Abstract
Systematic investigation of the lipidome will reveal new opportunities for disease diagnosis and intervention. However, lipidomic research has been hampered by the lack of molecular tools to track specific lipids of interest. Accumulating reports indicate lipid recognition can be achieved with properly constructed short peptides in addition to large proteins. This review summarizes the key developments of this area within the past decade. Select lantibiotic peptides present the best examples of low-molecular-weight probes of membrane lipids, displaying selectivity comparable to lipid-binding proteins. Designed peptides, through biomimetic approaches and combinational screening, have begun to demonstrate their potential for lipid tracking in cultured cells and even in living organisms. Biophysical characterization of these lipid-targeting peptides has revealed certain features critical for selective membrane binding, including preorganized scaffolds and the balance of polar and nonpolar interactions. The knowledge summarized herein should facilitate the development of molecular tools to target a variety of membrane lipids.
Needs for lipid-targeting molecules
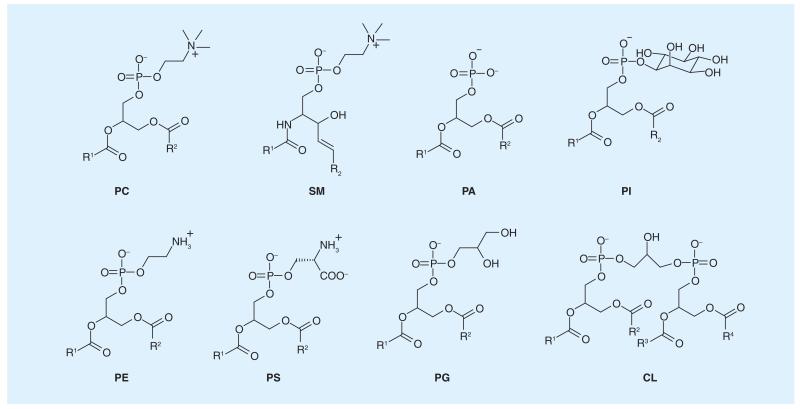
As a class of molecules essential to biology, lipids are no longer viewed only as amphipathic building blocks that constitute a membrane. Instead, they are observed to be actively involved in an expanding pool of processes in biology [1,2]. Metabolic profiling of the lipidome [3], largely using mass spectrometry, has revealed a surprisingly large diversity of membrane lipids: there are over 1000 different lipid species that are synthesized and metabolized by a living mammalian cell. This structural diversity (Figure 1), resulting from variations of both the headgroups and hydrophobic tails, naturally leads people to wonder why cells expend so much energy on acquiring this many different lipids and the roles for each individual lipid in cell physiology. Furthermore, biomembranes, which comprise assemblies of individual lipids, are now known to be heterogeneous in terms of their lipid composition across cell type and even at different subcellular locations. Biophysical experiments with model membranes have convincingly demonstrated the existence of lipid microdomains (also referred to as ‘lipid rafts’), yet their relevance in living cells and organisms needs to be further investigated [4]. It is also clear that the lipid composition can be quite different for the two leaflets of a cell membrane. For example, the aminophospholipids [5], including phosphatidylethanolamine (PE) and phosphatidylserine (PS), are known to preferentially reside in the inner (cytosol-facing) leaflet of a plasma membrane, while the outer leaflet is enriched with sphingomyelin (SM) and various glycolipids. This asymmetry of lipid distribution is believed to have significant ramifications to the fate of a cell (vide infra). The expanding knowledge of membrane lipids has also resulted in new questions that remain to be addressed in lipid biology: how does a cell control and regulate the homeostasis of these lipids? How does lipid homeostasis contribute to normal physiology and disease? These questions are of fundamental importance and will greatly benefit from novel molecular tools that allow one to track specific lipids in living cells and organisms. Importantly, the already known functions of select lipids suggest that they may serve as effective targets for disease diagnosis and intervention [6]. These biomedical applications will benefit greatly from synthetic molecules that target specific lipids as well.
Figure 1. Subset of common membrane lipids illustrating the diversity of lipid headgroups.
CL: Cardiolipin; PA: Phosphatidic acid; PC: Phosphatidylcholine; PE: Phosphatidylethanolamine; PG: Phosphatidylglycerol; PI: Phosphoinositol; PS: Phosphatidylserine; SM: Sphingomyelin.
Lipid targeting: proteins versus peptides
Consistent with the diverse roles that membranes play in biology, nature has evolved dedicated membrane-binding domains for a large number of proteins. For example, the recruitment of peripheral membrane proteins such as phospholipases and kinases to a membrane surface is a common mechanism to initiate signaling. Briefly, there are two mechanisms for membrane targeting: some proteins appear to recognize the physical properties of a membrane (e.g., electrostatic potential or membrane curvature), which result from certain lipid compositions; others are observed to interact with the targeted lipid headgroups in a stereospecific manner. Understanding the lipid specificity of these proteins has been a very active area of biochemical research. However, it will not be the main focus of this account and the readers are referred to an excellent review article that appeared in recent literature [7]. Although much work has been carried out in the field of lipid-binding proteins, it remains a challenge to predict the lipid preference for the sequence of a given protein. Similarly, engineering proteins to target specific lipids also remains nontrivial.
Naturally existing lipid-binding domains range from 150 to 400 amino acids. The large size makes them expensive to produce and difficult to label. For the purpose of tracking lipids in living animals, protein-based probes also suffer from limitations such as poor tissue penetration and slow clearance [8]. These problems have stimulated intense interest in developing low-molecular-weight markers of membrane lipids, which are anticipated to circumvent the limitations described above for protein-based probes. However, lipid-binding proteins have been evolved by nature to display a well-crafted pocket, around which the key residues are preorganized to achieve stereospecific recognition of the target lipid molecule (see later discussion for an example of lipid-binding proteins). Given the highly sophisticated structures behind protein function, an intriguing question naturally arises: to what extent can small molecules mimic large proteins for lipid targeting? Accumulating evidence in literature indicates that small molecules can do this job surprisingly well, with examples coming from both natural sources and synthetic endeavors.
Earlier work in this area, which has been nicely summarized by Lambert and Smith in their review [9], was largely limited to model membrane studies and had concentrated on cyclophane-based molecules designed to recognize the phosphatidylcholine (PC) headgroup. More recent work in the field has highlighted the promise of using peptides as low-molecular-weight markers for membrane lipids. Importantly, these peptide-based markers have demonstrated their effectiveness for lipid targeting not only in model membranes but also in cell cultures and even in living animals. This review intends to summarize such lipid-targeting peptides with an emphasis on the chemical principles underlying their specific recognition of lipids. Although beyond the scope of the current review, we note that the potential of other molecular scaffolds, such as aptamers, is beginning to be demonstrated for effective lipid recognition [10].
Lipid recognition by peptide natural products
Some of the most elegant designs of low-molecular-weight markers for membrane lipids come from nature. A number of peptide natural products from the lantibiotics family are observed to recognize lipids with protein-like specificities. Lantibiotics refer to a class of peptide antibiotics produced by a large number of Gram-positive bacteria, such as Streptococcus and Streptomyces, to attack other Gram-positive bacterium species [11]. A lantibiotic peptide is usually short in sequence (19-38 amino acids) and defined by the post-translational modifications that introduce the thioether amino acids lanthionine and methyllanthionine. Formation of thioether bonds yields the highly crosslinked scaffold for the lantibiotic peptides, which are capable of presenting well-crafted pockets to accept lipid molecules.
Nisin
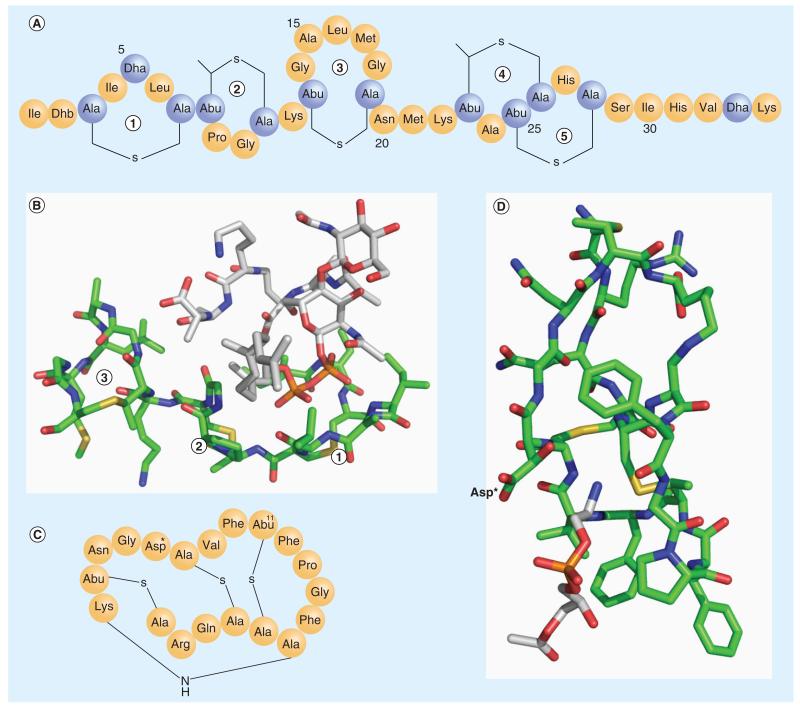
One of the best-characterized lantibiotics is nisin, which has been used commercially as a food preservative for the past several decades. Nisin displays nanomolar antimicrobial activity against a wide range of Gram-positive bacteria, including the methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci [12]. Similar to other lantibiotics, the primary structure of nisin features four thioether bonds and several dehydrated amino acids (Figure 2A). It was observed to cause membrane depolarization of and death of Micrococcus flavus, a Gram-positive bacterium, at low-nanomolar concentrations (minimal inhibitory concentration: 3.3 nM) [13]. This potency is rather impressive given that most antimicrobial peptides are only functional above micromolar concentrations. Although it possesses features commonly observed in pore-forming peptides, such as cationic charge and amphiphilicity, nisin requires the presence of lipid II in membranes in order to achieve its highly potent membrane depolarizing and cell-killing activity [12]. In fact, nisin is in active against vesicles made from the lipid extract of M. flavus, which is lipid II depleted; adding exogenous lipid II restores its nanomolar potency for membrane depolarization, which showcases the exquisite specificity of nisin-lipid II interaction. Furthermore, the membrane activity of nisin is inhibited by vancomycin, which is well known to bind lipid II to block cell wall biosynthesis.
Figure 2. Lipid-targeting lantibiotics.
(A) The primary structure of nisin. (B) The complex structure between nisin and lipid II (PDB: 1WCO). Only the N-terminal fragment (1, 2, 3 rings) of nisin interacts with lipid II. The color scheme used: C of nisin in green, C of lipid II in white, N in blue, O in red, S in yellow, and P in brown. (C) The primary structure of cinnamycin. Asp* represents β-hydroxyaspartate. (D) The NMR structure of the cinnamycin and lyso-phosphatidylethanolamine complex (PDB: 2DDE). The color scheme used: C of cinnamycin in green, C of lyso-phosphatidylethanolamine in white, N in blue, O in red and S in yellow. The lyso-phosphatidylethanolamine headgroup inserts into the hydrophobic pocket of cinnamycin. The key polar residue (Asp*), which interacts with the phosphatidylethanolamine headgroup, is labeled.
Abu: α-aminobutyric acid; Dha: Dehydroalanine; Dhb: Dehydrobutyrine.
Structural analysis with NMR spectroscopy reveals a 1:1 complex of nisin and lipid II (Figure 2B) [14]. As expected, the thioether crosslinks and the presence of dehydroalanines afford a rigid structure for nisin. Interestingly, lipid II forms molecular contacts only with the N-terminal fragment of nisin, binding into the concave surface created by the A, B and C rings. The three rings are each composed of a thioether bond. The A and B rings are bridged by a single amide bond, which is a flat and rigid structure. The B and C rings are connected through a single Lys residue. The complex appears to be primarily stabilized by the pyrophosphate moiety of lipid II binding into the concave face of the A ring, which displays multiple amide NH groups. In addition, the prenyl chain of lipid II forms hydrophobic contacts with Leu6 and Pro9, both residing on the rim of the A ring. The d-Ala-d-Ala motif forms molecular contacts with the C ring through a combination of hydrophobic and hydrogen bonding interactions. This presumably explains the inhibitory effect of nisin membrane activity by vancomycin.
Cinnamycin
The other well-characterized lantibiotic peptide known to target lipids is cinnamycin, which used to be referred to as Ro 09-1098 [15]. Cinnamycin has a closely related sibling duramycin, which is only different by an Arg→Lys mutation at the second position (Figure 2C). These two peptides share essentially identical properties [16]. Cinnamycin and duramycin were found to specifically recognize PE in cell membranes and the PE specificity tolerates conjugation with a variety of labels (biotin, 99mTc, Gd) onto the N-terminus of the peptides [17]. The labeled cinnamycin/duramycin has been frequently used to probe the physiological roles of PE in cell culture and in animal models. For example, PE is known to preferentially locate within the cytosol-facing leaflet of a cell; by using a fluorescently labeled cinnamycin, Emoto et al. reported that PE is externalized transiently and concentrated at the cleavage furrow in diving cells [18,19]. Similarly, targeting PE externalization with labeled cinnamycin also enabled detection of cell apoptosis [17,20] and protein C anticoagulation mechanisms. Recent research indicates that PE is also exposed in tumor vascular endothelium cells, which can serve as a target for tumor imaging or drug delivery [17].
Cinnamycin was found to bind membrane-embedded dialkyl-PE or lyso-PE with low-nanomolar affinities to form equimolar complexes [21,22]. The binding of cinnamycin to PE is highly specific. For example, the dissociation constant (Kd) of cinnamycin for PE is approximately 10 nM, which is in sharp contrast to that for PC (~10 mM) [22]. With a membrane leakage assay, Choung and co-workers demonstrated that cinnamycin caused membrane damage at nanomolar concentrations only when PE is present in the target membranes [23]. Substitution of PE with PS, phosphoinositol (PI) or cardiolipin (CL) completely abolishes the membrane-leakage ability of cinnamycin. Similarly, Zhao et al. demonstrated that duramycin binding into apoptotic Jurkat lymphocytes was inhibited only by PE-containing vesicles [24].
Cinnamycin belongs to the type B lantibiotics, which are globular and even more rigid than type A lantibiotics such as nisin. Cinnamycin is a 19-residue polypeptide and harbors rather exotic amino acids such as β-hydroxyaspartate (Asp*15; Figure 2C) [15]. Similar to nisin, cinnamycin displays multiple (three to be exact) thioether crosslinks. In addition, it displays a rather unique lysinoalanine ring, where the amino group of Lys19 covalently attaches to the β-carbon of Ala6. The highly crosslinked and, therefore, highly rigid conformation preorganizes cinnamycin for PE binding and is presumably responsible for its exclusive selectivity. The structure (Figure 2D) of the equimolar complex of cinnamycin and C12-lyso-PE was resolved in DMSO using NMR spectroscopy [25,26]. The NMR structure shows that the peptide folds into an amphiphilic cylinder with the polar residues on the upper end (Arg2, Gln3 and the C-terminal COOH) and hydrophobic residues on the bottom (Pro9, Phe10, Phe12 and Val13). The amphiphilic structure is expected to facilitate membrane insertion. The rigid fold of cinnamycin hosts a well-crafted cavity at the hydrophobic end of the cylinder. The size of the cavity perfectly matches with the ethanolamine moiety of the PE headgroup and does not even accommodate an additional methyl group on the amine. Interestingly, there is only one polar residue (Asp*15) that interacts with the PE headgroup. However, in this case, the carboxylate group of Asp*15 projects towards the exterior of the binding pocket, away from the ammonium group, and instead its hydroxyl group forms a hydrogen bond with the ammonium ion of PE. Therefore, the PE selectivity against PC originates primarily from two factors: the steric size of the cavity best matches with the PE headgroup and the hydrogen bonding interaction with the Asp*15 cannot be established by the PC headgroup. The NMR structure displays little hydrophobic contact between cinnamycin and the lipid tail of lyso-PE [25]. This, however, does not exclude the contribution of hydrophobic interactions to the membrane association of cinnamycin given the structure is determined in DMSO instead of model membranes. In fact, thermodynamic characterization with high sensitivity isothermal titration calorimetry reveals that the membrane-binding affinity of cinnamycin depends on the fatty acid chain length of PE in model membranes [21]. The C8 diacyl-PE elicits superior binding of cinnamycin (by at least one order of magnitude) than its shorter chain homologues, while extending the lipid chain beyond eight carbons provides little variation for cinnamycin-membrane association propensities. One possible explanation of this intriguing observation is that cinnamycin binds much more strongly to membrane-sequestered PEs than those in aqueous solution, as shortchain lipids partition less well into vesicles. In other words, the hydrophobic interaction between cinnamycin and the membrane contributes significantly to cinnamycin-PE recognition. Unfortunately, as no partition data were reported for those short-chain lipids, the physiochemical basis for the lipid chain length dependence of cinnamycin awaits further analysis. Nevertheless, given essentially all natural PEs have longer lipid chains (more than eight carbons), we anticipate cinnamycin to report on the total PE concentration in biomembranes.
These two examples from nature nicely demonstrate the feasibility of using small peptides to target specific membrane lipids. A shared feature of nisin and cinnamycin is the high degree of preorganization afforded by unnatural amino acids and intramolecular crosslinks. These small crosslinked scaffolds appear to be equally effective acceptors of lipid headgroups as the architecture of large proteins.
Lipid recognition by synthetic molecules
Inspired by the peptide natural products, there has been an increasing interest in developing synthetic receptors for membrane lipids. The efforts towards small-molecule receptors for lipids put our knowledge of peptide-membrane interactions to test, which, in turn, reveals new principles to guide the molecular-design efforts to target a variety of membrane lipids. Within the past decade, much effort has been paid towards targeting PS because of its known significance in cell death and blood coagulation pathways [6], as well as the lack of effective PS markers from natural sources. The following section will summarize recent activities on PS recognition, with an emphasis on the design principles and the chemical basis of lipid recognition.
As the most abundant acidic lipid in mammalian cell membranes, PS is believed to regulate the homeostasis of many proteins by modulating the electrostatic potential of membrane surfaces [27]. PS is also well known to play critical roles in apoptosis and blood coagulation. Similar to PE, the distribution of PS within the two leaflets of a plasma membrane is asymmetric as well. In fact, PS is exclusively confined in the cytosol-facing leaflet for a healthy cell. This asymmetric distribution is actively maintained by the ATP dependent flippase activity of the cell, and is, not surprisingly, disrupted when the cell is triggered to go through apoptosis, the programmed cell death pathway. As PS externalization is considered a hallmark event of apoptosis, cell biologists have been taking advantage of this phenomenon to detect apoptotic cells by imaging PS on surfaces. Another critical process in human physiology that PS actively contributes to is blood coagulation, during which PS is externalized on the surface of platelet cells [28]. Therefore, small molecules that target PS may serve as powerful tools to inhibit blood clotting and to image cell death in live animals, and perhaps ultimately even in patients.
Biomimetic designs for PS recognition
Both rational and screening approaches have been explored to develop small synthetic receptors for PS. Significant success has been achieved by using biomimetic approaches, in which the PS-binding mechanism of natural proteins is recreated by synthetic molecules. Here, several examples to highlight the design principles of lipid recognition uncovered from these studies will be discussed.
Mimicking annexin V
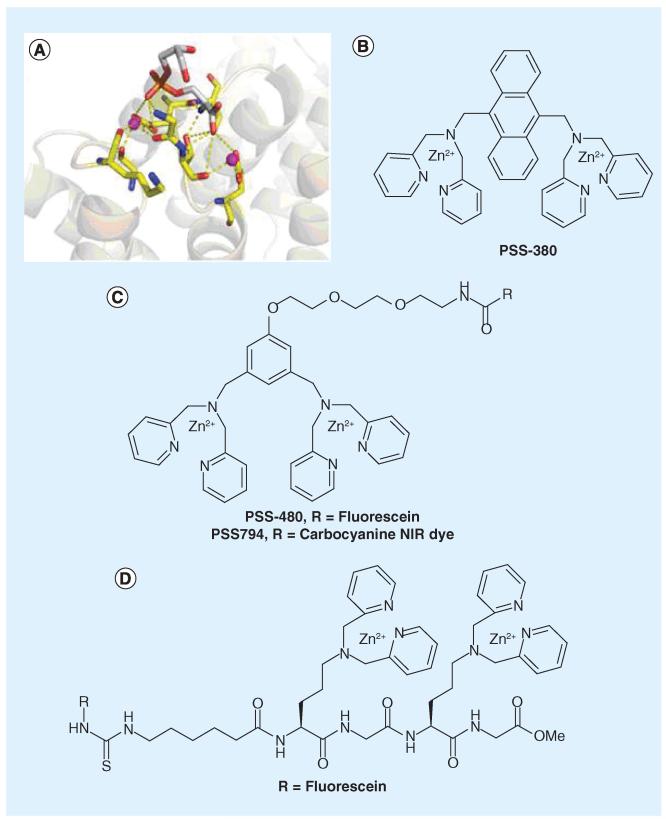
Annexin V is the perhaps the best-known marker of PS. In fact, fluorescently labeled annexin V is routinely used in cell biology experiments to stain apoptotic cells. Annexin V binds to PS-rich membranes with nanomolar affinity (Kd: 0.1-2 nM) [29,30]. The protein backbone carbonyl and side chain carboxylate groups coordinate with a number of Ca2+ ions, which bridge the protein and the PS headgroup [31]. More specifically, each PS headgroup engages with two Ca2+ ions: one interacts with the phosphate group and the other forms a salt bridge with the carboxylate group of the serine moiety (Figure 3A). Despite its frequent use, annexin V is less than ideal as a PS marker for several reasons. First, the unique mechanism of annexin–PS recognition necessarily results in high sensitivity to calcium concentration: annexin–PS binding is completely abolished in calcium free media and achieves maximum affinity at the calcium concentration of approximately 2.5 mM [29]. The Ca2+ dependence makes it hard to quantitatively depict PS distribution in living cells and organisms given the large variation of intracellular and extracellular calcium concentrations needed for Ca2+ signaling. Furthermore, the high calcium concentration needed for optimal annexin–PS binding might unintentionally activate the lipid scramblase activity, which causes lipid flip-flop and disturbs the asymmetric distribution of lipids across the two leaflets [32]. Finally, annexin V has approximately 400 amino acids and a molecular weight of approximately 36 kDa. The large size makes it problematic for imaging cell death in vivo due to poor tissue penetration and slow clearance [8,33,34].
Figure 3. Synthetic molecules mimicking annexin V.
(A) Phosphatidylserine (PS) headgroup by annexin V, highlighting the requirement of two Ca2+ ions, which, respectively, interact with the phosphate and carboxylate groups of PS. The purple spheres represent Ca2+ (PDB:1A8A). (B) PSS-380, the first Zn2+-DPA dimer shown to selectively bind anionic lipids. (C) Improved designs of the Zn2+-DPA dimer as PS-binding molecules. (D) Peptide presentation of the Zn2+-DPA dimer for imaging apoptosis.
Small molecules that mimic the function of annexin V, particularly those that do not require calcium for PS binding, would be highly beneficial to noninvasive imaging of apoptosis. With these considerations in mind, Smith and Hanshaw reported a series of elegantly designed small molecule mimics of annexin V [35]. These molecules display two Zn2+−2,2′-dipicolylamine (Zn2+–DPA, Figure 3B) subunits, which resemble the two calcium centers of the annexin V–PS complex. Originally reported by the Hamachi group for phosphotyrosine recognition [36], the compound PSS-380 is the first of the series to demonstrate selectivity towards apoptotic cells [37], which display PS on their surfaces. Importantly, PSS-380 was observed to compete with annexin V for apoptotic cell binding, suggesting that PS is the molecular target of PSS-380. Furthermore, apoptotic cell labeling by PSS-380 is insensitive to calcium concentration but inhibited by EDTA, which sequesters zinc ions. These observations indicate that PSS-380 mimics the mechanism of annexin to target PS. The design of the next generation places the two Zn2+–DPA moieties on the meta positions of a phenyl ring [38,39]. This design allows facile labeling with a variety of reporter groups including a number of fluorophores and biotin (Figure 3C). Titration experiments with synthetic vesicles demonstrate that these compounds bind to PS-presenting membranes with mid-micromolar affinity (Kd: 20–100 μM), while no binding was observed for vesicles that only contain PC. Further biophysical analysis of these Zn2+–DPA dimers reveals little selectivity against different anionic lipids (e.g., PS, PG, PA) [38], suggesting their membrane association is largely driven by the electrostatic attraction between the cationic zinc complex and the anionic phosphate groups of the lipids. Nevertheless, these Zn2+–DPA dimers were demonstrated to selectively stain apoptotic cells by using fluorescence microscopy and flow cytometry analysis [39]. This is perhaps not surprising given that PS is the most-abundant anionic lipid and no other anionic lipids exist in large quantities on the surface of healthy cells. More recently, the Zn2+–DPA dimer was conjugated to a near infrared dye to give PSS-794 (Figure 3C) [40-42]. Whole-body animal imaging has demonstrated that PSS-794 preferentially accumulates in radiation treated tumors than the untreated controls, although the signal-to-background ratio (~2) is modest.
In addition to the aromatic structures, peptides were demonstrated to present a viable scaffold to display Zn2+–DPA moieties for PS recognition [43]. Specifically, by using a lysine derivative in peptide synthesis, two Zn2+–DPA units are assembled onto a peptide backbone and separated from each other by a glycine residue (Figure 3D). The peptide-based Zn2+–DPA probe displays higher binding affinities to anionic lipids (Kd~20 μM) than to the zwitterionic lipid PC (Kd > 100 μM). These Kd values are very comparable to the afore-mentioned non-peptide based probes. Upon fluorescein labeling, the peptide derivative readily detects camptothecin-induced apoptosis in both Jurkat lymphocytes and HeLa cells.
These annexin mimics capitalize on the high affinity of the Zn2+–DPA dimers to phosphates and nicely circumvent the calcium dependence of lipid binding. It remains to be seen to what extent the requirement of zinc limits their applications in vivo and whether better designs can achieve selectivity within the group of anionic lipids.
Mimicking lactadherin
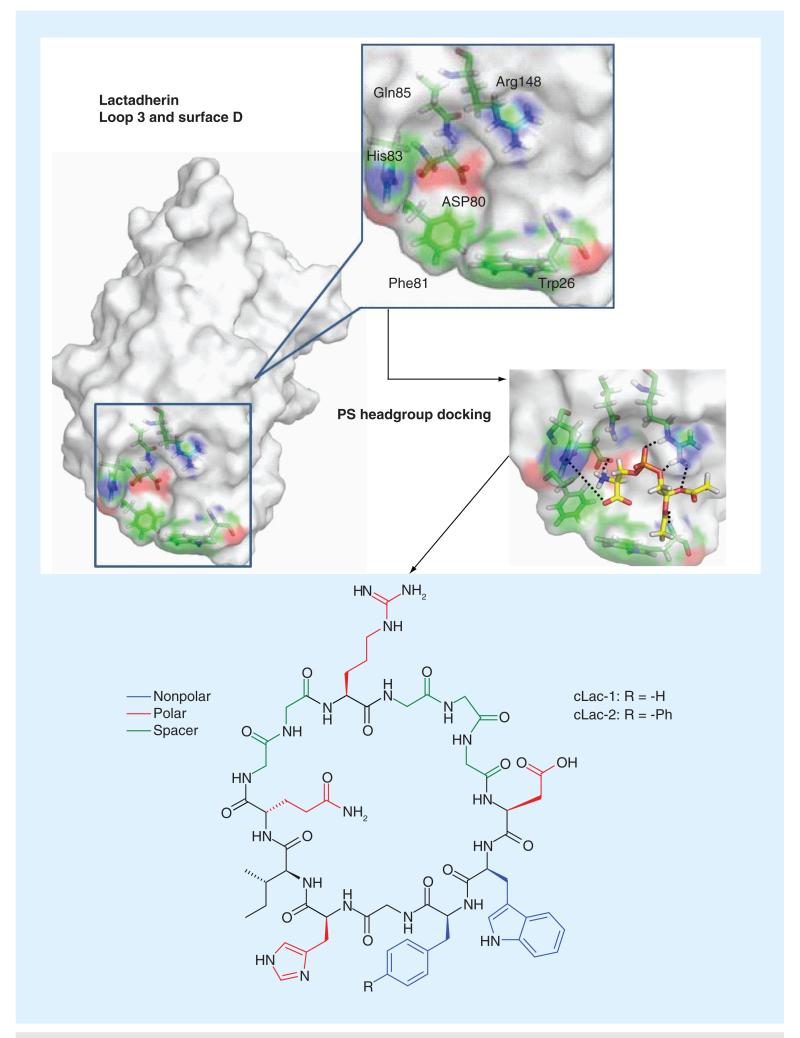
As one of the lipid-binding proteins, lactad-herin displays exquisite selectivity for PS and importantly it does not require any cofactors (such as calcium for annexin V) for PS recognition [44]. Lactadherin was originally identified in milk and later found to be secreted by macrophages. One of its native functions is to seek out PS-presenting cells, presumably apoptotic cells, and deliver them to macrophages for clearance [45]. Given this critical physiological role, it is perhaps expected that lactadherin recognizes PS with superb potency and specificity: it displays low nanomolar affinities to membranes with only 1% phosphatidyl-l-serine, the natural PS, yet does not bind to membranes with phosphatidyl-d-serine at all [44]. Specific recognition of PS is achieved through its C2 domain (Lact-C2). Lact-C2 has approximately 150 amino acids and its structure was resolved through x-ray crystallography [46,47]. Although there is no co-crystal structure of Lact-C2 and PS to date, molecular docking experiments have revealed a plausible binding site (Figure 4), which gives a binding free energy of −14.7 kcal/mol for diacetyl-PS. In contrast, the binding free energy for diacetyl-PC, −7.8 kcal/mol, is significantly smaller [48]. The energy-minimized structure of diacetyl-PS docking into Lact-C2 illustrates key interactions: Arg148 forms a salt bridge with the phosphate group of PS; Asp80 interacts with the PS carboxylate group via another salt bridge; and His83 possibly interacts with the serine ammonium moiety through hydrogen bonding. Importantly, there are two hydrophobic aromatic residues (Phe81 and Trp26) at the very bottom of Lact-C2, which presumably serve as membrane anchors to favor the insertion of the protein into lipid bilayers. Interestingly, this membrane insertion motif of Lact-C2 resembles the aromatic cluster of cinnamycin (Figure 2D) and is consistent with the fact that aromatic residues are enriched in the membrane-water interfacial region in membrane protein structures [49].
Figure 4. Mimicking the phosphatidylserine-binding site of lactadherin (PDB: 3BN6).
Computational modeling yields a plausible mechanism of PS binding into lactadherin, enabling the design of cyclic peptides to mimic lactadherin for phosphatidylserine recognition. PS: Phosphatidylserine.
Based on these docking experiments, it is hypothesized that this PS-binding pocket could be recreated by using a cyclic peptide. Most of the key residues responsible for PS binding are contiguous in the primary sequence of Lact-C2 except Trp26 and Arg148. To best mimic their spatial arrangement in the Lact-C2 structure, the authors positioned the Trp residue at the N-terminus of Phe81 to construct a membrane insertion motif and inserted the Arg residue in between Gln85 and the Trp residue (Figure 4) [48]. Molecular modeling suggested a minimum length of diglycine and triglycine linkages respectively on the N- and C-terminus of the Arg residue. These considerations led the design of cLac-1 as represented in Figure 4. However, when examined against vesicles of varied compositions, cLac-1 demonstrates no membrane insertion regardless of the presence of PS in the vesicles or not, suggesting that the combination of Trp and Phe is not sufficient to drive the partition of the peptide into membranes. It is likely that additional hydrophobic residues of Lact-C2 assist in its insertion into lipid bilayers. Further exploration of the membrane insertion motif resulted in cLac-2, which carries a biphenyl side chain in replacement of the Phe. The combination of biphenyl side chain and Trp appears to exhibit optimal hydrophobicity: cLac-2 does not partition into PC-only vesicles, but inserts readily into PS-containing vesicles. Titration experiments give a Kd of approximately 8 μM for cLac-2 binding into vesicles with 20% PS. Under the same conditions, the linear precursor peptide fails to bind any type of vesicles, showcasing the benefit of preorganized scaffolds as lipid receptors. The PS-binding affinity of cLac-2 is significantly lower than that of Lact-C2, suggesting that additional elements of Lact-C2 may be important for membrane insertion. An alternative explanation could be that the cLac peptide is more dynamic than the parent protein and, thus, is less well preorganized for lipid binding.
When subjected to fluorescence microscopy analysis, a fluorophore-labeled cLac-2 effectively differentiated apoptotic and healthy cells, presumably by targeting the surface-exposed PS. Furthermore, flow cytometry analysis demonstrated that cLac-2 identified the same percentages of cells that were undergoing apoptosis as annexin V, the current standard of apoptosis detection. It is suspected that the PS-binding mechanism of cLac-2 resembles that of Lact-C2, which, nevertheless, remains to be confirmed by structural characterizations of the cLac-2-PS complex.
Mimicking PS decarboxylase
Another example of functionally mimicking PS-binding proteins by peptides originates from the work of Iagarashi et al. in 1995 [50]. It was then proposed that the 12-amino acid consensus sequence shared by PKC and PSD is responsible for PS recognition. Indeed, a 14-residue synthetic peptide (FNFRLKAGQKIRFG) derived from the corresponding region of PSD was reported to effectively bind PS but not PC, PE, PI and CL. Even though this peptide has multiple cationic residues, the selectivity for PS over PI (both are anionic lipids) indicates that the peptide does not target PS purely through electrostatic mechanisms. Similarly the corresponding peptide (FVF-NLKPGDVERRL) from PKC was also reported to bind PS selectively, although with somewhat lower affinity. Recently, by using surface plasma resonance biosensor analysis, Li and co-workers quantitatively assessed the PS-binding affinity of the PSD peptide (known as PSBP-0) to give a Kd value of 1.38 μM [51]. Alanine scanning of the PSBP-0 sequence resulted in the Gln→Ala mutant (PSBPP-6: FNFRLKAGAKIRFG) that improves the PS-binding affinity by more than tenfold to give a Kd of 100 nM. Conjugation of a rhenium complex to the N-terminus of the PBSP-6 improved the binding affinity even further to 16 nM. Fluorescently labeled PSBP-6 was found to co-localize with annexin V on the surface of apoptotic cells. A radioactively labeled PSBP-6 (with 99mTc) was tested in mice that carry xenografted melanoma. When the animals were treated with paclitaxel, the radioactive peptide accumulated in the tumor tissue to a larger density than the untreated controls.
It is intriguing to ask the question whether PSBP-6 can be even further improved for PS imaging by strategies such as cyclization. Structural characterization of the peptide-PS complex will greatly facilitate efforts towards this goal. To date there is no structural information of these peptides that has yet been reported; a Protein Data Bank search gives no structures of PS decarboxylase. The homologous sequence in PKCα (PDB: 1DSY) displays a strand-helix-strand hairpin motif, which surprisingly resides on the opposite end of the β-sandwich protein from its PS-binding site. It is likely that the PBSP peptides fold into completely different structures for PS binding.
Combinatorial approaches to PS recognition
In addition to the biomimetic designs, combinatorial screening of peptide libraries has also been successfully applied to develop small-molecule receptors for PS. For example, Shao et al. screened a random 12-mer peptide library displayed on phage particles against liposomes prepared from PS and identified the PS3–10 peptide with the sequence of SVS-VGMKPSPRP [52]. PS3–10 was found to bind PS approximately four-times more effectively than PC. Furthermore, the PS3–10 phage was demonstrated to selectively associate with apoptotic cells and aged red blood cells, which are known to have surface exposed PS. Similarly, by screening a linear 6-mer peptide library, Burtea and co-workers discovered positive leads with sequences of PGDLSR and LIKKPF, which prefer binding to PS over PC by two-three-fold [53]. Interestingly, both peptides were found to compete with annexin V. Furthermore, a nine-residue cyclic peptide library displayed on M13 phage was screened for PS binding as well, from which a peptide with the sequence of CLSYY-PSYC was identified as a novel PS-recognizing moiety [54]. FITC-labeled CLSYYPSYC was reported to selectively stain apoptotic cells in vitro and accumulated in xenografted tumor tissues in mice upon treatments with anti-tumor drugs. While these peptides have demonstrated potential as medical imaging agents, mechanistic study remains to be done, perhaps through mutation studies or structural characterizations, to understand the chemical basis of their preference for PS.
Curvature sensing by synthetic molecules
In addition to specific signaling lipids, such as PS, membrane curvature is attracting increasing attention in medicinal chemistry as well. It has been long known that highly curved membranes are involved in membrane remodeling during cell growth, division and movement [55]. Membrane curvature is believed to alter the affinity of a battery of peripheral membrane proteins [56]. It is also conceivable that the function of transmembrane channels and transporters can be modulated by membrane curvature. More recently, negative membrane curvature of bacterial cells, resulting from the presence of PE and CL, has been reported to contribute to the selective toxicity of antimicrobial peptide mimetics [57]. On a different front, as a potentially powerful way of disease diagnosis, much attention has been paid to the detection and analysis of highly curved microvesicles that shed from tumor cells [58].
Analogous to the cLac peptide design discussed earlier, the biomimetic approach has recently been applied to membrane curvature sensing [59]. A variety of proteins have been found to selectively target regions of high curvature on membranes, with the C2B domain of synaptotagmin-I (Sty1-C2B) a prominent example. Sty1 is a critical component of the SNARE complex and is believed to mediate the calcium-dependent membrane trafficking and fusion. It exhibits much greater affinity to small and highly curved vesicles than to the larger ones [60]. Using the loop 3 of Sty1-C2B as a template, Yin and co-workers designed and synthesized a 12-residue cyclic peptide C2BL3C via the ‘click chemistry’ enabled by unnatural amino acids carrying azide and alkyne functionalities. C2BL3C displays more favorable binding to highly curved surfaces, with an Kd value of approximately 0.5 mM to liposomes of 30 nm diameters, but relatively weak binding to lipid vesicles of 100 nm (Kd > 1 mM). Although the selectivity is somewhat modest, the affinity of the peptide to high curvature membrane is cyclization dependent and sequence dependent: both the linear precursor and the control peptide with a scrambled sequence demonstrate reduced binding to 30-nm vesicle. Similar to the lactadherin mimic cLac-2, the membrane-binding affinity of C2BL3C is noticeably weaker than the parent protein. Regardless, the peptide C3BL3C was reported to preferentially bind to blood-borne microvesicles (d < 100 nm) in rat plasma samples, suggesting its affinity and specificity to curved membranes, although less than optimal, makes it potentially useful as molecular probes.
Although difficult to prove structurally, the authors propose that the peptide C2BL3C selects curved membranes due to its preferential insertion into lipid-packing defects. This study demonstrates the promise of using short peptides for curvature sensing. It also lends further support to the protein-mimetic approach to develop small-molecule probes of biomembranes. Indeed, more recently, the Yin group developed another high curvature binding peptide by modeling the protein MARCKS-ED, which is known to favor highly curved membrane surfaces [61].
Conclusion & future perspective
The diversity of membrane lipids, as well as their highly dynamic distribution, urgently calls for molecular tools that can trace each individual lipid in complex biological systems. As molecular probes in biology, peptides present intrinsic advantages over proteins in terms of ease of synthesis and labeling. Their small size may offer favorable pharmacokinetic properties over large proteins. These advantages fuel the intensifying efforts to develop peptide-based markers for membrane lipids. Examples from nature, and more recently from the peptide design efforts, have showcased the feasibility and potential of using peptides for specific lipid recognition, which nature usually accomplishes by using large proteins. The peptide design efforts within the past decade have concentrated on PS partly due to the potential of PS-targeting molecules to detect apoptotic cell death via the surface-exposed PS. Both biomimetic and combinational approaches have been explored with success, leading to peptide-based probes that allow for noninvasive imaging of cell death in animal models.
As pointed out throughout this article, a bottleneck of the field lies in the difficulty to obtain structural information of the membrane-bound peptides. The rapid developments in membrane-protein structural biology, enabled by solid-state NMR and crystallography, will surely catalyze activities towards peptide-based probes for membrane recognition. Although somewhat empirical, biochemical characterizations of these lipid-binding peptides, through peptide mutation and lipid selectivity studies, have, nevertheless, provided valuable insights into the chemical basis of lipid recognition by small peptides. For example, preorganization of a peptide scaffold (by cyclization and crosslinks) appears to be highly advantageous for membrane binding. On the other hand, tuning the balance between hydrophobicity and electrostatic interactions helps to achieve lipid-dependent membrane insertion of peptides. This new knowledge will serve as guidelines to speed up future efforts to develop small-molecule probes for a variety of other membrane lipids, including PI and its phosphorylated forms. Finally, it is worth noting that all work discussed above addresses lipid headgroup subtypes. Differentiating lipids according to their hydrophobic tails remains an essentially untapped area, which presents a challenge, as well as a huge opportunity for chemical biologists.
Executive summary.
-
■
Low-molecular-weight probes of membrane lipids can be developed by proper engineering of short peptides, which display protein-like affinities and specificities for lipid binding.
-
■
Targeting specific signaling lipids such as phosphatidylserine allows in vivo imaging of cell death, which is highly desirable for disease diagnosis and quick evaluation of cancer therapeutics.
-
■
Synthetic peptides are demonstrating promise to sense highly curved membranes.
-
■
Biomimetic approaches (modeling lipid-binding proteins) have been particularly powerful in creating lipid-targeting peptides.
-
■
Rigidification of the peptide scaffold and balancing polar/nonpolar interactions are essential in achieving high affinity and specificity for lipid recognition.
Acknowledgments
Financial & competing interests disclosure
The authors wish to thank Boston College and the NIH (R01 GM102735 to J Gao) for financial support of our research. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Key Terms
- Lipidome
Refers to the complete collection of lipids in a biological system.
- Phosphatidylserine
Minority membrane lipid that exclusively resides in the inner leaflet of a healthy cell. Externalization of phosphatidylserine is a critical step in cell apoptosis and blood coagulation.
- Membrane curvature
Relates to various aspect of membrane remodeling. Its interplay with various proteins regulates key biological processes including cell division and cell–cell communication.
- Lantibiotics
Class of peptide natural products displaying characteristic thioether crosslinks that afford highly rigid structures.
- Apoptosis
Major mechanism of programmed cell death, accurate detection of which is desirable in disease diagnosis and evaluation of cancer therapeutics.
References
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9(2):112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2■.Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 2010;11(8):593–598. doi: 10.1038/nrm2934. Provides a perspective on lipid diversity.
- 3.Wenk MR. Lipidomics: new tools and applications. Cell. 2010;143(6):888–895. doi: 10.1016/j.cell.2010.11.033. [DOI] [PubMed] [Google Scholar]
- 4.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327(5961):46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 5.Balasubramanian K, Schroit AJ. Aminophospholipid asymmetry: a matter of life and death. Annu. Rev. Physiol. 2003;65:701–734. doi: 10.1146/annurev.physiol.65.092101.142459. [DOI] [PubMed] [Google Scholar]
- 6.Schutters K, Reutelingsperger C. Phosphatidylserine targeting for diagnosis and treatment of human diseases. Apoptosis. 2010;15(9):1072–1082. doi: 10.1007/s10495-010-0503-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7■■.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008;9(2):99–111. doi: 10.1038/nrm2328. Describes mechanisms of lipid recognition by proteins.
- 8.Niu G, Chen X. Apoptosis imaging: beyond annexin V. J. Nucl. Med. 2010;51(11):1659–1662. doi: 10.2967/jnumed.110.078584. [DOI] [PubMed] [Google Scholar]
- 9.Lambert TN, Smith BD. Synthetic receptors for phospholipid headgroups. Coordination Chem. Rev. 2003;240(1-2):129–141. [Google Scholar]
- 10.Tseng CY, Ashrafuzzaman M, Mane JY, Kapty J, Mercer JR, Tuszynski JA. Entropic fragment-based approach to aptamer design. Chem. Biol. Drug Design. 2011;78(1):1–13. doi: 10.1111/j.1747-0285.2011.01125.x. [DOI] [PubMed] [Google Scholar]
- 11.Willey JM, Van Der Donk WA. Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 12.Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat. Rev. 2005;3(10):777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 13.Breukink E, Wiedemann I, Van Kraaij C, Kuipers OP, Sahl H, De Kruijff B. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science. 1999;286(5448):2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 14■■.Hsu ST, Breukink E, Tischenko E, et al. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 2004;11(10):963–967. doi: 10.1038/nsmb830. Describes the structural basis of lipid II recognition by nisin.
- 15.Kessler H, Steuernagel S, Will M, et al. The structure of the polycyclic nonadecapeptide Ro-09-0198. Helv. Chim. Acta. 1988;71(8):1924–1929. [Google Scholar]
- 16.Navarro J, Chabot J, Sherrill K, Aneja R, Zahler SA, Racker E. Interaction of duramycin with artificial and natural membranes. Biochemistry. 1985;24(17):4645–4650. doi: 10.1021/bi00338a025. [DOI] [PubMed] [Google Scholar]
- 17.Zhao M. Lantibiotics as probes for phosphatidylethanolamine. Amino Acids. 2011;41(5):1071–1079. doi: 10.1007/s00726-009-0386-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18■.Emoto K, Kobayashi T, Yamaji A, et al. Redistribution of phosphatidylethanolamine at the cleavage furrow of dividing cells during cytokinesis. Proc. Natl. Acad. Sci. USA. 1996;93(23):12867–12872. doi: 10.1073/pnas.93.23.12867. Illustrates the power of small-molecule probes for studying lipid homeostasis.
- 19.Emoto K, Umeda M. An essential role for a membrane lipid in cytokinesis. Regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine. J. Cell. Biol. 2000;149(6):1215–1224. doi: 10.1083/jcb.149.6.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maulik N, Kagan VE, Tyurin VA, Das DK. Redistribution of phosphatidylethanolamine and phosphatidylserine precedes reperfusion-induced apoptosis. Am. J. Physiol. 1998;274(1 Pt 2):H242–H248. doi: 10.1152/ajpheart.1998.274.1.H242. [DOI] [PubMed] [Google Scholar]
- 21.Machaidze G, Ziegler A, Seelig J. Specific binding of Ro 09-0198 (cinnamycin) to phosphatidylethanolamine: a thermodynamic analysis. Biochemistry. 2002;41(6):1965–1971. doi: 10.1021/bi015841c. [DOI] [PubMed] [Google Scholar]
- 22.Machaidze G, Seelig J. Specific binding of cinnamycin (Ro 09-0198) to phosphatidylethanolamine. Comparison between micellar and membrane environments. Biochemistry. 2003;42(43):12570–12576. doi: 10.1021/bi035225b. [DOI] [PubMed] [Google Scholar]
- 23.Choung SY, Kobayashi T, Takemoto K, Ishitsuka H, Inoue K. Interaction of a cyclic peptide, Ro09-0198, with phosphatidylethanolamine in liposomal membranes. Biochim. Biophys. Acta. 1988;940(2):180–187. doi: 10.1016/0005-2736(88)90193-9. [DOI] [PubMed] [Google Scholar]
- 24.Zhao M, Li Z, Bugenhagen S. 99mTc-labeled duramycin as a novel phosphatidylethanolamine-binding molecular probe. J. Nucl. Med. 2008;49(8):1345–1352. doi: 10.2967/jnumed.107.048603. [DOI] [PubMed] [Google Scholar]
- 25.Wakamatsu K, Choung SY, Kobayashi T, Inoue K, Higashijima T, Miyazawa T. Complex formation of peptide antibiotic Ro09-0198 with lysophosphatidylethanolamine: 1H NMR analyses in dimethyl sulfoxide solution. Biochemistry. 1990;29(1):113–118. doi: 10.1021/bi00453a013. [DOI] [PubMed] [Google Scholar]
- 26■■.Hosoda K, Ohya M, Kohno T, Maeda T, Endo S, Wakamatsu K. Structure determination of an immunopotentiator peptide, cinnamycin, complexed with lysophosphatidylethanolamine by 1H-NMR1. J. Biochem. 1996;119(2):226–230. doi: 10.1093/oxfordjournals.jbchem.a021226. Describes the structural basis of cinnamycin-phosphatidylethanolamine selectivity.
- 27.Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319(5860):210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 28.Shi J, Pipe SW, Rasmussen JT, Heegaard CW, Gilbert GE. Lactadherin blocks thrombosis and hemostasis in vivo: correlation with platelet phosphatidylserine exposure. J. Thromb. Haemost. 2008;6(7):1167–1174. doi: 10.1111/j.1538-7836.2008.03010.x. [DOI] [PubMed] [Google Scholar]
- 29.Meers P, Mealy T. Calcium-dependent annexin V binding to phospholipids: stoichiometry, specificity, and the role of negative charge. Biochemistry. 1993;32(43):11711–11721. doi: 10.1021/bi00094a030. [DOI] [PubMed] [Google Scholar]
- 30.Meers P, Mealy T. Relationship between annexin V tryptophan exposure, calcium, and phospholipid binding. Biochemistry. 1993;32(20):5411–5418. doi: 10.1021/bi00071a016. [DOI] [PubMed] [Google Scholar]
- 31.Swairjo MA, Concha NO, Kaetzel MA, Dedman JR, Seaton BA. Ca(2+)-bridging mechanism and phospholipid head group recognition in the membrane-binding protein annexin V. Nat. Struct. Biol. 1995;2(11):968–974. doi: 10.1038/nsb1195-968. [DOI] [PubMed] [Google Scholar]
- 32.Kamp D, Sieberg T, Haest CW. Inhibition and stimulation of phospholipid scrambling activity. Consequences for lipid asymmetry, echinocytosis, and microvesiculation of erythrocytes. Biochemistry. 2001;40(31):9438–9446. doi: 10.1021/bi0107492. [DOI] [PubMed] [Google Scholar]
- 33.Blankenberg FG, Katsikis PD, Tait JF, et al. In vivo detection and imaging of phosphatidylserine expression during programmed cell death. Proc. Natl. Acad. Sci. USA. 1998;95(11):6349–6354. doi: 10.1073/pnas.95.11.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ntziachristos V, Schellenberger EA, Ripoll J, et al. Visualization of antitumor treatment by means of fluorescence molecular tomography with an annexin V-Cy5.5 conjugate. Proc. Natl. Acad. Sci. USA. 2004;101(33):12294–12299. doi: 10.1073/pnas.0401137101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanshaw RG, Smith BD. New reagents for phosphatidylserine recognition and detection of apoptosis. Bioorg. Med. Chem. 2005;13(17):5035–5042. doi: 10.1016/j.bmc.2005.04.071. [DOI] [PubMed] [Google Scholar]
- 36.Ojida A, Oka Y, Mito-Inoue MA, Hamachi I. First artificial receptors and chemosensors toward phosphorylated peptide in aqueous solution. J. Am. Chem. Soc. 2002;124(22):6256–6258. doi: 10.1021/ja025761b. [DOI] [PubMed] [Google Scholar]
- 37■■.Koulov AV, Stucker KA, Lakshmi C, Robinson JP, Smith BD. Detection of apoptotic cells using a synthetic fluorescent sensor for membrane surfaces that contain phosphatidylserine. Cell. Death Differ. 2003;10(12):1357–1359. doi: 10.1038/sj.cdd.4401315. Describes the small-molecule mimic of annexin V for the first time.
- 38.Lakshmi C, Hanshaw RG, Smith BD. Fluorophore-linked zinc(II)dipicolylamine coordination complexes as sensors for phosphatidylserine-containing membranes. Tetrahedron. 2004;60(49):11307–11315. [Google Scholar]
- 39.Hanshaw RG, Lakshmi C, Lambert TN, Johnson JR, Smith BD. Fluorescent detection of apoptotic cells by using zinc coordination complexes with a selective affinity for membrane surfaces enriched with phosphatidylserine. ChemBioChem. 2005;6(12):2214–2220. doi: 10.1002/cbic.200500149. [DOI] [PubMed] [Google Scholar]
- 40.Smith BA, Akers WJ, Leevy WM, et al. Optical imaging of mammary and prostate tumors in living animals using a synthetic near infrared zinc(II)-dipicolylamine probe for anionic cell surfaces. J. Am. Chem. Soc. 2010;132(1):67–69. doi: 10.1021/ja908467y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith BA, Xiao S, Wolter W, Wheeler J, Suckow MA, Smith BD. In vivo targeting of cell death using a synthetic fluorescent molecular probe. Apoptosis. 2011;16(7):722–731. doi: 10.1007/s10495-011-0601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith BA, Gammon ST, Xiao S, et al. In vivo optical imaging of acute cell death using a near-infrared fluorescent zinc-dipicolylamine probe. Mol. Pharm. 2011;8(2):583–590. doi: 10.1021/mp100395u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Divittorio KM, Johnson JR, Johansson E, Reynolds AJ, Jolliffe KA, Smith BD. Synthetic peptides with selective affinity for apoptotic cells. Org. Biomol. Chem. 2006;4(10):1966–1976. doi: 10.1039/b514748d. [DOI] [PubMed] [Google Scholar]
- 44.Shi J, Heegaard CW, Rasmussen JT, Gilbert GE. Lactadherin binds selectively to membranes containing phosphatidyl-l-serine and increased curvature. Biochim. Biophys. Acta. 2004;1667(1):82–90. doi: 10.1016/j.bbamem.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 45.Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S. Identification of a factor that links apoptotic cells to phagocytes. Nature. 2002;417(6885):182–187. doi: 10.1038/417182a. [DOI] [PubMed] [Google Scholar]
- 46.Lin L, Huai Q, Huang M, Furie B, Furie BC. Crystal structure of the bovine lactadherin C2 domain, a membrane binding motif, shows similarity to the C2 domains of factor V and factor VIII. J. Mol. Biol. 2007;371(3):717–724. doi: 10.1016/j.jmb.2007.05.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47■.Shao C, Novakovic VA, Head JF, Seaton BA, Gilbert GE. Crystal structure of lactadherin C2 domain at 1.7A resolution with mutational and computational analyses of its membrane-binding motif. J. Biol. Chem. 2008;283(11):7230–7241. doi: 10.1074/jbc.M705195200. Describes the structural basis of phosphatidylserine specificity of lactadherin.
- 48 ■■.Zheng H, Wang F, Wang Q, Gao J. Cofactor-free detection of phosphatidylserine with cyclic peptides mimicking lactadherin. J. Am. Chem. Soc. 2011;133(39):15280–15283. doi: 10.1021/ja205911n. Reports the successful development of phosphatidylserine-targeting peptide mimicking lactadherin.
- 49.Killian JA, von Heijne G. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 2000;25(9):429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- 50.Igarashi K, Kaneda M, Yamaji A, et al. A novel phosphatidylserine-binding peptide motif defined by an anti-idiotypic monoclonal antibody. Localization of phosphatidylserine-specific binding sites on protein kinase C and phosphatidylserine decarboxylase. J. Biol. Chem. 1995;270(49):29075–29078. doi: 10.1074/jbc.270.49.29075. [DOI] [PubMed] [Google Scholar]
- 51■.Xiong C, Brewer K, Song S, et al. Peptide-based imaging agents targeting phosphatidylserine for the detection of apoptosis. J. Med. Chem. 2011;54(6):1825–1835. doi: 10.1021/jm101477d. Describes the optimization of a low-affinity probe for in vivo imaging of apoptosis.
- 52.Shao R, Xiong C, Wen X, Gelovani JG, Li C. Targeting phosphatidylserine on apoptotic cells with phages and peptides selected from a bacteriophage display library. Mol. Imaging. 2007;6(6):417–426. [PubMed] [Google Scholar]
- 53.Burtea C, Laurent S, Lancelot E, et al. Peptidic targeting of phosphatidylserine for the MRI detection of apoptosis in atherosclerotic plaques. Mol. Pharm. 2009;6(6):1903–1919. doi: 10.1021/mp900106m. [DOI] [PubMed] [Google Scholar]
- 54.Thapa N, Kim S, So IS, et al. Discovery of a phosphatidylserine-recognizing peptide and its utility in molecular imaging of tumour apoptosis. J. Cell. Mol. Med. 2008;12(5A):1649–1660. doi: 10.1111/j.1582-4934.2008.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mcmahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438(7068):590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 56.Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nature Struct. Mol. Biol. 2007;14(2):138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 57.Yang L, Gordon VD, Trinkle DR, et al. Mechanism of a prototypical synthetic membrane-active antimicrobial: efficient hole-punching via interaction with negative intrinsic curvature lipids. Proc. Natl Acad. Sci. USA. 2008;105(52):20595–20600. doi: 10.1073/pnas.0806456105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balaj L, Lessard R, Dai L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59■■.Saludes JP, Morton LA, Ghosh N, et al. Detection of highly curved membrane surfaces using a cyclic peptide derived from synaptotagmin-I. ACS Chem. Biol. 2012;7(10):1629–1635. doi: 10.1021/cb3002705. Reports the first biomimetic design of curvature-sensing peptides.
- 60.Hui E, Johnson CP, Yao J, Dunning FM, Chapman ER. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca(2+)-regulated fusion. Cell. 2009;138(4):709–721. doi: 10.1016/j.cell.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morton LA, Yang H, Saludes JP, et al. MARCKS-ED peptide as a curvature and lipid sensor. ACS Chem. Biol. 2013;8(1):218–225. doi: 10.1021/cb300429e. [DOI] [PMC free article] [PubMed] [Google Scholar]