabori Y, Yoshida K, Nakano K, et al. Preparation of a single prostate needle biopsy specimen for histological diagnosis and RNA analysis. J Urol. 2006;176:1204–1207. doi: 10.1016/j.juro.2006.04.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.