Abstract
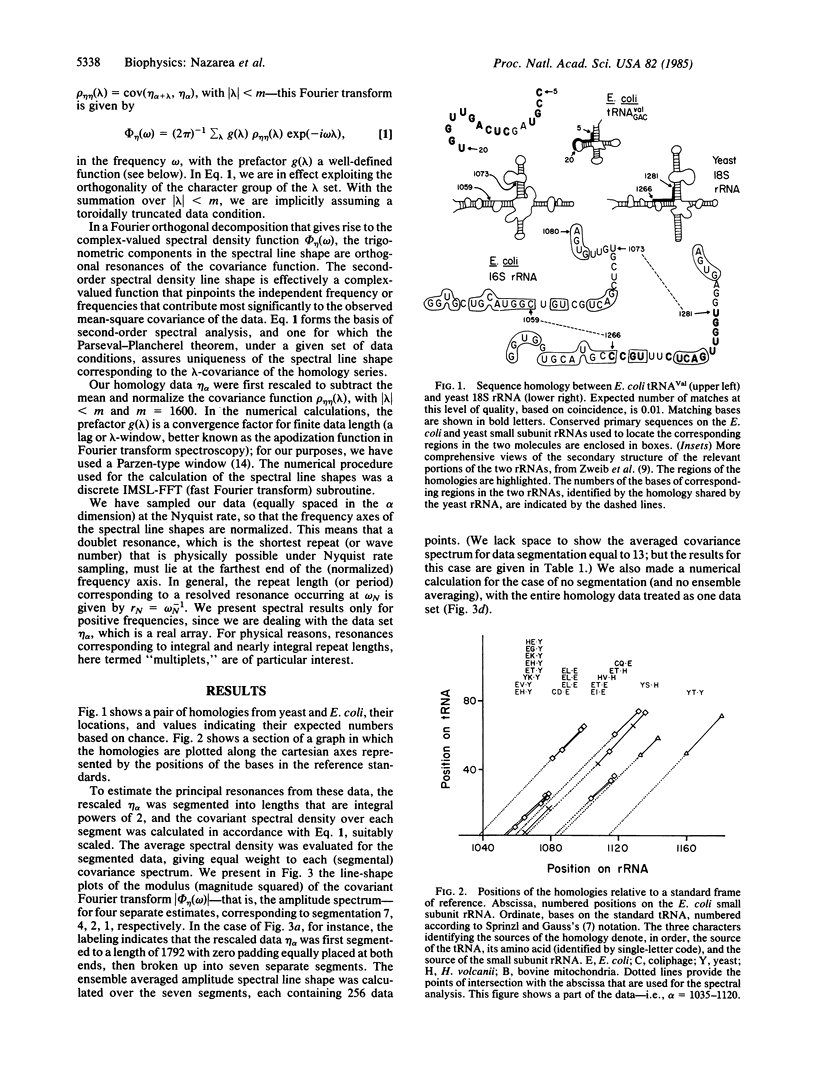
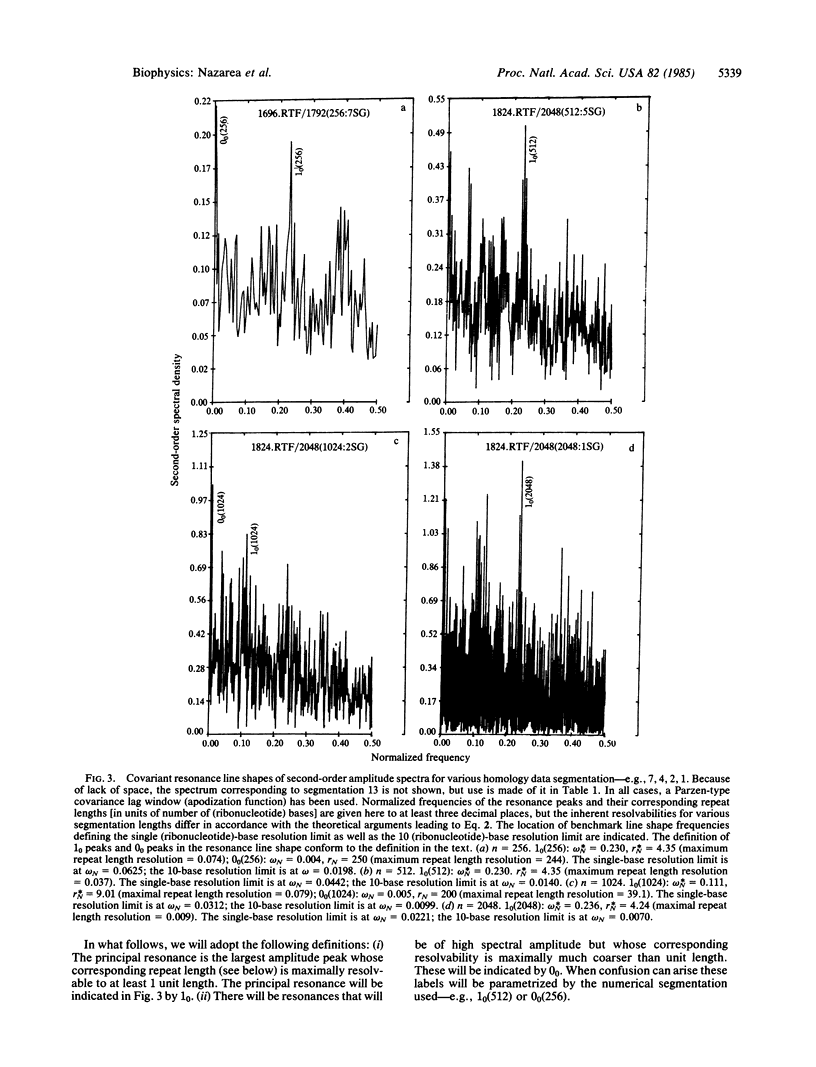
Second-order spectral analysis is used to detect rigorously and to characterize the principal periodicities in the positions of conserved sequences common to tRNAs and rRNAs. It is shown that the shared periodicity having the largest spectral amplitude is 9, followed by 8 and 10, thus forming a closed triad of significant multiplets centered at 9 bases. This conclusion is proposed to reflect a closed triadic set of fundamental tandem repeat lengths in a class of ancestral macromolecules possessing a restricted sequence symmetry. The terms "remanent" and "archeomodular" are used to describe a relic modular format, traces of which are shown here to persist despite the changes that have occurred in the primary structures of ribonucleic acids during the course of their evolution.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., de Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., Young I. G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- Bloch D. P., McArthur B., Mirrop S. tRNA-rRNA sequence homologies: evidence for an ancient modular format shared by tRNAs and rRNAs. Biosystems. 1985;17(3):209–225. doi: 10.1016/0303-2647(85)90075-9. [DOI] [PubMed] [Google Scholar]
- Bloch D. P., McArthur B., Widdowson R., Spector D., Guimaraes R. C., Smith J. tRNA-rRNA sequence homologies: evidence for a common evolutionary origin? J Mol Evol. 1983;19(6):420–428. doi: 10.1007/BF02102317. [DOI] [PubMed] [Google Scholar]
- Bloch D., McArthur B., Widdowson R., Spector D., Guimaraes R. C., Smith J. tRNA-rRNA sequence homologies: a model for the origin of a common ancestral molecule, and prospects for its reconstruction. Orig Life. 1984;14(1-4):571–578. doi: 10.1007/BF00933706. [DOI] [PubMed] [Google Scholar]
- Cedergren R. J., LaRue B., Sankoff D., Lapalme G., Grosjean H. Convergence and minimal mutation criteria for evaluating early events in tRNA evolution. Proc Natl Acad Sci U S A. 1980 May;77(5):2791–2795. doi: 10.1073/pnas.77.5.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A. Geomagnetic reversals. Science. 1969 Jan 17;163(3864):237–245. doi: 10.1126/science.163.3864.237. [DOI] [PubMed] [Google Scholar]
- Fitch W. M. Distinguishing homologous from analogous proteins. Syst Zool. 1970 Jun;19(2):99–113. [PubMed] [Google Scholar]
- Goad W. B., Kanehisa M. I. Pattern recognition in nucleic acid sequences. I. A general method for finding local homologies and symmetries. Nucleic Acids Res. 1982 Jan 11;10(1):247–263. doi: 10.1093/nar/10.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Lanter J. M., Woese C. R. Sequence of the 16S Ribosomal RNA from Halobacterium volcanii, an Archaebacterium. Science. 1983 Aug 12;221(4611):656–659. doi: 10.1126/science.221.4611.656. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. Use of statistical criteria for screening potential homologies in nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):203–213. doi: 10.1093/nar/12.1part1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Wilbur W. J., Smith T. F., Waterman M. S. On the statistical significance of nucleic acid similarities. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):215–226. doi: 10.1093/nar/12.1part1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. W., Remington S. J., Grütter M. G., Anderson W. F. Relation between hen egg white lysozyme and bacteriophage T4 lysozyme: evolutionary implications. J Mol Biol. 1981 Apr 25;147(4):545–558. doi: 10.1016/0022-2836(81)90399-5. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Gauss D. H. Compilation of tRNA sequences. Nucleic Acids Res. 1982 Jan 22;10(2):r1–55. [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Fox G. E. Archaebacteria. J Mol Evol. 1978 Aug 2;11(3):245–251. doi: 10.1007/BF01734485. [DOI] [PubMed] [Google Scholar]
- Zwieb C., Glotz C., Brimacombe R. Secondary structure comparisons between small subunit ribosomal RNA molecules from six different species. Nucleic Acids Res. 1981 Aug 11;9(15):3621–3640. doi: 10.1093/nar/9.15.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]


