Abstract
N6-methyladenosine (m6A) is the most abundant mRNA modification, and has important links to human health. While recent studies have successfully identified thousands of mammalian RNA transcripts containing the modification, it is extremely difficult to identify the exact location of any specific m6A. Here we have identified a polymerase with reverse transcriptase activity (from Thermus thermophilus) that is selective by up to 18-fold for incorporation of thymidine opposite unmodified A over m6A. We show that the enzyme can be used to locate and quantify m6A in synthetic RNAs by analysis of pausing bands, and have used the enzyme in tandem with a nonselective polymerase to locate the presence and position of m6A in high-abundance cellular RNAs. By this approach we demonstrate that the long-undetermined position of m6A in mammalian 28S rRNA is nucleotide 4190.
In the quest to understand cellular function at the molecular level, the study of post-transcriptional modification of RNA is of vital interest. In particular, N6-methyladenosine (m6A, 1) is a relatively abundant modification in the messenger RNA of higher eukaryotes and some viruses.1 Although its discovery in mRNA occurred decades ago,2 there has been renewed interest in m6A due to the finding that it acts as a substrate for fat mass and obesity-associated protein (FTO),3 an oxidative demethylase which has been linked to obesity and the regulation of homeostasis;4–6 and for AlkBH5, an oxidative demethylase in the same family.7
Identifying the function of m6A modifications has proved challenging.8–12 Early work using enzymatic digestion and radiolabelling led to the discovery of the consensus sequence RAC (R=A or G) for m6A13–18 and the identification of specific modified sites in Rous Sarcoma Virus mRNA19–21 and in bovine prolactin mRNA.22,23 This research also showed that methylation at any particular site can be incomplete, with a methylation extent of 20–90% at one site. Modern RNA sequencing techniques have advanced our ability to identify transcripts modified by m6A. Using massively parallel sequencing and m6A-selective antibodies, two groups recently identified thousands of modified mRNAs and ncRNAs from mice and humans and confirmed the general consensus sequence of RRACU for the location of methylated adenines.24,25 After completion of the present work, Liu et al. reported a new method for detecting and quantifying m6A at a specific site using multiple enzymatic steps.26 However, the ability to interrogate the methylation status of any specific adenine at nucleotide resolution, without painstaking digestion analysis, has remained elusive.
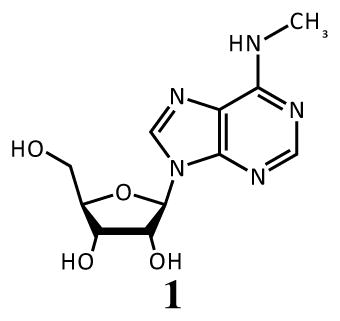
The ability to locate m6A modifications in RNAs at nucleotide resolution will no doubt aid in understanding their function. Polymerase enzymes offer a possible mechanism for locating modifications due to their sterically sensitive active sites. Notably, polymerase selectivity has previously been harnessed to detect m6A in DNA via single-molecule sequencing.27 An early attempt at a related single-molecule sequencing technique for RNA has also been described,28 but employed an enzyme with low selectivity (HIV-RT; see below), and will need further development before it is practical. Another class of DNA-processing enzymes, ligases, can also be sensitive to structure, and Dai et al. describe a technique in which ligation of complementary DNAs occurs more favorably in the presence of A than m6A.29 However, the conditions were tuned carefully for the specific reaction, and no attempt was made to detect m6A in an actual sample of cellular RNA.
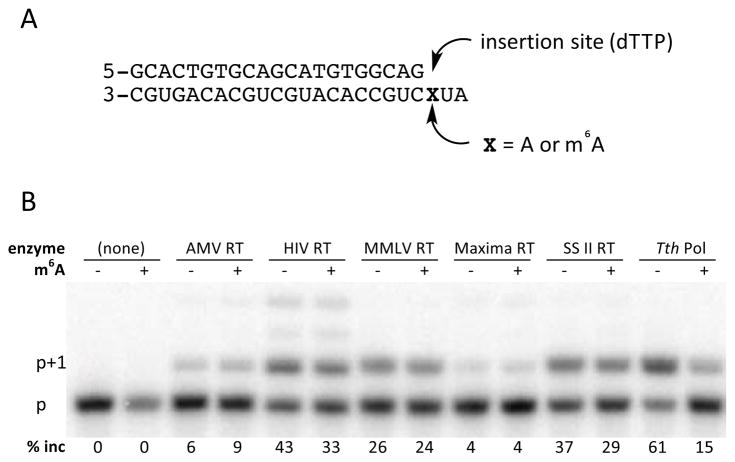
We postulated that there might exist a polymerase enzyme with substantial selectivity against m6A, and that such an enzyme might be harnessed for site-specific detection of the modification. We carried out a screen of enzymes with reverse transcriptase activity, monitoring their ability to extend a radiolabeled DNA primer by incorporating thymidine triphosphate (dTTP) opposite either A or m6A in an RNA template (Figure 1). The data showed that only recombinant Thermus thermophilus DNA polymerase I (Tth DNA pol) showed strong selectivity (61% vs. 15% primer extension) among the enzymes tested. This DNA polymerase is known to act as a reverse transcriptase in the presence of Mn2+.30
Figure 1.
Screen of polymerase selectivity for incorporation of dTTP opposite A or m6A in an RNA template. (A) Sequences of RNA template/DNA primer used in screen. (B) Autoradiogram showing primer extension (p+1 band) in the presence of A or m6A. Products were resolved on a 20% polyacrylamide denaturing gel using a 32P-5′-labeled primer.
Since Tth DNA pol showed selectivity in the context of one specific template sequence under one set of conditions, we tested if variations in temperature, time, and buffer composition might enhance selectivity (see Supporting Information (SI)). In particular, Mn2+ is known to decrease enzyme selectivity;31 however, we found that Mn2+ was required for reverse transcriptase activity in Tth DNA pol.
We next investigated whether this selectivity would extend to other sequence contexts. A 24mer template sequence was chosen from the 3′ untranslated region (3′-UTR) of the eef2 gene, which was found to be highly expressed and highly modified in mouse tissue and mouse embryonic stem cells.25,32 The template was synthesized containing either A or m6A, and the bases on either side of the A/m6A were varied systematically to allow a comparison of sequence context effects (see SI for details). In addition, several of the native sequence contexts in which m6A has been reported to occur were also synthesized. Single nucleotide incorporation kinetics were determined for each sequence containing A and m6A using steady-state methods33 (Table 1).
Table 1.
Steady-state incorporation efficiency for insertion of dTTP opposite A or m6A in synthetic RNAs by Tth DNA pol in varied sequence contexts.
| sequence context | efficiency, X=A (Vmax/Km) | efficiency, X=m6A (Vmax/Km) | ratio |
|---|---|---|---|
| UXC | 0.86±0.17 | 0.10±0.04 | 8.6 |
| GXC | 0.58±0.15 | 0.14±0.03 | 4.0 |
| (G)GXC | 1.22±0.17 | 0.19±0.04 | 6.4 |
| GXU | 0.95±0.61 | 0.10±0.10 | 9.1 |
| CXU | 1.13±0.77 | 0.14±0.14 | 8.2 |
| CXG | 1.39±0.26 | 0.13±0.03 | 10.4 |
| AXG | 1.51±0.61 | 0.10±0.02 | 15.6 |
| AXA | 1.87±1.25 | 0.11±0.06 | 17.5 |
| (G)AXC | 1.24±0.32 | 0.10±0.05 | 13.1 |
| UXA | 3.52±0.94 | 0.33±0.22 | 10.6 |
The RNA templates containing A show 4- to 18-fold better enzyme efficiency with Tth pol than the corresponding sequence containing m6A. Overall, then, differences in context produce moderate to negligible differences in selectivity. For example, for T incorporation opposite unmodified A, the UAA sequence context is processed with higher efficiency than other sequences. Additionally, 5′ G and 3′ C both appear to decrease enzyme efficiency to a small degree. Most notably, the selectivity of the Tth polymerase in the consensus methylation site context (GAC) is 4 to 6.4-fold, thus supporting the notion that the enzyme’s selectivity may be useful for identifying the most common occurrences of m6A in naturally occurring RNAs.
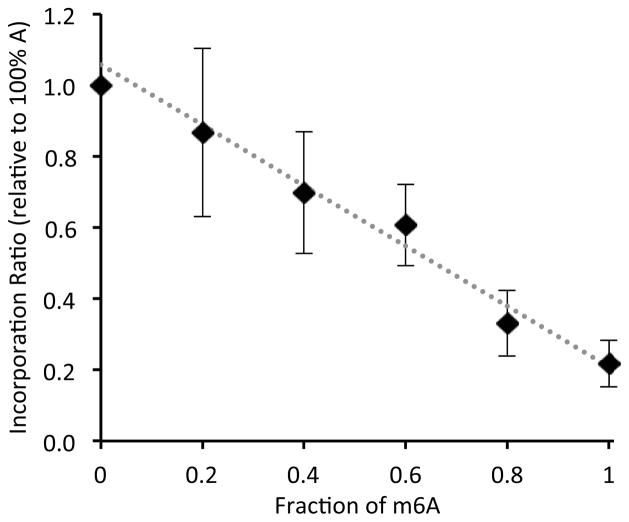
Next we asked whether the Tth enzyme could be used in a quantitative sense, to evaluate the degree of methylation at a specific site. To test this, we mixed known ratios of m6A-containing RNAs with their A-containing counterparts, and measured the yield of dTTP incorporation at a fixed timepoint. The percent extension of the primer in this RNA context was linearly proportional to the amount of m6A present (Figure 2), suggesting that the polymerase can be used in quantitative evaluations of the extent of methylation at one position.
Figure 2.
T insertion is correlated to the relative amount of m6A at target position. AGXCUGCCACAUGCUGCA CAGUGC was used as the template RNA at 1 3M concentration with varied ratios of m6A:A at the target position. Error bars show standard deviations from 5 trials.
We proceeded to carry out experiments to test whether this enzyme could be employed in probing methylation in RNAs extracted from mammalian cells. Since the amount of RNA and its secondary structure are likely to affect primer extension efficiencies, we introduced two control strategies. The first of these involves the use of two simultaneous primers (one adjacent to the probed methylation site, and one nearby but adjacent to a nucleotide with known methylation status). Comparison of these two should account for the amount of a given RNA present in a sample. The second control makes use of the same primers with a non-selective enzyme, avian mye-loblastosis virus reverse transcriptase (AMV RT). AMV RT was selected because it showed consistent ratios relative to Tth DNA pol across a range of RNA concentrations. Insertion of T at a site of interest could then be compared both to the control site and to incorporation by a control nonselective enzyme.
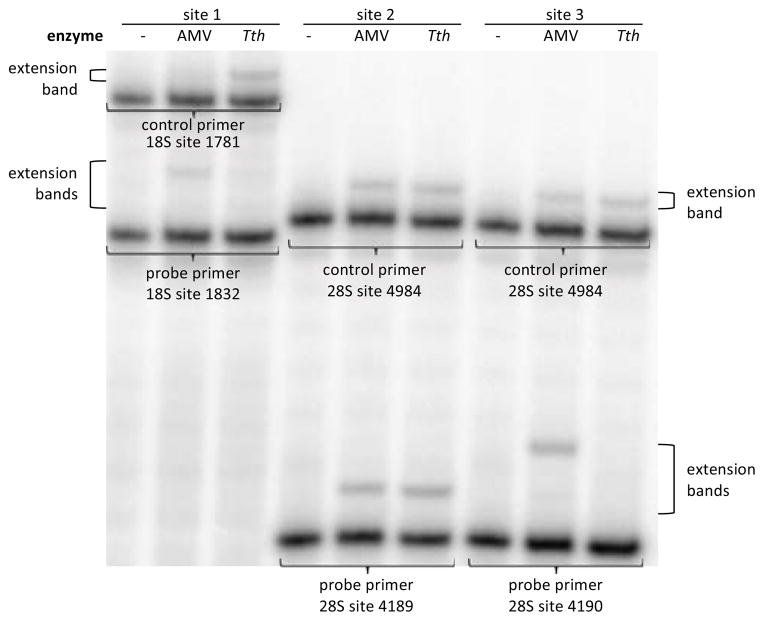
Human ribosomal RNA is known to contain two m6A modifications, one at position 1832 in the 18S subunit, and one at position 4189 or 4190 in the 28S subunit.34 Based on ratios to nearby unmodified bases after enzymatic digestion, these sites are reported to consist only of the modified base (100% m6A).35,36 Primers were designed to interrogate these three sites, as well as two other sites known to contain A (1781 in 18S; 4984 in 28S). Total RNA was extracted from 293T cells, and the RNAs probed with primer sets (see SI). As seen in Figure 3, results showed less than 20% incorporation of T by Tth DNA pol at the 1832 m6A site and the 4190 m6A site relative to controls, consistent with complete methylation in our quantitative experiments with synthetic RNAs. In contrast, we observed a high degree of incorporation (~120%) by Tth pol relative to AMV RT at the 4189 site. As a result, we can assign the previously-undetermined site of methylation in human 28S RNA as 4190 and not at the neighboring adenine at 4189.
Figure 3.
Identification of methylated adenine sites in human ribosomal RNA. Primers were designed to probe for m6A at three sites in rRNA. Incorporation at these sites by Tth pol was compared to incorporation by Tth pol at known non-methylated control sites and to incorporation by nonselective AMV RT.
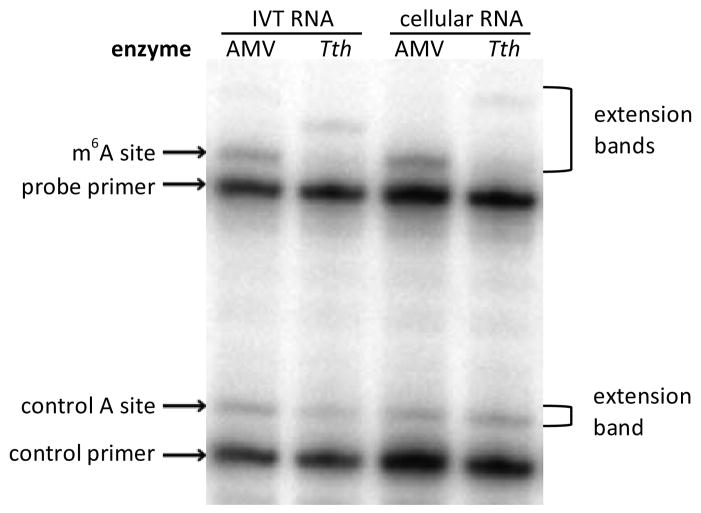
Finally, we attempted to detect m6A in a cellularly expressed messenger RNA (Figure 4). We chose a known site in the 3′-UTR of the bovine prolactin (bPRL) transcript.23 This is the only precisely mapped m6A site in a mammalian mRNA; native levels of modification are estimated to be ca. 20%, and this presents a challenging case for detection.23 The 3′-UTR was cloned into a plasmid and overexpressed in 293T cells. Two primers were designed, one to detect the known modification site and one to detect a nearby adenosine (see SI). The putative m6A site showed significantly lower incorporation than the nearby control A site. As an additional control, we performed in vitro transcription of the bPRL transcript in the absence of m6A, allowing us to compare the primer incorporation at the site of interest when only A was present. At the high enzyme concentrations initially used, no difference was seen between the total cellular RNA and the in vitro transcribed RNA. Use of a lower enzyme concentration, however, did reveal a significant difference in the m6A:A ratio (0.75±0.09 for IVT RNA vs. 0.43±0.04 for total cellular RNA; all extension products were included in quantification), confirming the presence of m6A in the mRNA expressed in 293T cells.
Figure 4.
Identification of the m6A site in the bPRL 3′-UTR. Incorporation of dTTP with in vitro transcribed (IVT) RNA was compared to incorporation templated by cellular bPRL RNA to differentiate the site of methylation. Incorporation with nonselective AMV RT is shown as an additional control.
In summary, we have characterized a commercially available polymerase that discriminates m6A from A in all tested sequence contexts. We have used it to detect m6A in abundant cellular RNAs. While further development is needed before this method is robust enough for detection and quantification of m6A in lower abundance mRNAs, it seems likely that the inherent selectivity of this enzyme will prove useful in the development of future m6A analysis techniques.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (GM068122) for support. EMH is supported by a Graduate Research Fellowship (DGE-1147470) from the NSF. TE is supported by a Postdoctoral Research Fellowship from the DAAD. PJB is the Kenneth G. and Elaine A. Langone Fellow of the Damon Runyon Cancer Research Foundation; HYC is an Early Career Scientist of Howard Hughes Medical Institute.
Footnotes
Supporting Information. Experimental methods and materials, additional data, and characterization of synthetic intermediates. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Sources
No competing financial interests have been declared.
References
- 1.Bokar JA. The Biosynthesis and Functional Roles of Methylated Nucleosides in Eukaryotic mRNA. In: Grosjean H, editor. Fine-tuning of RNA Functions by Modification and Editing. Springer-Verlag; Berlin Heidelberg: 2005. pp. 141–177. Topics in Current Genetics 12. [Google Scholar]
- 2.Desrosiers R, Friderici K, Rottman F. Proc Natl Acad Sci U S A. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JRB, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJF, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CNA, Doney ASF, Morris AD, Smith GD, Hattersley AT, McCarthy MI. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerken T, Girard CA, Tung YCL, Webby CJ, Saudek V, Hewitson KS, Yeo GSH, McDonough MA, Cunliffe S, McNeill LA, Galvanovskis J, Rorsman P, Robins P, Prieur X, Coll AP, Ma M, Jovanovic Z, Farooqi IS, Sedgwick B, Barroso I, Lindahl T, Ponting CP, Ashcroft FM, O’Rahilly S, Schofield CJ. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Brüning JC, Rüther U. Nature. 2009;458:894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 7.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RPG, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachellerie J, Amalric F, Caboche M. Nucleic Acids Res. 1978;5:2927–2944. doi: 10.1093/nar/5.8.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoltzfus CM, Dane RW. J Virol. 1982;32:918–931. doi: 10.1128/jvi.42.3.918-931.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkel D, Groner Y. Virology. 1983;131:409–425. doi: 10.1016/0042-6822(83)90508-1. [DOI] [PubMed] [Google Scholar]
- 11.Camper SA, Albers RJ, Coward JK, Rottman FM. Mol Cell Biol. 1984;4:538–543. doi: 10.1128/mcb.4.3.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll SM, Narayan P, Rottman FM. Mol Cell Biol. 1990;10:4456–4465. doi: 10.1128/mcb.10.9.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schibler U, Kelley DE, Perry RP. J Mol Biol. 1977;115:695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]
- 14.Wei C, Moss B. Biochemistry. 1977;16:1672–1676. doi: 10.1021/bi00627a023. [DOI] [PubMed] [Google Scholar]
- 15.Dimock K, Stoltzfus CM. Biochemistry. 1977;16:471–478. doi: 10.1021/bi00622a021. [DOI] [PubMed] [Google Scholar]
- 16.Canaani D, Kahana C, Lavi S, Groner Y. Nucleic Acids Res. 1979;6:2879–2899. doi: 10.1093/nar/6.8.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nichols JL, Welder L. Plant Sci Lett. 1981;21:75–81. [Google Scholar]
- 18.Narayan P, Ludwiczak RL, Goodwin EC, Rottman FM. Nucleic Acids Res. 1994;22:419–426. doi: 10.1093/nar/22.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beemon K, Keith J. J Mol Biol. 1977;113:165–179. doi: 10.1016/0022-2836(77)90047-x. [DOI] [PubMed] [Google Scholar]
- 20.Kane SE, Beemon K. Mol Cell Biol. 1985;5:2298–2306. doi: 10.1128/mcb.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Csepany T, Lin A, Baldick CJ, Beemon K. J Biol Chem. 1990;265:20117–20122. [PubMed] [Google Scholar]
- 22.Horowitz S, Horowitz A, Nilsen TW, Munns TW, Rottman FM. Proc Natl Acad Sci U S A. 1984;81:5667–5671. doi: 10.1073/pnas.81.18.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narayan P, Rottman FM. Science. 1988;242:1159–1162. doi: 10.1126/science.3187541. [DOI] [PubMed] [Google Scholar]
- 24.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 25.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. RNA. 2013 doi: 10.1261/rna.041178.113. published online in advance of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vilfan ID, Tsai YC, Clark TA, Wegener J, Dai Q, Yi C, Pan T, Turner SW, Korlach J. J Nanobiotechnology. 2013;11:8. doi: 10.1186/1477-3155-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai Q, Fong R, Saikia M, Stephenson D, Yu Y, Pan T, Piccirilli JA. Nucleic Acids Res. 2007;35:6322–6329. doi: 10.1093/nar/gkm657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers TW, Gelfand DH. Biochemistry. 1991;30:7661–7666. doi: 10.1021/bi00245a001. [DOI] [PubMed] [Google Scholar]
- 31.Hillebrand GG, Beattie KL. Nucleic Acids Res. 1984;12:3173–3184. doi: 10.1093/nar/12.7.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batista PJ, Chang H. unpublished data. [Google Scholar]
- 33.Goodman MF, Creighton S, Bloom LB, Petruska J. Crit Rev Biochem Mol Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 34.Maden BEH. Prog Nucleic Acid Res Mol Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 35.Maden BEH. J Mol Biol. 1986;189:681–699. doi: 10.1016/0022-2836(86)90498-5. [DOI] [PubMed] [Google Scholar]
- 36.Maden BEH. J Mol Biol. 1988;201:289–314. doi: 10.1016/0022-2836(88)90139-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.