Abstract
Studies on the origin of species have focused largely on anatomy, yet animal populations are generally limited by energy. Animals can adapt to available energy resources at three levels: (1) evolution of different anatomical forms between groups of animals through nuclear DNA (nDNA) mutations, permitting exploitation of alternative energy reservoirs and resulting in new species with novel niches, (2) evolution of different physiologies within intraspecific populations through mutations in mitochondrial DNA (mtDNA) and nDNA bioenergetic genes, permitting adjustment to energetic variation within a species’ niche, and (3) epigenomic regulation of dispersed bioenergetic genes within an individual via mitochondrially generated high-energy intermediates, permitting individual adjustment to environmental fluctuations. Because medicine focuses on changes within our species, clinically relevant variation is more likely to involve changes in bioenergetics than anatomy. This may explain why mitochondrial diseases and epigenomic diseases frequently have similar phenotypes and why epigenomic diseases are being found to involve mitochondrial dysfunction. Therefore, common complex diseases may be the result of changes in any of a large number of mtDNA and nDNA bioenergetic genes or to altered epigenomic regulation of these bioenergetic genes. All of these changes result in similar bioenergetic failure and consequently related phenotypes.
BIOENERGETICS AND THE ORIGIN OF SPECIES
Darwin and Wallace (1858; Darwin 1859) proposed that natural selection acts on random variation in plants and animals to shape new species. Natural selection encompasses the environmental constraints acting on the organism, one of the most important being energy availability.
Life exists in a nonequilibrium thermodynamic state requiring the constant flow of energy to sustain its complex structures and to permit the accumulation and transmission of biological information. In the absence of energy flow, complex systems decay. Therefore, life exists through the interplay among structure, energy, and information.
The source of most biological energy is the Sun. The high-energy photons collected by photosynthetic cyanobacteria and their chloroplast descendants are used to split water into hydrogen and oxygen. The oxygen is released into the atmosphere and the hydrogen is used to reduce CO2 to generate glucose. Plant glucose is consumed by herbivores and the energy therein sequentially passed through the animal and fungal food chains. Ultimately, the degraded energy is dissipated as infrared radiation into space.
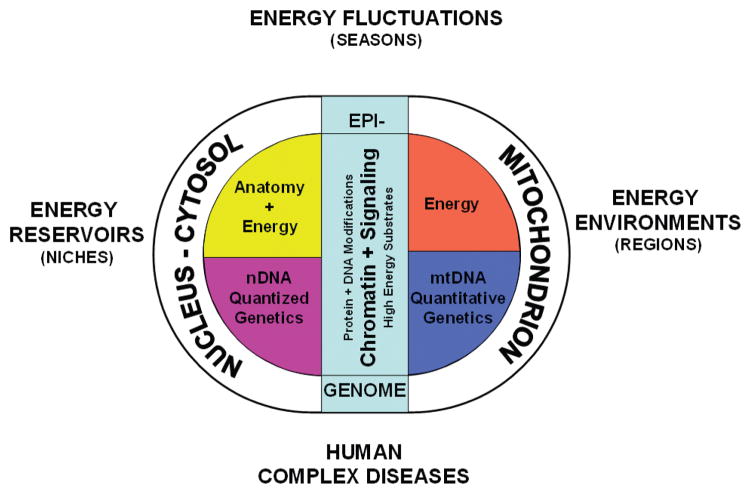
Animal populations grow and multiply until energy becomes limiting. Animals can adapt to available energy resources at three levels: interspecific exploitation of different energy reservoirs that define the species’ niche, intraspecific exploitation of differences in regional energy resources, and individual responses to oscillating environmental energy resources (Fig. 1).
Figure 1.
Three hypothesized levels of animal eukaryotic cell adaptation to varying energy resources. The original eukaryotic symbiosis brought together the glycolytic nucleus-cytosol with the oxidative mitochondrion. Most of the mitochondrial genome was then transferred to the nDNA, such that the current animal cell nucleus encodes the genes for determining cellular and organismal structure plus the genes for glycolysis and most of the genes for oxidative metabolism, all inherited according to the laws of Mendel. Maternally inherited mtDNA retains the core genes for generating, maintaining, and using the mitochondrial inner membrane potential, ΔP, which links the calories metabolized with cellular energy metabolism. The epigenome evolved to coordinate nDNA gene expression in relation to the availability of environmental calories. This is mediated by the modification of proteins and DNA elements of the epigenome via intracellular levels of the high-energy intermediates: ATP, acetyl-CoA, SAM, plus the redox status of the cell. When calories are abundant, the bioenergetic intermediates increase, chromatin is modified and decondensed, gene expression increases, and growth and reproduction are stimulated. When calories are limited, the reverse is true. Between species and higher taxa, mutations in the nDNA developmental genes change anatomy and permit the exploitation of different energy reservoirs. This creates species and defines niches. Within a species, mutations in the mtDNA change the cellular physiology to permit adaptation of regional populations to consistent regional differences in energy resources. Frequent fluctuations in energy resources of a species are addressed by changes in the epigenome that modulate the coordinate expression of cis- and trans-distributed nDNA bioenergetic genes. De novo mutations in nDNA bioenergetic genes, mutations and polymorphisms in mtDNA bioenergetic genes, and mutational or environmentally induced alterations in the epigenomic regulation of bioenergetic genes can all perturb bioenergetic homeostasis and contribute to the pathophysiology of common diseases, cancer, and aging.
Interspecific energy resource adaptation involves exploitation of distinct energy reservoirs through anatomical variation. This helps to delineate species and genera. Energy reservoirs are the distinct source of calories used by a species throughout its existence, perhaps over hundreds of thousands of years. Factors that can define an energy reservoir include being an herbivore versus a carnivore. For herbivores, these could involve eating fruits versus nuts or leaves versus sap. For carnivores, they could encompass being a predator versus a scavenger or consuming blood versus bugs, etc. To exploit alternative energy reservoirs, selection acts on nDNA genetic variation to alter animal anatomical traits, permitting access to novel energy reservoirs. This interaction between the species’ anatomy and the energy reservoir helps to define the species’ niche (Fig. 1).
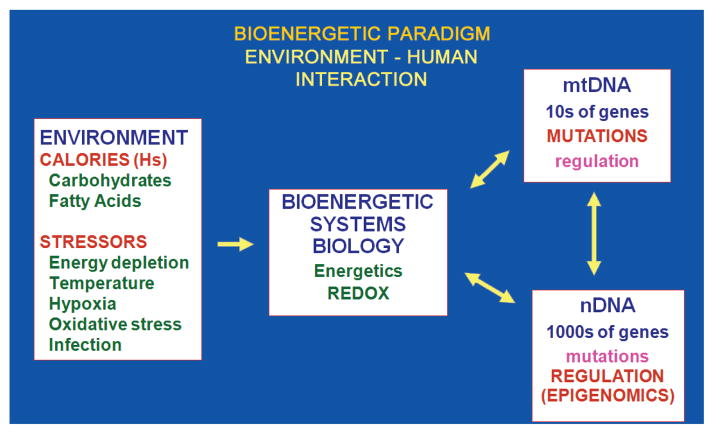
Intraspecific energy resource adaptation involves the exploitation of regional energy environments through changes in physiology. Within a species’ niche, regional energy resources can vary in energy type (carbohydrate, fat, protein), amount, and demand (activity level, stress, etc.). These regional differences can be stable over thousands of years and lead to development of distinct geographically constrained populations within the species. To exploit different regional energy environments, genetic variants in mtDNA and nDNA bioenergetic genes are selected that adjust the bioenergetic metabolism to be in balance with local energy resources (Fig. 2).
Figure 2.
Bioenergetic interface with the environment explains the importance of mtDNA and epigenomic variation of intraspecific animal adaptation. Energy availability and demand are the central factors in an animal’s environment, the energy environment. Primary sources of available energy for omnivores, such as humans, are dietary calories generally obtained as carbohydrates and fats. Demands for calories include physical activity, thermal stress, hypoxia, oxidative stress, infection, body maintenance, and reproduction. Available calories are processed through cellular and mitochondrial bioenergetic pathways. The bioenergetic system is assembled from both mtDNA and nDNA genes. The mtDNA encodes core genes of OXPHOS. It has a very high mutation rate, resulting in the continual generation of functional variants, thus providing the genetic variation to permit animals to adapt to regional variation in the energetic environment. The nDNA encodes all of the genes for glycolysis, most of the genes for mitochondrial biogenesis and energy production, and all of the genes for the energetic- and redox-regulated signal transduction systems. These nDNA genes have a low mutation rate, within the time range for speciation. However, expression of the ~2000 nDNA-encoded energy genes is regulated by the production of high-energy intermediates by glycolysis and OXPHOS including ATP, acetyl-CoA, and SAM. These cellular bioenergetic substrates then drive the modification of the chromatin by phosphorylation, acetylation, and methylation, thus coordinating gene expression in relation to short-term fluctuations in the individual’s energetic environment.
Individual energy resource adaptation involves adjustments within the individual to short-term oscillations in energy availability and demand. These changes can occur within days to years and include seasonal changes and inflammation. Adjustment to these changes is accomplished through epigenomic modulation of nDNA and mtDNA bioenergetic gene expression (Fig. 2).
These three levels of energy adaptation strategy result from the fact that nDNA has a low mutation rate and encodes all of the genes for determining anatomy. Hence, changes in nDNA genes will define the species’ structure and thus the energy reservoir that it can exploit. In contrast, the mtDNA has a very high mutation rate and encodes the central mitochondrial bioenergetic genes, augmented by hundreds of nDNA bioenergetic genes. Hence, mtDNA plus nDNA variation in bioenergetic genes has a dominant role in adapting to regional energy environments. Finally, the bioenergetic genes of mtDNA and nDNA must be coordinately regulated in response to environmental energetic oscillations. This is accomplished by modification of the epigenome, mediated by high-energy intermediates generated by the flow of calories through cellular bioenergetic systems (Figs. 1 and 2).
Interspecific variation leading to species, genera, families, orders, and classes is therefore the product of genetic variation in anatomical genes of nDNA. Alternatively, intraspecific variation resulting in population and individual adaptation to energy environments is dominated by mutations in bioenergetic genes, particularly those of the mtDNA, plus alterations in bioenergetic gene regulation through modifications of the epigenome (Fig. 1).
Since Darwin and Wallace proposed natural selection, considerable attention has been focused on changes in the genes and processes that dictate animal structures. However, little consideration has been given to the role of energy flow through the biosphere. Yet it is the flow of energy that drives the generation of complexity, the accumulation of information, and animal radiation. Indeed, it is precisely the intraspecific adaptation to regional energy resources that determines whether a population will survive, reproduce, and radiate.
The role of intraspecific energy adaptation is particularly pertinent for medicine, which is concerned with the health and well being of a single species, Homo sapiens. Therefore, genetic and epigenetic adaptations affecting energy metabolism are more likely to mediate environmental interactions than are changes in anatomy. Hence, the strong anatomical and nDNA focus of Western medicine may account for the reason that it has been so difficult to understand genetics and environmental inter actions associated with common “complex diseases” that have a bioenergetic component to their etiology (Fig. 2).
BIOENERGETICS IN CELLULAR EVOLUTION
The duality of structure and energy for our cells became delineated with the symbiosis that created the original eukaryotic cell ~2 × 109 years ago. This symbiosis is thought to have combined a glycolytic motile microorganism, the protonucleus-cytosol, and an oxidative α-protobacterium, the protomitochondrion. One impetus for this association was the increase in atmospheric oxygen generated by cyanobacteria. Oxygen provides the terminal electron acceptor for the efficient burning of the reducing equivalents (hydrogens) from calories consumed by animals.
For the first 1.2 × 109 years after symbiosis, the two symbiotic organisms consolidated their metabolic pathways and exchanged genes, with natural selection enriching for more efficient forms. Ultimately, one genetic and metabolic combination proved to be sufficiently efficient to permit the advent of multicellularity. During the ensuing intersymbiont reorganization, most of the genes of the mitochondrial genome were transferred sequentially into nDNA. As a result, mitochondrial genes became randomly dispersed across all of the nuclear chromosomes (Wallace 2007).
In the eukaryotic cell that gave rise to multicellular organisms, all of the polypeptide genes for mitochondrial growth, reproduction, metabolism, and energy production came to reside in nDNA. These include genes for mitochondrial biogenesis such as the mtDNA polymerase γ, mtDNA RNA polymerase, the Twinkle helicase, ribosomal proteins, elongation factors, and tRNA synthetases; intermediate metabolism proteins including those for the tricarboxylic acid cycle (TCA), amino acid metabolism, folate metabolism, and nucleotide biogenesis; structural and assembly proteins including import complex proteins, chaperones, proteases; and ~80 mitochondrial OXPHOS proteins including all four of the complex II polypeptides and polypeptides for other dehydrogenases that feed electrons into the electron transport chain (ETC) (Wallace et al. 2007).
It has been estimated that mammalian nDNA encompasses ~1500 genes of the mitochondrial genome. These mitochondrial genes were added to the already existing nDNA anaerobic energy metabolism genes. Therefore, the genes involved in energy metabolism encoded by nDNA must number in the thousands.
By the time that the fungal–animal lineage was established, mtDNA retained only 13 polypeptide genes. Although small in number, these genes are by no means inconsequential because these are core polypeptides for the mitochondrial energy–generating system, oxidative phosphorylation (OXPHOS). mtDNA polypeptides include seven (ND1, 2, 3, 4L, 4, 5, 6) of the 45 subunits of complex I, one (cytochrome b, cytb) of the 11 subunits of complex III, three (COI, II, III) of the 13 subunits of complex IV, and two (ATP 6 and 8) of the ~17 subunits of complex V. Because these mtDNA polypeptides must be translated within the mitochondrion, the animal mtDNA also retains rRNAs and tRNA genes for mitochondrial protein synthesis (Wallace 2007).
The retention of these polypeptide genes by mtDNA is likely the product of their being central electron- and/or proton-carrying polypeptides for complexes I, III, IV, and V. These are the only OXPHOS complexes that transport protons. Consequently, they are critical for the generation, maintenance, and use of the mitochondrial inner membrane electrochemical gradient (ΔP = Δψ + ΔμH+). ΔP, in turn, is central to the conversion of dietary calories into ATP to perform work or generate heat to maintain body temperature (Wallace 2007).
ΔP is generated by the ETC through the oxidation of reducing equivalents recovered from carbohydrates and fats, the electrons passing sequentially through complexes I, III, and IV to reduce ½ O2 into H2O. The energy that is released is used by these three complexes to pump protons out of the mitochondrial matrix into the intermembrane space, generating ΔP. The energy stored in ΔP is then used by complex V to condense ADP + Pi to ATP, which is exported to the cytosol by adenine nucleotide translocators (ANTs). ΔP can also be used to import molecules and ions including Ca2+ into the mitochondrial matrix. If ΔP increases to its maximum, the ETC stalls and the electron carriers become saturated with electrons. In the presence of oxygen, electrons in complexes I and III can be donated directly to O2 to give superoxide anion, the first of the reactive oxygen species (ROS). Mitochondrial ROS provides an important signal transduction system from the mitochondrion to the nucleus cytosol. However, excessive ROS can damage mitochondrial and cellular lipids, proteins, and DNA, ultimately resulting in cell death (Wallace et al. 2009).
The efficiency of complexes I, III, and IV at producing ΔP is determined by the ratio of the number of protons pumped out of the mitochondrial matrix relative to the number of electrons that move down the ETC. The efficiency of complex V, the ATP synthase, is defined by the number of protons that pass back into the matrix relative to ATP generated. Taken together, these two parameters make up the OXPHOS coupling efficiency. Tightly coupled OXPHOS will generate the maximum ATP per calorie burned, whereas a more loosely coupled mitochondria will require the oxidation of more calories to generate the same amount of ATP, the differential energy being dissipated as increased heat production. Therefore, the mitochondrial coupling efficiency determines the relative allocation of the calories consumed to generate ATP versus heat produced.
Because all mitochondrial energy production is contingent on ΔP, proton permeability of complexes I, III, IV, and V must be balanced to ensure that one complex is not more permeable to protons than the others. Otherwise, the leaky complex will short circuit the capacitor (ΔP) and negate the coupling efficiency of the other complexes. Having the core proton and electron carrier protein genes linked together in the mtDNA ensures that the genes of the complexes will be selected as a unit and thus work optimally with one another (Wallace 2007).
Assurance that complex I, III, IV, and V mtDNA polypeptides will coevolve is achieved by maternal inheritance of the mtDNA. Exclusive maternal inheritance prohibits mixing within the same cell of the mtDNAs from two individuals with different coupling efficiencies, thus blocking inter-mtDNA recombination. Therefore, the only way that mtDNA genes can change is by the sequential accumulation of mutations along radiating maternal lineages. Each new mutation is then tested by selection against the background of the previously existing mtDNA genetic variants (Wallace 2007).
Because each cell has hundreds to thousands of copies of mtDNA, when a new mtDNA mutation arises within an oocyte or cell, it generates an intracellular mixture of mutant and normal mtDNAs, a state known as heteroplasmy. Because mtDNAs are distributed randomly between daughter cells during mitotic division, the percentage of mutant and normal mtDNAs can drift during mitosis and meiosis, the process of replicative segregation. Thus, neutral, beneficial, or mildly deleterious mtDNA mutations can become fixed within a maternal lineage through intracellular genetic drift. However, as the percentage of deleterious mutations increases, the energy output of the cell declines until it drops below the minimum energy output required for that cell type to function, the bioenergetic threshold, and symptoms ensue (Wallace 2005, 2007).
BIOENERGETICS IN HUMAN ORIGINS AND DISEASE
The most important functions of the mitochondrion for animal cellular and tissue physiology and thus environmental adaptation and health are its (1) production of most of the cellular energy in the form of ATP and heat, (2) generation of much of the endogenous ROS, (3) uptake of cytosolic Ca2+ thus maintaining Ca2+ homeostasis, and (4) regulation of cell death through activation of the mitochondrial permeability transition pore (mtPTP). The mtPTP monitors mitochondrial ΔP, adenine nucleotides, ROS, and Ca2+ levels, and when energy production is too low or ROS production and matrix Ca2+ levels are too high, mtPTP opens a channel through the mitochondrial membrane, shorting ΔP. This causes the release of proapoptotic proteins from the mitochondrial intermembrane space into the cytosol, initiating cell death. Thus, mitochondrially mediated apoptosis removes energetically impaired cells from the tissue, eliminating their disruption of tissue function (Wallace 2005).
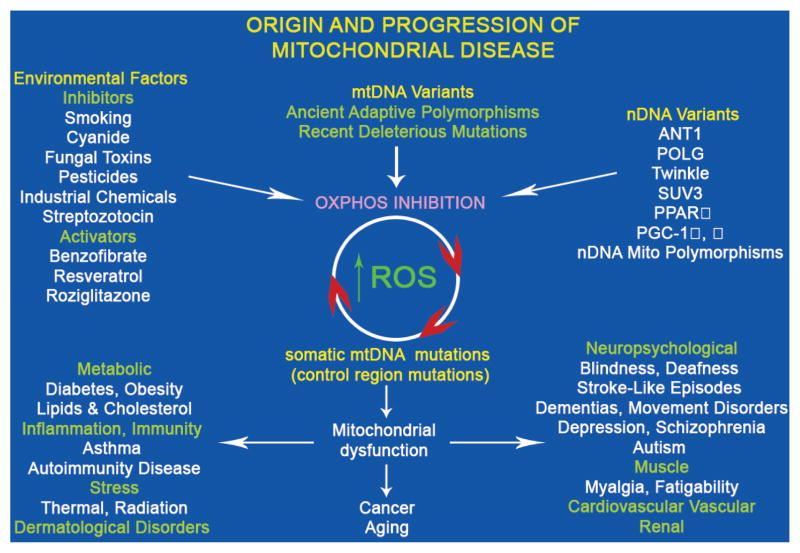
All four of these critical physiological functions of the mitochondrion can be modulated by variation in mitochondrial genes, both nDNA and mtDNA (Fig. 3). Phenotypically relevant nDNA-encoded mitochondrial gene variants alter the structural and assembly genes for building functional mitochondrial OXPHOS complexes. Mutations in the heart muscle ANT (ANT1) result in myopathy and cardiomyopathy (Palmieri et al. 2005; Wallace et al. 2007). Functional polymorphisms in nDNA genes can alter transcriptional and translational systems of nDNA or mtDNA mitochondrial biogenesis. For example, polymorphisms in peroxisome proliferation-activated receptor γ (PPARγ) (Altshuler et al. 2000) and PGC-1α (Ek et al. 2001; Muller et al. 2003) genes have been associated with regional predilection to diabetes. Finally, mutations in nDNA-encoded genes involved in mitochondrial biogenesis, such as mtDNA polymerase γ (POLG) and the Twinkle helicase, can inhibit mtDNA replication resulting in multiple deletions and/or depletion of mtDNA (Wallace et al. 2007).
Figure 3.
Classes of human mitochondrial gene mutations in the origin of metabolic and degenerative diseases, cancer, and aging. The “mitochondrial genome” encompasses ~1500 nDNA genes dispersed across the chromosomes plus 37 critical energetic genes within mtDNA. Genetic variation in any of these mitochondrial genes may perturb the mitochondrial OXPHOS. An array of common environmental agents and pharmacological agents can also modulate mitochondrial bioenergetics and/or biogenesis. Inhibition of OXPHOS can increase mitochondrial ROS production, which will damage mtDNA, gradually erode the cellular capacity to generate energy, and create the clock central to aging and adult cancers. OXPHOS dysfunction will have the greatest effect on tissues having the highest energy demand (brain, heart, skeletal muscle, kidney, endocrine system) to cause degenerative diseases. Altered mitochondrial energy production will also perturb caloric sensing and use, resulting in common metabolic diseases such as diabetes and obesity. Finally, altered mitochondrial ROS production and redox biology will precipitate inflammatory disease and change mitochondrial coupling efficiency to affect thermal modulation and sensitivity to radiation-induced cellular toxicity.
Phenotypically relevant inherited mtDNA sequence variants include recent deleterious mutations and ancient adaptive polymorphisms. Deleterious mtDNA mutations can cause an array of human degenerative and metabolic diseases, primarily affecting the more oxidative tissues: brain, heart, muscle, kidney, and endocrine system. Adaptive mutations permitted ancestral human populations to adapt to different energetic environments as they migrated throughout the world. Although an adaptive mutation can be beneficial in one energetic environment, it can be deleterious in another. Hence, population-specific variants have been found to be risk factors for a variety of clinical problems (Khusnutdinova et al. 2008; Wallace 2008). The mitochondrial energy production system is also specifically inhibited by a wide range of environmental agents including cyanide, MPTP, and rotenone. Therefore, the mitochondria provide a direct link among the environment, cellular physiology, and genes (Fig. 3).
Because mtDNAs within the cell are continuously turning over, mtDNA mutations accumulate in tissue cells. An important source of mtDNA mutations is ROS damage. The resulting somatic mtDNA mutations accumulate with age in postmitotic tissues, eroding cellular energetics, exacerbating inherited mitochondrial defects, and producing the “aging clock.” The accumulation of these somatic mutations may explain the delayed onset and progressive course of multiple age-related diseases and certain forms of cancer (Fig. 3) (Wallace 2005).
Because mitochondria provide a direct link between the energetic environment and the bioenergetics of an individual’s cells and tissues, they must also be central to diseases influenced by environmental variation. Accordingly, mitochondrial dysfunction has been associated with a wide range of common metabolic and generative disease symptoms (Wallace 2005; Wallace et al. 2007).
That mitochondrial dysfunction is sufficient to cause disease has been demonstrated by the introduction of mtDNA mutations into the mouse. Mice harboring mutant mtDNAs developed the same symptoms as observed in common age-related diseases (Fan et al. 2008; Wallace and Fan 2009b). Thus, mitochondrial dysfunction must have a central role in the pathophysiology of metabolic and degenerative diseases, cancer, and aging (Wallace 2005).
The discovery that human mtDNAs can harbor adaptive mutations offers a novel perspective on how species adapt to the regional energetic environment. Unlike nDNA variation, mtDNA variation correlates strongly with indigenous populations and their geographic location. This is the result of both maternal inheritance and energetic environmental selection. Mutations in mtDNA accumulated sequentially on radiating maternal lineages as women migrated out of Africa to colonize Eurasia and the Americas. Those mutations that resulted in an energy metabolism better suited for the new energy environment became enriched, creating region-specific branches of the mtDNA tree. These branches represent clusters of related mtDNA haplotypes constituting a haplogroup. Haplogroups are frequently founded by one or more adaptive mutations. Moreover, the same adaptive mutation can be observed on different mtDNA backgrounds, indicating that the mutation has arisen several independent times and in each case has been enriched in the population by natural selection. This convergent evolution provides direct evidence that mtDNA mutations can be adaptive.
The greatest degree of mtDNA diversity resides in Africa and constitutes macrohaplogroup L. This high African mtDNA diversity demonstrates an African origin for the mtDNA tree (Johnson et al. 1983; Cann et al. 1987; Merriwether et al. 1991). However, of all African mtDNA variation, only two mtDNAs successfully left Africa and colonized all of Eurasia, founding macrohaplogroups M and N (Wallace et al. 1999; Mishmar et al. 2003; Ruiz-Pesini et al. 2004, 2007). The mtDNA that founded the macrohaplogroup N lineage harbored two missense mutations, ND3 10398 A114T and ATP6 8701 A59T, which have been shown to alter several mitochondrial physiological parameters (Kazuno et al. 2006). From Africa, the N lineage radiated into Europe, generating the European-specific lineages H, I, J, Uk, T, U, V, W, and X. Both the M and N mtDNA founders radiated into Asia, generating a plethora of mtDNA lineages. Of these, A, C, and D became enriched in northeastern Siberia and were in position to cross the Bering land bridge to colonize the Americas (Wallace et al. 1999; Ruiz-Pesini et al. 2007).
The ROS-induced high mtDNA mutation rate rapidly produces new variants within a population that can be acted on by selection. Alterations in mtDNA genes can affect OXPHOS coupling efficiency, changing the allocation of calories between ATP versus heat, thus optimizing energy metabolism to warmer versus colder climates. Other mtDNA alterations can increase or decrease mitochondrial ROS production at particular caloric loads, modulate Ca2+ buffering, or regulate the rate of apoptosis (Wallace 1994; Mishmar et al. 2003; Ruiz-Pesini et al. 2004; Ruiz-Pesini and Wallace 2006). The contribution of mtDNA variation to climate adaptation has been supported by demonstrating that mtDNA variation but not nDNA variation correlates with regional temperature extremes (Balloux et al. 2009). Therefore, mtDNA variation is of central importance in animal and human adaptation to the changing energy environments.
The mtDNA functional mutations that have permitted human populations to adapt to regional environmental differences have accumulated over thousands of years. These functional changes in mtDNA polypeptide and structural RNA genes impart a shift in the range of environmental conditions that the individual can tolerate, in effect defining the outer limits of individual environmental tolerance. The changes in mtDNA structural genes can be further modulated by mutations in the mtDNA regulatory control region, which can alter both mtDNA transcription levels and copy number (Suissa et al. 2009). Thus, control region variation may permit the fine-tuning of an individual’s mitochondrial energetics (Fig. 3).
BIOENERGETIC ORIGIN OF THE EPIGENOME
Because nDNA has a much lower mutation rate than mtDNA, the thousands of nDNA energy genes must adjust to changes in the energetic environment by alterations in gene expression, not mutation (Fig. 2).
Calories are the limiting factor for cellular and organismal growth and reproduction, and thus the expression of nDNA genes for energy metabolism and cell growth must be modulated according to the availability of calories. For unicellular organisms, a direct link is required among calories, growth, and gene expression. For multicellular organisms, with specialized organs and tissues, this more primitive connection between calories and gene expression can be overlaid with other genetic programs that regulate gene expression and growth within the context of specific tissue requirements (Wallace and Fan 2009a).
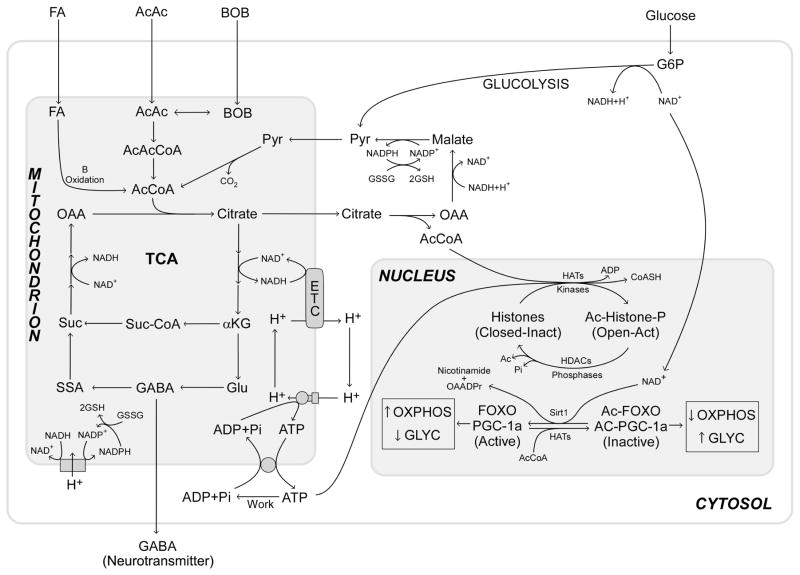
In humans, calories can come from either carbohydrates or fatty acids and ketone bodies (acetoacetate and β-hydroxybutyrate). Carbohydrates such as glucose are initially processed through cytosolic glycolysis to generate pyruvate, ATP, and reduced NAD+, NADH + H+. The pyruvate then enters the mitochondrion where it is converted to acetyl-CoA. Acetyl-CoA condenses with oxaloacetate (OAA) to give citrate that fuels the tricarboxylic acid cycle (TCA). In contrast, fatty acids and ketone bodies are transferred directly into the mitochondrion where they are converted to acetyl-CoA and intramitochondrial reducing equivalents in the form of NADH + H+ and FADH2 (Fig. 4) (Wallace and Fan 2009a; Wallace et al. 2009).
Figure 4.
Mitochondrial bioenergetic coupling with the epigenome. The flow of reducing equivalents (calories) through glycolysis and mitochondrial oxidation regulates levels of cellular ATP and acetyl-CoA. High-calorie intake increases ATP and acetyl-CoA levels, thereby increasing histone phosphorylation and acetylation. This opens the chromatin to stimulate transcription, growth, and replication. Acetylation also regulates major signal transduction pathways, including the insulin signaling pathway. Forkhead box class O (FOXO) transcription factors that regulate expression of PGC-1α, the key transcription factor to regulate mitochondrial OXPHOS and biogenesis, can be acetylated and inactivated. During carbohydrate metabolism by glycolysis (Glyc), the cytosolic NADH/NAD+ ratio increases, thus limiting NAD+ availability for Sirt1-mediated FOXO and PGC-1α deacetylation. This inhibits FOXOs and PGC-1α, which down-regulate mitochondrial OXPHOS and biogenesis. Fatty acid and ketone body oxidation within the mitochondrion leaves the cytosolic NADH/NAD+ more oxidized, thereby stimulating Sirt1-mediated deacetylation and activation of FOXOs and PGC-1α, causing up-regulation of mitochondrial OXPHOS. Thus, the availability and nature of calories directly regulates the epigenome and modulates the bioenergetic pathways required for optimal caloric exploitation. (AcCoA) Acetyl-CoA, (AcAc) acetylaldehyde, (BOB) β-hydroxybutyrate, (OAA) oxaloacetate, (Pyr) pyruvate, (suc) succinate, (Suc-CoA) succinyl-CoA, (SSA) succinate semialdehyde, (GABA) γ-aminobutyric acid, (αKG) α-ketoglutarate, (G6P) glucose-6-phosphate, (HATs) histoacetyltransferases, (HDAC) histone deacetylase, (GSSG and GSH) oxidized and reduced glutathione.
At the level of the unicellular eukaryote, the connection between calorie availability and gene expression is mediated through high-energy intermediates of glycolysis and OXPHOS, because these intermediates can reflect caloric availability for the cell. The high-energy intermediates used are ATP, acetyl-CoA, and S-adenosylmethionine (SAM). These three bioenergetic intermediates are the primary substrates for modifying and modulating the epigenome (Wallace and Fan 2009a).
nDNA is packaged in nucleosomes that encompass ~146–147 nucleotides of DNA wrapped around two molecules each of histones H2A, H2B, H3, and H4. Each of these histones has an amino-terminal tail that is basic and thus has high affinity for the negatively charged sugar-phosphate backbone of DNA. When these amino-terminal tails are unmodified, they bind to the acidic DNA, stabilize its packaging, and shut down transcription. However, when the tails are modified by phosphorylation via kinases using ATP, acetylation via histone acetylases (HATs) using acetyl-CoA, and methylation via methyltransferases using SAM, these modifications increase the negative charge, decrease the positive charge, and alter the molecular interactions of histone tails with the negatively charged DNA, thus opening the chromatin and increasing transcription (Figs. 3 and 4) (Wallace and Fan 2009a).
ATP can be generated by both glycolysis and OXPHOS. However, acetyl-CoA is generated predominantly within the mitochondrion. After condensation with OAA, it is exported to the cytosol within citrate. Once in the cytosol, citrate can be cleaved back to acetyl-CoA and OAA by ATP-citrate lyase (Wallace and Fan 2009a).
SAM is generated from methyl groups recovered from the export of serine from the mitochondrion, the methyl group being transferred to folate by serine hydroxymethyl-transferase and then transferred to homocysteine to generate methionine. This methionine is subsequently converted to SAM by condensation with ATP. In addition to modifying histones, SAM is also used to add methyl groups to the cytosines of CpG dinucleotides, modulating DNA–protein interactions (Wallace and Fan 2009a).
When calories are abundant, cytosolic and nuclear ATP, acetyl-CoA, and SAM therefore increase. This drives the modification of the histone tails and opens the chromatin, thus turning on transcription and stimulating growth and reproduction. When calories become limiting, ATP, acetyl-CoA, and SAM decline and phosphatases remove the phosphate groups, histone deacetylases (HDACs) remove the acetyl groups, and demethylases remove the methyl groups. The chromatin then condenses, shutting down transcription until calories are once again available. Thus, the major factor in the evolution of the epigenome must have been the need to provide a global mechanism for coordinating nDNA gene expression in response to energy availability, mediated through cellular energy metabolism (Wallace and Fan 2009a).
Pangenetic transcriptional regulation is particularly pertinent for the regulation of the thousands of dispersed nDNA energy-generating genes. These genes need to be up-regulated when calories are plentiful and down-regulated when calories are limited. However, their scattered distribution would require both regulation in cis for genes relatively close to one another on the same chromosome and in trans for genes that are dispersed on different chromosomes. The former could have provided the impetus for the formation of transcriptional loops and the latter for the development of nDNA transcriptional islands, where genes from multiple chromosomes come together for coordinate gene transcription (Wallace and Fan 2009a). Once established in unicellular eukaryotes, these systems could be generalized for the coordinate regulation of other interrelated genes.
Although changes in chromatin could provide an effective means of linking caloric availability to gene expression and growth, this approach does not discriminate between type of calorie, carbohydrate versus fat, and ketone bodies. In multicellular animals, switching reliance between glycolysis and OXPHOS must be controlled both systemically and intracellularly.
Systemic control is achieved through hormonal secretion by pancreatic α and β cells. When carbohydrates are abundant, insulin is released from mammalian β cells. Insulin binds to insulin receptors on target cells, activating the phosphoinositol-3 kinase (PI3K) pathway. This activates Akt/PKB (protein kinase B) to phosphorylate and inactivate FOXO (Forkhead box class O) transcription factors. When active, FOXOs bind to insulin response elements (IREs) of DNA, some of which are upstream of the transcriptional coactivator PPARγ–coactivator-1α (PGC-1α). PGC-1α, in turn, binds to a range of transcription factors in various tissues including PPARγ and up-regulates mitochondrial biogenesis and OXPHOS. Therefore, when carbohydrates are abundant, FOXOs are inactive, PGC-1α is down-regulated, OXPHOS declines, and the animal relies increasingly on glycolysis for ATP production. The excess dietary calories are then stored as fat (Wallace 2005, 2007).
During starvation, when carbohydrates are limited, the animal mobilizes its stored fat and up-regulates mitochondrial OXPHOS, because only OXPHOS can burn fat to generate ATP. In the absence of carbohydrates, insulin production declines, inactivating Akt/PKB, and resulting in the dephosphorylation of FOXOs. Activated FOXOs then induce PGC-1α and this up-regulates OXPHOS. Concurrently, the mammalian pancreatic α cells sense the low glucose and respond by secreting glycagon. This hormone binds to the glucagon receptors of target cells, activating adenylylcyclase. The increased cAMP activates PKA (protein kinase A) to phosphorylate and activates CREB (cAMP response element binding). Phospho-CREB then enters the nucleus and binds to cAMP response elements (CRE), one of which is also upstream of PGC-1α. Therefore, during fasting and starvation, PGC-1α and thus OXPHOS is doubly induced by dephosphorylated FOXOs and phosphorylation of CREB, thus shifting metabolism toward OXPHOS to burn stored fat (Wallace 2005, 2007).
This systemic regulation is complemented and extended by regulation of the activity of FOXOs and PGC-1α transcription factors. FOXOs and PGC-1α can be acetylated by HATs using acetyl-CoA and deacetylated by the class III deacetylase Sirt1. Sirt1 requires NAD+ as a coreactant for deacetylation; it cannot use NADH. When carbohydrate or fat calories are abundant, acetyl-CoA is abundant and FOXOs and PGC-1α are acetylated. When carbohydrates are metabolized by glycolysis, cytosolic NAD+ is reduced to NADH + H+, rendering Sirt1 inactive. As a result, FOXOs and PGC-1α stay acetylated, the transcription and activity of PGC-1α remains suppressed, OXPHOS is inhibited, and glycolysis is favored. In contrast, when fatty acids or ketones are metabolized, cytosolic NAD+ remains oxidized, Sirt1 is activated to deacetylate FOXOs and PGC-1α, OXPHOS is up-regulated, and fats and ketones are oxidized (Fig. 4) (Wallace and Fan 2009a; Wallace et al. 2009).
BIOENERGETICS AND EPIGENOMIC DISEASE
Although mtDNA variation determines the outer limits of the environmental challenge that an individual can tolerate, animals are also subjected to daily, monthly, and annual changes in their energetic environment (e.g., available calories, temperature, and activity). These short-term changes in energy availability and demand must be accommodated by changes in the expression of nDNA-encoded energetic genes (Fig. 2).
It has been proposed that the epigenome provides the interface between the environment and nDNA gene expression (Feinberg 2007, 2008). Yet the mechanism by which the epigenome perceives the environment has not been specified. The fact that the epigenome is modulated by high-energy intermediates (ATP, acetyl-CoA, and SAM) immediately suggests that the primary interface between the environment and the epigenome is the cellular bioenergetic system (Fig. 2). Not only would the availability and demand for calories require coordinate changes in the nDNA genes for energy metabolism, but changes in the energetic status of the cell and individual would also determine which developmental pathways would be optimal for growth, differentiation, and death. Therefore, the primary interface between the environment and the individual must be the cytosolic and mitochondrial bioenergetic systems (Fig. 2) (Wallace and Fan 2009a; Wallace et al. 2009).
The role of the epigenome in regulating bioenergetics is supported by the observations that (1) pathogenic mutations in the mtDNA result in symptoms that overlap with those attributed to the epigenome disease and (2) several epigenomic diseases have been found to be associated with mitochondrial dysfunction. This leads to a bioenergetic hypothesis for common diseases, which is based on six key tenets:
Energy is derived from the calories in our diet.
Energy is required by every organ, but in different ways and to different extents.
Hundreds of critical bioenergetic genes are dispersed throughout mtDNA and nDNA.
The expression of bioenergetic genes is coordinated in relation to calories via modification of the epigenome.
High-energy intermediates generated by cellular bioenergetic systems mediate changes in the epigenome.
Defects in either bioenergetic genes or the epigenome can disrupt energy flow and cause multisystem disease.
“Epigenomic diseases” affect imprinting, methylation, and chromatin organization (Feinberg 2007, 2008) and alter transcription regulatory proteins such as PPARγ and PGC-1α (Fig. 5). Thus, the epigenome must be regulating bioenergetic gene expression in the cis configuration for adjacent bioenergetic genes within individual chromatin loop domains and in the trans configuration for genetic elements on multiple chromosomes, possibly through transcriptional islands.
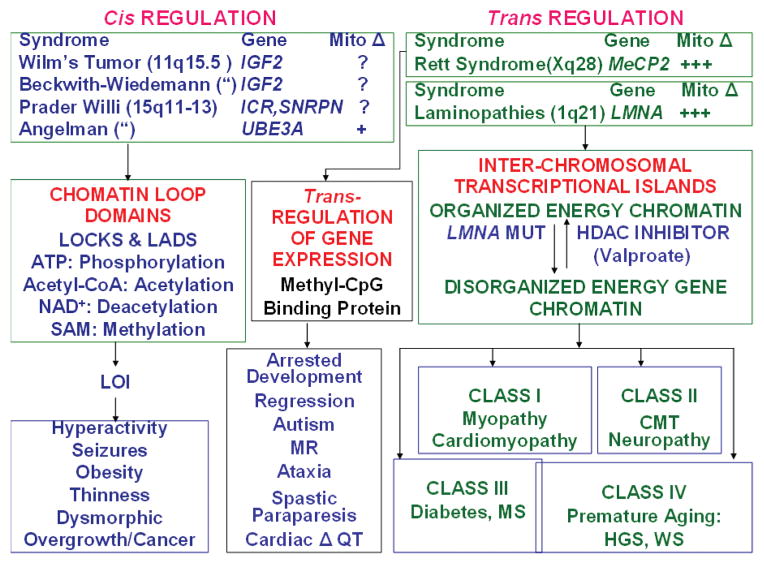
Figure 5.
Epigenomic disease association with mitochondrial dysfunction. (Left side) Diseases associated with loss of imprinting (LOI) potentially alter chromatin loop domain structure and cause aberrant cis interactions of bioenergetic genes. Chromatin domains affected by LOI can correlate with LOCHs (large organized chromatin K9 modifications blocks [Wen et al. 2009]) and LADs (lamina-associated domains [Guelen et al. 2008]). (Right side) Diseases associated with aberrations in the interaction of chromatin domains on different chromosomes or chromosomal domains resulting in aberrant trans interactions on bioenergetic genes. (Mito Δ) Extent of evidence for mitochondrial dysfunction associated with epigenetic disease. (IGF2) insulin-like growth factor 2 gene, (SNRPN) small nuclear ribonuclear polypeptide N, (ICR) imprinting control region, (UBE3A) ubiquitin-protein ligase E3A, (MeCP2) methyl C binding protein 2, (LMNA) lamin A gene, (MUT) mutant, (HDAC) histone deacetylase.
Imprinting diseases are examples of cis-acting diseases affecting transcriptional chromatin domains. Alterations in DNA methylation and methylation-binding proteins or in overall chromatin organization are examples of trans-acting factors (Fig. 5).
The perturbation of imprinting seen in Angelman and Prader-Willi syndromes involves inactivation of the active allele on chromosome 15q11-13 in the context of an imprinted and inactive maternal or paternal allele. In a mouse model of Angelman syndrome, hippocampal neurons were found to have reduced synaptic vesicle density and shrunken mitochondria in association with a reduction of brain mitochondrial complex II + III (Su et al. 2009). In Beckwith-Wiedemann syndrome and Wilm’s tumor, there is loss of imprinting (LOI) on chromosome 11q15.5 within a chromatin loop domain encompassing the insulin-like growth factor 2 (IGF2) gene (Bjornsson et al. 2007; Feinberg 2007, 2008). IGF2 could act through the PI3K-Akt-FOXO pathway to modulate energy metabolism (Kaneda et al. 2007; Wallace and Fan 2009a).
Alterations in trans-acting transcription regulatory factors affecting mitochondrial function are seen in Rett syndrome and the laminopathies. Rett syndrome is caused by mutations in methyl-CpG-binding protein 2 (MeCP2) (Loat et al. 2008). Yet multiple studies have reported abnormal mitochondria and mitochondrial function in Rett patients (Eeg-Olofsson et al. 1989; Heilstedt et al. 2002; Kriaucionis et al. 2006; Wallace and Fan 2009a). The laminopathies are caused by mutations in the laminin A/C gene (LMNA), with different mutations encompassing most of the clinical phenotypes observed in mitochondrial diseases (Stewart et al. 2007; Liu and Zhou 2008). Class I laminopathies result in myopathy and cardiomyopathy. Class II laminopathies result in the peripheral neuropathy Charcot-Marie-Tooth. Class III laminopathies result in diabetes and metabolic syndrome, and Class IV laminopathies result in premature aging syndromes including Hutchinson-Guilford progeria and Werner syndrome. That these phenotypes are generated by bioenergetic dysfunction is supported by the report that cells harboring LMNA mutations exhibit mitochondrial biochemical defects (Caron et al. 2007; Wallace and Fan 2009a).
Data on “epigenomic diseases,” although limited, therefore support their association with mitochondrial dysfunction. If confirmed, then epigenomic and mitochondrial diseases share a common pathophysiology, explaining their similar clinical phenotypes. This would establish the central role of energy metabolism in modulating the epigenome, thus providing the direct link among the nucleus-cytosol, mitochondrion, and environment (Wallace and Fan 2009a).
BIOENERGETIC PARADIGM IN ANIMAL EVOLUTION AND HUMAN DISEASE
The bioenergetic paradigm now provides a framework within which to link animal evolution and human disease (see Figs. 1 and 2). Nuclear gene mutations, both anatomical and bioenergetic, permit speciation. The mutation rate of nDNA is low. Hence, it may take hundreds of thousands of years for sufficient mild nDNA mutations to accumulate in anatomical genes to result in a shift in animal anatomy to permit exploitation of a new energy reservoir and, thus, speciation. For species in established niches, more severe anatomical mutations will render the individual incompatible with its niche, resulting in elimination by selection. In humans, such mutations cause classical Mendelian diseases such as osteogenesis imperfecta. Relatively mild nDNA mutations in bioenergetic genes can be compatible with survival within a niche because there is some latitude for differences in energy efficiency within a species. Functional variants in important energy regulatory genes such as PPARγ and PGC-1α can reach polymorphic frequencies in regional populations. These variants may be beneficial in one local energy environment. However, when found outside the appropriate environment, they become risk factors for disease. Thus, PPARγ (Altshuler et al. 2000) and PGC-1α (Ek et al. 2001; Muller et al. 2003) polypeptide polymorphisms show very high correlations with diabetes in certain regional populations, but no association with that phenotype in large interpopulation studies (Saxena et al. 2007; Scott et al. 2007; Sladek et al. 2007; Zeggini et al. 2007). More significant bioenergetic variants might also permit exploitation of adjacent energy reservoirs, ultimately leading to speciation. For example, changes in certain nDNA-encoded complex I genes have been associated with branches in primate radiation (Mishmar et al. 2006).
In contrast to nDNA, mtDNA mutations occur at a very high rate. Because mtDNA encodes only genes related to OXPHOS, mtDNA mutations primarily alter bioenergetics. Because mtDNA genes are vital and highly conversed, most functional mtDNA mutations are deleterious. However, the most severe mtDNA mutations are eliminated by intraovarian selection before ovulation and thus do not contribute to the genetic load of the population (Fan et al. 2008; Stewart et al. 2008). This explains why there are relatively few common pathogenic mtDNA mutations (Wallace et al. 2007). As a consequence of intraovarian selection, most mtDNA mutations are mildly deleterious, neutral, or beneficial variants. Still, the ovarian threshold for functional defects is relatively high, ensuring that enough mildly deleterious mutations are introduced into the population that at least some variants will be available to permit adaptation to sustained energy environmental change.
Because of the lack of recombination, beneficial mtDNA mutations must be continually regenerated through de novo mutation. There are a finite number of physiologically beneficial mtDNA-coding gene changes, and thus the same mutations are observed repeatedly in different regional populations on different mtDNA backgrounds. This convergent evolution confirms the beneficial nature of these variants (Wallace et al. 2003; Ruiz-Pesini et al. 2004; Ruiz-Pesini and Wallace 2006). Because beneficial coding region mutations are relatively rare, mtDNA bioenergetic flexibility is expanded through mtDNA control region mutations. Because control region mutations can alter mtDNA transcription and copy number, they can have a significant effect on mitochondrial function without disrupting bioenergetic pathways. Consequently, polymorphic mtDNA control region variants are much more common than coding region variants (Suissa et al. 2009). Therefore, the mtDNA provides a powerful system for bioenergetic adaptation to environmental change. However, the negative consequence of this system is that both recent deleterious and ancient polymorphic mtDNA variants contribute substantially to the etiology of common metabolic and degenerative diseases.
Changes in the epigenome permit modulation of bioenergetics in response to short-term fluctuations in the energy environment. This results from alterations in gene expression mediated by the modification of histones, signal transduction proteins, and DNA, driven by the availability of high-energy intermediates (Wallace and Fan 2009a) and the cellular redox potential (Wallace et al. 2009). These cellular metabolic intermediates and parameters, in turn, reflect the flux of reducing equivalents through mitochondrial and glycolytic bioenergetic pathways of the cell. Thus, environmental conditions are linked to coordinate gene expression through bioenergetics. Daily and weekly changes can be accommodated by activation of signal transduction systems (e.g., insulin and glucagon) or the expression and activity of transcription factors that regulate bioenergetics (e.g., FOXOs and PGC-1α) (Wallace 2005, 2007). More long-term regulation can occur through cis-acting processes, as is seen for imprinted loci associated with chromatic loop domains (e.g., Angelman, Prader-Willi, and Beckwith-Weidemann syndromes). Longer-term regulation can also be mediated by trans-acting factors, as is observed for MeCP2 protein and perhaps lamin A. DNA methylation is another way that bioenergetic gene expression could be modulated over prolonged periods, and DNA methylation patterns can be altered in respiratory-deficient cancer cells (Feinberg 2007, 2008; Smiraglia et al. 2008). Genetic diseases affecting cis-acting imprinted loci can involve either inactivation or activation of a functional chromosomal allele and have been documented to be associated with mitochondrial dysfunction (Wallace and Fan 2009a). Similarly, genetic diseases affecting trans-acting systems involving DNA-binding factors or chromatin structural elements have also been associated with mitochondrial dysfunction.
Perturbation of cellular bioenergetics is therefore a common feature of metabolic and degenerative diseases. Bioenergetic defects can be caused by an array of genetic and epigenetic perturbations, including mutations in nDNA and mtDNA-encoded bioenergetic genes and in the epigenetic regulation of bioenergetic genes. Consequently, the complexity of “complex diseases” is the product of the large number of ways that bioenergetic genes can be perturbed. However, the pathophysiology of common diseases is the same, bioenergetic dysfunction, which explains why all of these diseases affect the more metabolically active tissues: brain, heart, muscle, renal, endocrine, and hepatic systems. Although this analysis confirms the complexity of the factors that can cause common diseases, it implies that the clinical symptoms of a wide range of common diseases might be treated by addressing the same target, the bioenergetic systems of the cell.
Acknowledgments
The author thanks Dr. Weiwei Fan, Ms. Marie T. Lott, and Kate Hartshorn for their assistance in assembling this document. The work has been supported by National Institutes of Health grants NS21328, AG24373, DK73691, AG13154, AG16573, a CIRM comprehensive grant RC1-00353-1, and a Doris Duke Clinical Interfaces award 2005 to D.C.W.
References
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, et al. The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- Balloux F, Handley LJ, Jombart T, Liu H, Manica A. Climate shaped the worldwide distribution of human mitochondrial DNA sequence variation. Proc Biol Sci. 2009;276:3447–3455. doi: 10.1098/rspb.2009.0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson HT, Brown LJ, Fallin MD, Rongione MA, Bibikova M, Wickham E, Fan JB, Feinberg AP. Epigenetic specificity of loss of imprinting of the IGF2 gene in Wilms tumors. J Natl Cancer Inst. 2007;99:1270–1273. doi: 10.1093/jnci/djm069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann RL, Stoneking M, Wilson AC. Mitochondrial DNA and human evolution. Nature. 1987;325:31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- Caron M, Auclair M, Donadille B, Bereziat V, Guerci B, Laville M, Narbonne H, Bodemer C, Lascols O, Capeau J, Vigouroux C. Human lipodystrophies linked to mutations in A-type lamins and to HIV protease inhibitor therapy are both associated with prelamin A accumulation, oxidative stress and premature cellular senescence. Cell Death Differ. 2007;14:1759–1767. doi: 10.1038/sj.cdd.4402197. [DOI] [PubMed] [Google Scholar]
- Darwin C. On the origin of species by means of natural selection (or the preservation of favoured races in the struggle for life) Murray; London: 1859. [PMC free article] [PubMed] [Google Scholar]
- Darwin CR, Wallace AR. On the tendency of species to form varieties; and on the perpetuation of varieties and species by natural means of selection. J Proc Linn Soc Lond Zool. 1858;3:46–50. [Google Scholar]
- Eeg-Olofsson O, al-Zuhair AG, Teebi AS, al-Essa MM. Rett syndrome: Genetic clues based on mitochondrial changes in muscle. Am J Med Genet. 1989;32:142–144. doi: 10.1002/ajmg.1320320131. [DOI] [PubMed] [Google Scholar]
- Ek J, Andersen G, Urhammer SA, Gaede PH, Drivsholm T, Borch-Johnsen K, Hansen T, Pedersen O. Mutation analysis of peroxisome proliferator-activated receptor-γ coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to type II diabetes mellitus. Diabetologia. 2001;44:2220–2226. doi: 10.1007/s001250100032. [DOI] [PubMed] [Google Scholar]
- Fan W, Waymire K, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, MacGregor GR, Wallace DC. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–962. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. Epigenetics at the epicenter of modern medicine. J Am Med Assoc. 2008;299:1345–1350. doi: 10.1001/jama.299.11.1345. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Heilstedt HA, Shahbazian MD, Lee B. Infantile hypotonia as a presentation of Rett syndrome. Am J Med Genet. 2002;111:238–242. doi: 10.1002/ajmg.10633. [DOI] [PubMed] [Google Scholar]
- Johnson MJ, Wallace DC, Ferris SD, Rattazzi MC, Cavalli-Sforza LL. Radiation of human mitochondria DNA types analyzed by restriction endonuclease cleavage patterns. J Mol Evol. 1983;19:255–271. doi: 10.1007/BF02099973. [DOI] [PubMed] [Google Scholar]
- Kaneda A, Wang CJ, Cheong R, Timp W, Onyango P, Wen B, Iacobuzio-Donahue CA, Ohlsson R, Andraos R, Pearson MA, et al. Enhanced sensitivity to IGF-II signaling links loss of imprinting of IGF2 to increased cell proliferation and tumor risk. Proc Natl Acad Sci. 2007;104:20926–20931. doi: 10.1073/pnas.0710359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazuno AA, Munakata K, Nagai T, Shimozono S, Tanaka M, Yoneda M, Kato N, Miyawaki A, Kato T. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS Genet. 2006;2:e128. doi: 10.1371/journal.pgen.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khusnutdinova E, Gilyazova I, Ruiz-Pesini E, Derbeneva O, Khusainova R, Khidiyatova I, Magzhanov R, Wallace DC. A mitochondrial etiology of neurodegenerative diseases: Evidence from Parkinson’s disease. Ann NY Acad Sci. 2008;1147:1–20. doi: 10.1196/annals.1427.001. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Paterson A, Curtis J, Guy J, Macleod N, Bird A. Gene expression analysis exposes mitochondrial abnormalities in a mouse model of Rett syndrome. Mol Cell Biol. 2006;26:5033–5042. doi: 10.1128/MCB.01665-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zhou Z. Lamin A/C, laminopathies and premature ageing. Histol Histopathol. 2008;23:747–763. doi: 10.14670/HH-23.747. [DOI] [PubMed] [Google Scholar]
- Loat CS, Curran S, Lewis CM, Duvall J, Geschwind D, Bolton P, Craig IW. Methyl-CpG-binding protein 2 polymorphisms and vulnerability to autism. Genes Brain Behav. 2008;7:754–760. doi: 10.1111/j.1601-183X.2008.00414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriwether DA, Clark AG, Ballinger SW, Schurr TG, Soodyall H, Jenkins T, Sherry ST, Wallace DC. The structure of human mitochondrial DNA variation. J Mol Evol. 1991;33:543–555. doi: 10.1007/BF02102807. [DOI] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini EE, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, et al. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Mondragon-Palomino M, Procaccio V, Gaut B, Wallace DC. Adaptive selection of mitochondrial complex I subunits during primate radiation. Gene. 2006;378:11–18. doi: 10.1016/j.gene.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Muller YL, Bogardus C, Pedersen O, Baier L. A Gly482Ser missense mutation in the peroxisome proliferator-activated receptor γ coactivator-1 is associated with altered lipid oxidation and early insulin secretion in Pima Indians. Diabetes. 2003;52:895–898. doi: 10.2337/diabetes.52.3.895. [DOI] [PubMed] [Google Scholar]
- Palmieri L, Alberio S, Pisano I, Lodi T, Meznaric-Petrusa M, Zidar J, Santoro A, Scarcia P, Fontanesi F, Lamantea E, Ferrero I, Zeviani M. Complete loss-of-function of the heart/muscle-specific adenine nucleotide translocator is associated with mitochondrial myopathy and cardiomyopathy. Hum Mol Genet. 2005;14:3079–3088. doi: 10.1093/hmg/ddi341. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Wallace DC. Evidence for adaptive selection acting on the tRNA and rRNA genes of the human mitochondrial DNA. Hum Mutat. 2006;27:1072–1081. doi: 10.1002/humu.20378. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Lott MT, Procaccio V, Poole J, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res (database issue) 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- Smiraglia DJ, Kulawiec M, Bistulfi GL, Gupta SG, Singh KK. A novel role for mitochondria in regulating epigenetic modification in the nucleus. Cancer Biol Ther. 2008;7:1182–1190. doi: 10.4161/cbt.7.8.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CL, Kozlov S, Fong LG, Young SG. Mouse models of the laminopathies. Exp Cell Res. 2007;313:2144–2156. doi: 10.1016/j.yexcr.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JB, Freyer C, Elson JL, Wredenberg A, Cansu Z, Trifunovic A, Larsson NG. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008;6:e10. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Fan W, Coskun PE, Vesa J, Gold JA, Jiang YH, Potluri P, Procaccio V, Acab A, Weiss JH, Wallace DC, Kimonis VE. Mitochondrial dysfunction in CA1 hippocampal neurons of the UBE3A deficient mouse model for Angelman syndrome. Neurosci Lett. 2009 doi: 10.1016/j.neulet.2009.06.079. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suissa S, Wang Z, Poole J, Wittkopp S, Feder J, Shutt TE, Wallace DC, Shadel GS, Mishmar D. Ancient mtDNA genetic variants modulate mtDNA transcription and replication. PLoS Genet. 2009;5:e1000474. doi: 10.1371/journal.pgen.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial DNA sequence variation in human evolution and disease. Proc Natl Acad Sci. 1994;91:8739–8746. doi: 10.1073/pnas.91.19.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Why do we have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu Rev Biochem. 2007;76:781–821. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria as chi. Genetics. 2008;179:727–735. doi: 10.1534/genetics.104.91769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Fan W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2009a doi: 10.1016/j.mito.2009.09.006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Fan W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 2009b;23:1714–1736. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Brown MD, Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–230. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- Wallace DC, Ruiz-Pesini E, Mishmar D. mtDNA variation, climatic adaptation, degenerative diseases, and longevity. Cold Spring Harbor Symp Quant Biol. 2003;68:479–486. doi: 10.1101/sqb.2003.68.471. [DOI] [PubMed] [Google Scholar]
- Wallace DC, Lott MT, Procaccio V. Mitochondrial genes in degenerative diseases, cancer and aging. In: Rimoin DL, et al., editors. Emery and Rimoin’s principles and practice of medical genetics. 5. Churchill Livingstone Elsevier; Philadelphia: 2007. pp. 194–298. [Google Scholar]
- Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol Mech Dis. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;49:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]