Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression post-transcriptionally. Previous studies, which characterized miRNA function, revealed their involvement in fundamental biological processes. Importantly, miRNA expression is deregulated in many human diseases. Specific inhibition of miRNAs using chemically modified anti-miRNA oligonucleotides (AMOs) can be a potential therapeutic strategy for diseases in which a specific miRNA is overexpressed. 2′-O-Methyl (2′-OMe)-4′-thioRNA is a hybrid type of chemically modified oligonucleotide, exhibiting high binding affinity to complementary RNAs and high resistance to nuclease degradation. Here, we evaluate 2′-OMe-4′-thioribonucleosides for chemical modification on AMOs. Optimization of the modification pattern using a variety of chemically modified AMOs that are perfectly complementary to mature miR-21 revealed that the uniformly 2′-OMe-4′-thioribonucleoside–modified AMO was most potent. Further investigation showed that phosphorothioate modification contributed to long-term miR-122 inhibition by the 2′-OMe-4′-thioribonucleoside–modified AMO. Moreover, systemically administrated AMOs to mouse using a liposomal delivery system, YSK05-MEND, showed delivery to the liver and efficient inhibition of miR-122 activity at a low dose in vivo.
INTRODUCTION
MicroRNAs (miRNAs) are a class of endogenously expressed small noncoding RNAs (18 ∼ 25 nt), which regulate gene expression post-transcriptionally. In animals, single-stranded mature miRNAs hybridize to the 3′ untranslated region (3′UTR) of the target mRNA through complete base paring with positions 2–8 of the miRNA, known as the seed region. Binding on the seed region nucleates miRNA–mRNA association, and causes translational inhibition or mRNA degradation (1,2). Because complementarity of the seed region consists of only 7 nt, a single miRNA may regulate multiple genes, and a single mRNA can be modulated by several different miRNAs (3–5). To date, >1000 miRNAs have been identified in humans and regulate up to 60% of protein coding genes (6,7). MiRNAs are implicated in important functions in the biological process, including cell differentiation, proliferation, development, metabolism and apoptosis. Furthermore, up- or downregulation of miRNA expression is correlated with a variety of human diseases such as cancer, viral infection and cardiovascular disorders (8). Thus, regulation of specific miRNA function is a promising therapeutic strategy for treatment of such diseases.
Among the approaches to modulate the function of miRNAs, anti-miRNA oligonucleotide (AMO)-based inhibition has been most widely used not only to exploit the biological function of miRNAs but also as candidates for therapeutic agents (9). To develop oligonucleotide (ON)-based therapeutic strategies, there are several issues to overcome, namely poor stability of ONs in biological fluids, weak binding affinity to target RNA, poor cellular uptake and unfavorable immunostimulatory activity. Thus far, a wide variety of chemically modified ONs have been developed to date, including 2′-O-methyl (2′-OMe) (10–13), 2′-O-methoxyethyl (14) and locked nucleic acid (LNA) to overcome these disadvantages (15–17). These chemical modifications have successfully been applied to an antisense technology as well as ON-based therapeutic technologies (e.g. siRNAs, aptamers, ribozymes). Two successful reports of 2′-OMe–modified AMOs were reported in 2004 (18,19), lending credence to the idea of using chemically modified AMOs. Furthermore, considerable efforts in optimization have been dedicated to develop AMOs as a new therapeutic agent, and several chemically modified AMOs are currently undergoing clinical trials (20). Therefore, further optimization of chemical modifications for AMO-based miRNA suppression will continue to improve this therapeutic approach.
We developed a novel chemically modified ON, 2′-OMe-4′-thioRNA (21) (Figure 1), which can be considered a hybrid chemical modification based on 2′-OMe RNA (13) and 4′-thioRNA (22–28). In our previous study, we reported the development of 2′-OMe-4′-thioribonucleoside–modified siRNA (29). Optimization of both the number and position of modification by 2′-OMe-4′-thioribonucleoside afforded modified siRNAs with more potent and persistent RNAi activity. In addition, investigation of the duration of RNAi activity resulted in long-lasting gene silencing in vitro owing to the improved intracellular stability of 2′-OMe-4′-thioribonucleoside–modified siRNA. Like many other chemically modified ONs that can successfully be applied to AMO as well as siRNA, we expected that 2′-OMe-4′-thioribonucleoside modification acts as a promising AMO. Therefore, we set out to evaluate the utility of 2′-OMe-4′-thioribonucleoside for chemical modification on AMOs. In this study, we investigated the modification pattern of AMOs by 2′-OMe-4-thioribonucleoside in terms of potency and duration of activity in two kinds of target miRNA (miR-21 and miR-122) in vitro. Moreover, systemically administrated AMOs to mouse liver using a liposomal delivery system, a multifunctional envelope-type nano device (MEND) with a pH-sensitive cationic lipid, YSK05 (YSK05-MEND) (30), showed efficient inhibition of miR-122 activity at a low dose in vivo, implicating potential use of 2′-OMe-4′-thioribonucleoside–modified AMO in nucleic acid therapy.
Figure 1.
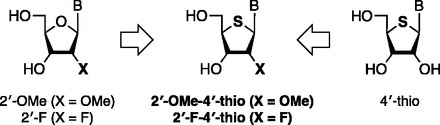
Structure of 2′-modified-4′-thioribonucleoside.
MATERIALS AND METHODS
Oligonucleotides
The chemically modified AMOs used in this study were synthesized on an Applied Biosystem 3400 DNA synthesizer according to our previous report (21). Thus, support bound chemically modified AMOs were synthesized using the corresponding phosphoramidite units at a 1.0 µmol scale following the standard procedure described for oligoribonucleotides. Each of the phosphoramidite units was used at a concentration of 0.1 M in dry acetonitrile, and the coupling time was extended to 10 min for each step. AMOs with phosphorothioate (PS) backbone were achieved by oxidation with 3H-1,2-benzodithiol-3-one-1,1-dioxide (Beaucage reagent) during ON synthesis. After completion of the synthesis, the CPG support was treated with concentrated NH4OH or NH4OH/EtOH (3:1) at 55°C for 16 h. In the case of CPG supports containing either 2′-F or 2′-F-4′-thioribonucleoside modification, these were treated with methanolic ammonia (saturated at 0°C) at room temperature for 24 h. Then, the support was filtered off. The filtrate was concentrated and the ON protected by a DMTr group at the 5′-end was chromatographed on a C-18 silica gel column with a linear gradient of acetonitrile (from 5 to 40%) in 0.1 N TEAA buffer (pH 7.0). The fractions containing the full-length ON were combined and concentrated. The residue was treated with aqueous acetic acid (70%) for 20 min at room temperature. The solution was concentrated and the residue was purified on reversed-phase high performance liquid chromatography, using a J’sphere ODS-M80 column (4.6 × 150 mm, YMC) with a linear gradient of acetonitrile (from 10 to 40%) in 0.1 N TEAA buffer (pH 7.0). The structures of each RNA were confirmed by measurement of MALDI-TOF/MASS spectrometry on Ultraflex TOF/TOF (Buruker Daltonics). The analytical data of synthetic AMOs are summarized in Supplementary Table S1.
Tm measurement
Thermally induced transitions were monitored at 260 nm on a Beckman DU 650 spectrophotometer. Samples were prepared as follows: AMO and target miRNA (3 µM each) were mixed in a phosphate buffer (10 mM, pH 7.0) containing 0.1 mM EDTA and 1 mM NaCl, heated at 90°C for 5 min, cooled gradually to room temperature and used in thermal denaturation studies. The sample temperature was increased 0.5°C/min.
Cell culture
HeLa cells and Huh-7 cells were cultured at 37°C under 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific), 100 units ml−1 penicillin, 100 µg ml−1 streptomycin, respectively. Cells were regularly passaged to maintain exponential growth.
Construction of miRNA reporter plasmids and luciferase reporter assay
The miRNA reporter plasmids were generated by cloning the synthetic 5′-phosphorylated ONs corresponding to tandem perfect-match target sited for human miR-21 or miR-122 into the 3′UTR of firefly luciferase gene (luc2) in the pmirGLO Vector (Promega). HeLa cells or Huh-7 cells were plated into 96-well plates, at 10 000 cells/well in DMEM supplemented with 10% FBS, 100 units ml−1 penicillin, 100 µg ml−1 streptmycin. After 24 h of plating, reporter plasmid and AMOs were co-transfected into the cells in triplicate using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer’s instructions in 100 µl/well Opti-MEM (Invitrogen). AMO concentrations varied as indicated, whereas plasmid concentrations remained constant at 0.1 µg/well. After 6 h of transfection, cells were re-fed with complete media. Cells were lysed at indicated hours after transfection, and luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega). Relative firefly luciferase signals were normalized against Renilla luciferase signals. Results are expressed as relative Fluc/Rluc ratios to that of mirGLO-treated cells. Each experiment was performed at least three times.
Preparation of AMO encapsulated liposome
As a liposomal delivery system, YSK05-MENDs encapsulating AMOs were prepared by a t-BuOH dilution procedure (30,31). Thus, each AMO was mixed with 90% (v/v) t-BuOH containing YSK05 (synthesized as previously described) (30), cholesterol (Avanti Polar Lipid), 1,2-dimyristoyl-sn-glycerol and methoxyethyleneglycol 2000 ether (Avanti Polar Lipid) at a molar ratio of 70:30:3 in 20 mM citrate buffer (pH 4.0) at AMO/lipids of 0.1 (wt/wt) under strong agitation to a t-BuOH concentration of 60% (v/v). Then, lipids/AMO mixture was added into 20 mM citrate buffer (pH 4.0) under strong agitation to a t-BuOH concentration of <12% (v/v). Ultrafiltration was performed to remove t-BuOH, replace external buffer with phosphate buffered saline (PBS, pH 7.0) and concentrate a resulting YSK05-MEND encapsulating AMO (AMO-YSK05-MEND). The average diameter and zeta-potential of the AMO-YSK05-MENDs were determined using a Zetasizer Nano ZS ZEN3600 (MALVERN Instrument, Worchestershire).
In vivo experiments
Female BALB/c mice (8 weeks old) were purchased from Japan SLC. All in vivo experiments were approved by the Institutional Animal Care and Use Committee. One day before the administration, serum was collected for measuring cholesterol concentration. Each AMO-YSK05-MEND was diluted to the appropriate concentrations in PBS (pH 7.4), followed by intravenous administration via the tail vein at a dose of 1 mg AMO/kg in 10 ∼ 15 ml/kg given once a day, every other day, for three times. At 48 h after the last injection, liver and blood were collected. Blood sample was centrifuged at 8g at 4°C for 5 min to obtain plasma. To obtain serum, blood sample was stored overnight at 4°C, followed by centrifugation (10 000 rpm, 4°C, 10 min). Cholesterol concentration in plasma and alanine aminotransferase level in serum were determined using a cholesterol E-test WAKO and Transaminase CII-test WAKO (Wako) according to the manufacturer’s protocols. Total RNAs in liver were isolated using TRIzol (Invitrogen) according to the manufacturer’s protocols. Then, isolated RNAs were reverse transcribed using a High Capacity RNA-to-cDNA kit (Applied Biosystems) according to manufacturer‘s protocol. A quantitative polymerase chain reaction (PCR) analysis was performed with 20 ng of cDNA using Fast SYBR Green Master Mix (Applied Biosystems) on the Lightcycler480 System II (Roche Applied Science). All reactions were performed at a volume of 15 μl. The primers for qRT-PCR are as follows: mouse AldoA (forward) 5′-GATGGGTCCAGCTTCAAC-3′ and (reverse) 5′-GTGCTTTCCTTTCCTAACTCTG-3′; mouse Bckdk (forward) 5′-AGGACCTATGCATGGCTTTG-3′ and (reverse) 5′-CCGTAGGTAGACATCCGTG-3′; mouse Ndrg3 (forward) 5′-ATGGGCTACATACCATCTGC-3′ and (reverse) 5′-TCTGACTGATTGCTGGTCAC-3′; mouse Hprt1 (forward) 5′-CGTGATTAGCGATGATGAAC-3′ and (reverse) 5′-GCAAGTCTTTCAGTCCTGTC-3′. Each data was normalized by Hprt1 expression. All experiments were performed in triplicate and data show mean values from at least three assays.
Statistical analysis
Comparisons between multiple treatments were made using one-way analysis of variance (ANOVA), followed by the SNK test. Pair-wise comparisons between treatments were made using a student’s t-test. P < 0.05 was considered significantly different.
RESULTS AND DISCUSSION
Evaluation of inhibitory activity of AMOs against miR-21
We first synthesized modified AMOs against miR-21, a miRNA that is overexpressed in many tumors and thus is considered to be a potential therapeutic target in oncology (32). The sequences and modification patterns of AMOs were shown in Table 1. Because 2′-F-4′-thioRNA showed highest hybridization ability with its complementary RNA among the chemically modified ONs tested (21,33), we synthesized chimeric 2′-OMe-4′-thio/2′-F-4′-thio–modified AMOs (AM21SMF1 and AM21SMF2) along with 2′-OMe-4′-thio–modified AMO (AM21SM), which are complementary and the same length (22-mer) as mature miR-21. For comparison, 4′-oxo congeners (AM21M, AM21MF1 and AM21MF2) were also prepared. Because Hutvagner et al. reported that AMOs possessing a complementary sequence core with 5 nt flanking sequences on both 5′- and 3′-ends showed more potent anti-miRNA activity (19), we also designed a 32-mer uniformly 2′-OMe-4′-thioribonucleoside–modified AMO (AM21SM-L), which exhibits perfect complimentarity to miR-21 and 5 nt flanking regions on both ends, as well as that of 2′-OMe (AM21M-L).
Table 1.
Sequence and modification patterns of anti-miR-21 AMOs
| AMO | Sequencea | Tm (°C)b |
|---|---|---|
| AM21SM | 5′-UCAACAUCAGUCUGAUAAGCUA-3′ | 64.1 |
| AM21SMF1 | 5′-ucAAcAucAGucuGAuAAGcuA-3′ | 69.2 |
| AM21SMF2 | 5′-UCaacaucagucugauaagcUA-3′ | 71.1 |
| AM21M | 5′-UCAACAUCAGUCUGAUAAGCUA-3′ | 59.9 |
| AM21MF1 | 5′-ucAAcAucAGucuGAuAAGcuA-3′ | 64.2 |
| AM21MF2 | 5′-UCaacaucagucugauaagcUA-3′ | 67.9 |
| AM21SM-L | 5′-UCUUAUCAACAUCAGUCUGAUAAGCUAACCUU-3′ | 62.1 |
| AM21M-L | 5′-UCUUAUCAACAUCAGUCUGAUAAGCUAACCUU-3′ | 58.0 |
aUppercase letters represent 2′-OMe; lowercase letters represent 2′-F; bold uppercase letters are 2′-OMe-4′-thioribonucleotides; bold lowercase letters are 2′-F-4′-thioribonucleotides, bTms were measured versus with target miR-21 (22-mer) in a phosphate buffer (10 mM, pH 7.0) containing 0.1 mM EDTA and 1 mM NaCl, 3 µM strand concentration. Values were given as an average of three independent experiments.
To evaluate anti-miRNA activity of the synthetic AMOs, we constructed a miR-21 luciferase reporter plasmid (34), which has two perfectly matched miR-21 binding sites arranged in tandem on 3′UTR of firefly luciferase gene (pmirGLO21). The firefly luciferase activity was completely suppressed (3.7%) when the pmirGLO21 was transfected into HeLa cells, a cell line known to have high endogenous levels of miR-21 (Figure 2). The activity of pmirGLO, which has no miR-21 binding sites, was high (normalized as 100%) on transfection into HeLa cells. Our hypothesis is that introduction of miR-21 AMO can prevent miR-21 binding to the pmirGLO21 target sites, resulting in increased luciferase expression by either causing translational inhibition or cleavage of the mRNA. We then tested anti-miRNA activity of AMOs. As shown in Figure 2, all our AMOs showed miR-21 inhibitory activity in a dose-dependent manner. In general, 4′-thio–modified AMOs (AM21SM, AM21SMF1 and AM21SMF2) showed much higher activity than 4′-oxo congeners (AM21M, AM21MF1 and AM21MF2). The activities of chimera AMOs (AM21SMF1, AM21SMF2, AM21MF1 and AM21MF2) were less effective compared with uniformly modified AMOs (AM21SM and AM21M). Contrary to the results reported by Hutvanger et al. (19), no significant improvement was observed in 32-mer AM21M-L (30.0% at 50 nM) compared with 22-mer AM21M (27.9% at 50 nM). Furthermore, among the AMOs tested, the shorter AMO, namely AM21SM, was more potent than AM21SM-L (74.3 and 45.9%, respectively) in the case of 4′-thio–modified AMOs. The highest activity was observed with AM21SM, with a luciferase level of 74.3% at 50 nM, which was >2.5 times as much as that of AM21M (27.9% at 50 nM).
Figure 2.
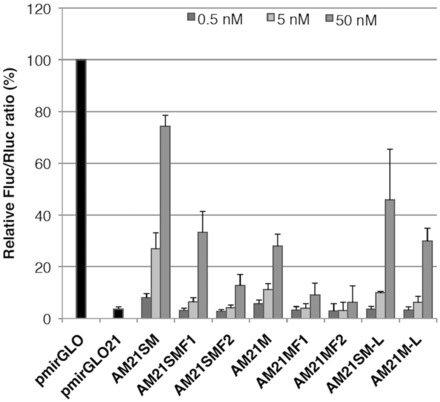
Anti-miR-21 activity of modified AMOs. HeLa cells were co-transfected with the miR-21 reporter plasmid (mirGLO21) and the indicated modified anti-miR-21 AMOs at the indicated concentrations. The dual luciferase assay was performed at 24 h after transfection. Data are shown as mean with standard deviation (SD).
It is known that there is a good correlation between Tm value of AMOs and in vitro potency (35). Therefore, we compared the thermal stability of the AMOs with target miR-21 based on their Tm values. As can be seen in Table 1, the Tm values of chimera AMOs, which contain either 2′-F or 2′-F-4′-thioribonucleoside modification, were higher than those of the uniformly modified AMOs. Although AM21SMF2 showed the highest Tm value, its anti-miR-21 activity was lowest among 4′-thio–modified AMOs. A similar trend was observed in a series of AMOs consisting of 4′-oxo congeners. Thus, no correlation was observed between activity and binding affinity in our case. Meanwhile, as we reported previously (21), 2′-OMe-4′-thioRNA showed higher nuclease stability compared with 2′-OMe RNA, 2′-F-4′-thioRNA and 2′-F RNA. Therefore, chimera AMOs might be more susceptible to degradation than the uniformly modified AMOs, serving as a potential explanation why AM21SM showed the highest activity amongst all AMOs tested. From these results, it can be concluded that the 2′-OMe-4′-thioribonucleoside modification is useful for inhibition of miRNA function. Also, uniform modification is simple and seems to be the most promising choice for subsequent studies.
Improvement in duration of anti-miRNA activity of AMOs by backbone substitution
To examine if 2′-OMe-4′-thioribonucleoside–modified AMO is capable of inhibiting other miRNAs, we chose miR-122 as another miRNA target. MiR-122 is a liver-specific miRNA, known to be involved in cholesterol metabolism, fatty acid metabolism (36) and hepatitis C virus replication (37). Many successful studies of miR-122 inhibition by AMOs both in vitro and in vivo have been reported (20,36–42), and some of them are currently under investigation in clinical trials (20). We hypothesized that 2′-OMe-4′-thioribonucleoside–modified AMO could have high potency to inhibit miR-122 as well as miR-21.
We synthesized a uniformly 2′-OMe-4′-thioribonucleoside–modified AMO, which is complementary to and the same length (23-mer) as mature miR-122, with unmodified phosphodiester (PO) backbone (AM122SM, Table 2). Because ONs can be degraded by nuclease by cleaving a phosphodiester linkage in biological fluid, substitution of a PO linkage for a PS linkage is ideal to prevent such nuclease cleavage reactions. In fact, PS modification has successfully been applied to many previously reported AMOs. Because our end goal of this study is in vivo application, we synthesized 2′-OMe-thioribonucleoside–modified AMOs with PS backbones (AM122SM-PS). For comparison, we also synthesized uniformly 2′-OMe–modified AMOs with either PO or PS backbones (AM122M and AM122M-PS, respectively). Recently, Obad et al. developed seed targeting 8-mer tiny LNAs, which can simultaneously inhibit miRNA families that share the same seed region (42). According to this report, we also prepared a seed targeting 8-mer (AM122SM-PS 8 nt) along with two shorter sequences (AM122SM-PS 15 nt and AM122SM-PS 20 nt) to examine their potency as well as to find an optimal length of AMOs.
Table 2.
Sequence and modification pattern of anti-miR-122 AMOs
| AMO | Sequencea | Tm (°C)b |
|---|---|---|
| AM122SM | 5′-ACAAACACCAUUGUCACACUCCA-3′ | 65.8 |
| AM122SM-PS | 5′-ACAAACACCAUUGUCACACUCCA-3′ | 64.8 |
| AM122SM-PS 20 nt | 5′-AAACACCAUUGUCACACUCC-3′ | 60.7 |
| AM122SM-PS 15 nt | 5′-CCAUUGUCACACUCC-3′ | 54.6 |
| AM122SM-PS 8 nt | 5′-CACACUCC-3′ | 36.3 |
| AM122M | 5′-ACAAACACCAUUGUCACACUCCA-3′ | 62.9 |
| AM122M-PS | 5′-ACAAACACCAUUGUCACACUCCA-3′ | 58.0 |
aUpper case letters represent 2′-OMe; bold upper case letters are 2′-OMe-4′-thioribonucleotides; underlined are PS backbone modification., bTms were measured versus miR-122 in a phosphate buffer (10 mM, pH 7.0) containing 0.1 mM EDTA and 1 mM NaCl, 3 µM strand concentration. Values were given as an average of three independent experiments.
Each AMO and a miR-122 reporter plasmid (pmirGLO122), constructed in a same manner to pmirGLO21, were co-transfected into Huh-7 cells at various AMO concentrations (0.5–5 nM). At 24 h after transfection, we harvested the cells and carried out dual luciferase reporter assay. The resulting AMO activities were shown in Figure 3. As was the case of miR-21 inhibition, all AMOs showed miR-122 inhibitory activity in a dose-dependent manner, and AM122SM gave higher activity than AM122M (63.9 versus 36.6% at 5 nM). No obvious difference was observed in the activity between AMOs with PO and PS backbones. Concerning of the length of AMO, anti-miRNA activity decreased as AMO length became shorter, and tiny AMO, namely AM122SM PS 8 nt, did not show any activity. Thus, the 2′-OMe-4′-thioribonucleoside–modified AMO possessing complementary matched sequence and length seems to be suitable for miRNA inhibition.
Figure 3.
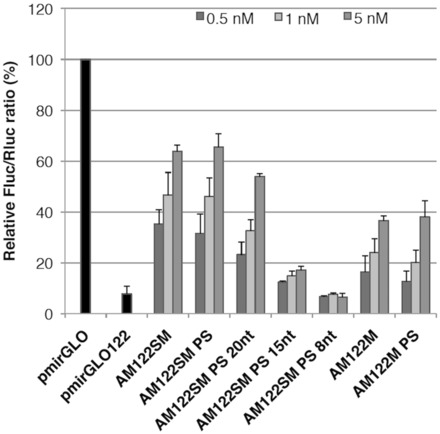
Anti-miR-122 activity of the modified AMOs. Huh-7 cells were co-transfected with the miR-122 reporter plasmid (mirGLO122) and the anti-miR-122 AMOs at the indicated concentrations. The dual luciferase assay was performed at 24 h after transfection. Data are shown as mean with SD.
In our previous study, we showed prolonged activity of 2′-OMe-4′-thioribonucleoside–modified siRNA (29). Thus, we expected that the 2′-OMe-4′-thioribonucleoside modification can prolong the activity of AMOs. We next performed a time-course experiment. As described above, no difference between PO and PS AMOs was observed in the activity after 24 h. However, considerable differences were observed after 48 h, where the activities of PO AMOs (AM122SM and AM122M) declined (Figure 4), while activities of PS AMOs (AM122SM-PS and AM122M-PS) increased. AM122SM-PS showed dramatic increase of the activity throughout the assay, as much as 90% at 72 h. From these results, it can be concluded that 2′-OMe-4′-thioribonucleoside–modified AMO are superior compared with 2′-OMe–modified AMO, and that PS modification conferred long-term activity on AM122SM.
Figure 4.
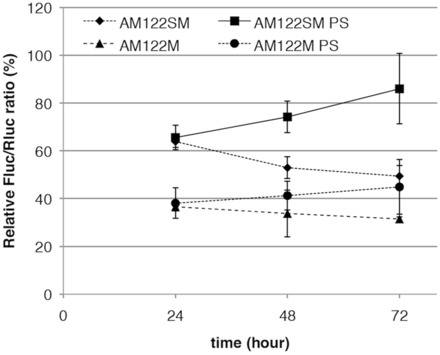
Duration of inhibition by anti-miR-122 AMOs. Huh-7 cells were co-transfected with the miR-122 reporter plasmid (mirGLO122) and the anti-miR-122 AMOs at 5 nM. The dual luciferase assay was performed at the indicated hours. Data are shown as mean with SD.
In the case of the miR-21 study described above, we suggested that nuclease resistance is more closely related to AMO potency than the Tm values. We wished to explore further which of these two factors correlated more strongly with the potency. To explore a correlation between the potency and physical properties of 2′-OMe-4′-thioribonucleotide–modified miR-122 AMOs in more detail, we first measured the Tm values of duplexes with a target miR-122. As can be seen in Table 2, AM122M had a Tm value of 62.9°C and that of AM122M-PS was lower (Tm = 58.0°C and ΔTm = 4.9°C). On the other hand, AM122SM showed a higher Tm value (65.8°C) compared with that of AM122M, and AM122SM-PS had the Tm value of 64.8°C with only a slight decrease from that of AM122SM (ΔTm = 1.0°C). It is well known that the PS modification lowers the Tm of ONs (∼0.5–0.7°C per modification) (43,44). This decrease of the binding affinity would cause loss of inhibitory activity of AMOs. However, replacement of the PO backbone of AM122SM with a PS had no effect on the binding affinity of the AMO, a possible explanation for the retained AMO activity.
We then investigated the nuclease stability of the modified AMOs. A series of AMOs were labeled at the 5′-end with 32P and incubated in 50% human plasma. The reactions were analyzed by denaturing PAGE (Supplementary Figure S1). As expected, PS AMOs exhibited higher stability than that of PO AMOs, and as we reported previously, 2′-OMe-4′-thioribonucleoside–modified AMOs were slightly more stable than that of the corresponding 2′-OMe–modified AMOs. It is worth noting that AM122SM-PS was totally intact under these conditions, while AM122M-PS showed some mild degradation. The rank order of stability in plasma was AM122SM-PS > AM122M-PS > AM122SM > AM122M, indicating that AM122SM-PS is extremely stable against nuclease degradation in human plasma. These results suggested that the Tm value of the AMO had relatively little impact on AMO potency, whereas nuclease resistance seemed to have much greater effect.
For therapeutic use, nuclease resistance in biological fluid, tissues and cells is a prerequisite feature of chemically modified ONs. Our previous comprehensive study of nuclease stability revealed that 2′-OMe-4′-thioRNA was significantly stable against both exo- and endonucleases in spite of consisting of PO backbones (21). Here, we showed PS modification on the 2′-OMe-4′-thioribonucleoside–modified AMO can further improve its nuclease resistance. As a more specific scenario, the endonuclease activity of Ago2 cleaves the unselected strand of RNA duplexes loading into the RISC. It is thought that AMOs act primarily by hybridizing with mature miRNAs that have been loaded into the RISC. Wang et al. explained the molecular mechanisms of target RNA cleavage by a crystal structure of Ago protein (45). They found that both 2′-OMe and PS substitution at the cleavage site (positions 10′–11′) disrupted Ago slicer activity, owing to configurations of the sulfur atoms in Ago protein. Thus, we suggest that the potency and long-term activity of the AM122SM-PS would be due in part to its resistance to cleavage by Ago. Also, PS modification could increase intracellular stability of AMOs, consequently anti-miRNA activity AMOs increased over time (Figure 4). Taken together, combinatorial use of 2′-OMe-4′-thioribonucleosides and a PS backbone is an attractive choice for therapeutic AMOs.
Targeted delivery and anti-miR-122 activity of 2′-OMe-4-thioribonucleoside–modified AMO in mouse liver
In vitro studies showed that AM122SM-PS possessed the most favorable properties. Hence, we next carried out in vivo studies to assess whether AM122SM-PS could also inhibit miR-122 in mice.
Two aspects of successful nucleic acid therapeutics are delivery, followed by ON stability. We have developed a new liposomal nucleic acid delivery system, YSK05-MEND, which contains a pH-sensitive cationic lipid for efficient release of siRNAs from the endosome into the cytoplasm (29). Thus, we prepared YSK05-MEND for in vivo delivery of AMOs to the liver. YSK05-MENDs encapsulating AM122M-PS and AM122SM-PS represented comparable size, charge, polydispersity and encapsulation efficiency (Supplementary Table S2). We assessed in vivo efficacy of the modified miR-122 AMOs by treating mice three times with intravenous injection of 1 mg/kg YSK05-MEND formulated AMO122SM-PS or AMO122M-PS every 2 days.
As described above, a single miRNA may control the levels of multiple mRNAs (3–6). MiR-122 is no exception, as many mRNAs whose expressions are directly controlled by miR-122 have been identified. Elmén et al. demonstrated inhibition of miR-122 using LNA-modified AMOs, LNA-antimiR (41). In their experiments, derepressions of four mRNAs, AldoA, Bckdk, Ndrg3 and Cd320, all of which are direct targets of miR-122 in the mouse liver, were observed after treatment of mice with LNA-antimiR. Therefore, we conducted expression analysis of three miR-122 target mRNAs in the mouse liver, AldoA, Bckdk and Ndrg3 (Figure 5A), by real-time PCR 48 h after the last injection. The levels of all three mRNAs were higher in the AMO-treated mice compared with those treated with saline. It is worth noting that AM122SM-PS induced higher mRNA expression levels than AM122M-PS in all three mRNAs examined. We also observed statistically significant differences between AM122SM-PS and AM122M-PS in the expressions of both AldoA and Bckdk (P < 0.05).
Figure 5.
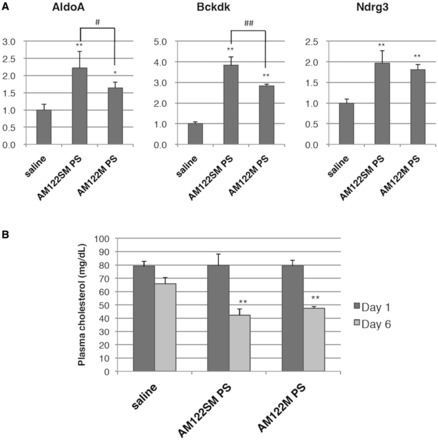
Inhibition of miR-122 in mice. (A) Levels of miR-122 target genes in liver RNA analyzed by qRT-PCR. Each mRNA level was normalized to the mean of saline control group. Data are shown as mean with SD (n = 4, 48 h). (B) Plasma cholesterol levels in mice. The data were normalized to the mean of saline control group at each time point (Day 1 and Day 6). Data are shown as mean with SD (n = 4). *P < 0.05, **P < 0.01 versus saline, #P < 0.05, ##P < 0.01 determined by one-way ANOVA followed by SNK test.
We next examined the change in plasma cholesterol level associated with the increase in expression of these mRNAs. There was no obvious difference between the AMO- and saline-treated mice at 1 day after the last dose, while drastic changes were observed at day 6, where the serum cholesterol levels were reduced by 57.7% for AM122SM-PS and 52.5% for AM122M-PS (Figure 5B) without any hepatotoxicity (Supplementary Figure S2). These results suggest that AM122SM-PS was properly delivered to the mouse liver by YSK05-MEND and elicited an anti-miR-122 effect.
In the previous reports (20,36–38,40–42), AMOs have been administered either as naked/saline formulated or as small molecule conjugates (e.g. cholesterol, a cell-penetrating peptide) to assist in vivo delivery. These approaches often required relatively high doses (ranging from 10 to 80 mg/kg for mice). Instead, our YSK05-MEND dosing strategy achieves efficient miR-122 AMO delivery to the liver. Although the mouse strains used for our experiments versus other researchers’ previous studies were different (e.g. we used inbred strain mice, BALB/c, which are considered to be genetically identical, whereas others used outbred strains, such as NMRI), we successfully increased miR-122 target mRNA expression, followed by a decrease in serum cholesterol level with three intravenous doses of 1 mg/kg AM122SM-PS in mice.
CONCLUSION
We demonstrated the inhibition of miRNAs with 2′-OMe-4′-thioribonucleoside–modified AMOs in vitro as well as in vivo. We first evaluated a variety of chemically modified AMOs that are complementary to mature miR-21, and found the potency of the uniformly 2′-OMe-4′-thioribonucleoside–modified AMO to be the best in our series. Further investigation revealed that PS modification contributes to long-term miR-122 inhibition by the 2′-OMe-4′-thioribonucleoside–modified AMO.
For in vivo studies, we took advantage of an efficient delivery technology, YSK05-MEND, and successfully increased three miR-122 target mRNA levels in the AMO-treated mice liver, followed by a decline of serum cholesterol levels. Although many successful in vivo AMO studies have been reported so far, to our knowledge, none of them have used liposome-like delivery systems. In our previous study, we confirmed that YSK05-MEND efficiently releases siRNAs into the cytoplasm in a pH-dependent manner (30). It is with this property that we believe we were able to achieve efficient AMO delivery in mice.
Together, here we showed not only the potency of 2′-OMe-4′-thioribonucleoside–modified AMOs but also the utility of YSK05-MEND for AMO delivery. Further in vivo studies leading to the understanding of the mechanism of the 2′-OMe-4′-thioribonucleoside–modified AMO function as well as their potential side effect are currently under way.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-Aid for Scientific Research [23249008]; Grant-in-Aid for Scientific Research on Innovative Area “Nanomedicine Molecular Science” [2306] from the Ministry of Education, Culture, Sports, Science and Technology in Japan. Funding for open access: Grant-in-Aid for Scientific Research [23249008].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
The authors would like to thank Ms Y. Misawa (Hokkaido University) for technical assistance.
REFERENCES
- 1.Liu Z, Sall A, Yang D. MicroRNA: an emerging therapeutic target and intervention. Int. J. Mol. Sci. 2008;9:978–999. doi: 10.3390/ijms9060978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 3.Lewis B, Shih I, Jones-Rhoades M, Bartel D, Burge C. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 4.Krek D, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 5.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol. Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 9.Esau CC. Inhibition of microRNA with antisense oligonucleotides. Methods. 2008;44:55–60. doi: 10.1016/j.ymeth.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Inoue H, Hayase Y, Iwai S, Ohtsuka E. Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett. 1987;215:327–330. doi: 10.1016/0014-5793(87)80171-0. [DOI] [PubMed] [Google Scholar]
- 11.Inoue H, Hayase Y, Imura A, Iwai S, Miura K, Ohtsuka E. Sythesis and hybridization studies on two complementary nona(2′-O-methyl)ribonucleotides. Nucleic Acids Res. 1987;15:6131–6148. doi: 10.1093/nar/15.15.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lesnik EA, Guinosso CJ, Kawasaki AM, Sasmor H, Zounes M, Cummins LL, Ecker DJ, Cook PD, Freier SM. Oligodeoxynucleotides containing 2′-O-modifies adenosine: synthesis and effects on stability of DNA: RNA duplexes. Biochemistry. 1993;32:7832–7838. doi: 10.1021/bi00081a031. [DOI] [PubMed] [Google Scholar]
- 13.Cummins LL, Owens SR, Risen LM, Lesnik EA, Freier SM, McGee D, Guinosso CJ, Cook PD. Characterization of fully 2′-modified oligonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995;23:2019–2024. doi: 10.1093/nar/23.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teplova M, Minasov G, Tereshko V, Inamati GB, Cook PD, Manoharan M, Egli M. Crystal structure and improved antisense properties of 2′-O-(2-methoxyethyl)-RNA. Nat. Struct. Biol. 1999;6:535–539. doi: 10.1038/9304. [DOI] [PubMed] [Google Scholar]
- 15.Obika S, Nanbu D, Hari Y, Morio K, In Y, Ishida T, Imanishi T. Synthesis of 2′-O,4′-C-methyleneuridine and-cytidine: novel bicyclic nucleosides having a fixed C3’-endo sugar puckering. Tetrahedron Lett. 1997;38:8735–8738. [Google Scholar]
- 16.Christensen NK, Petersen M, Nielsen P, Jacobsen JP, Olsen CE, Wengel J. A novel class of oligonucleotide analogues containing 2′-O,3’-C-linked [3.2.0]bicycloarabinonucleoside monomers: synthesis, thermal affinity studies, and molecular modeling. J. Am. Chem. Soc. 1998;120:5458–5463. [Google Scholar]
- 17.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 18.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of micro-RNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi M, Minakawa N, Matsuda A. Synthesis and characterization of 2′-modified-4′-thioRNA: a comprehensive comparison of nuclease stability. Nucleic Acids Res. 2009;37:1353–1362. doi: 10.1093/nar/gkn1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naka T, Minakawa N, Abe H, Kaga D, Matsuda A. The stereoselective synthesis of 4′-β-thioribonucleosides via the Pummerer reaction. J. Am. Chem. Soc. 2000;122:7233–7243. [Google Scholar]
- 23.Hoshika S, Minakawa N, Matsuda A. Synthesis and physical and physiological properties of 4′-thioRNA: application to post-modification of RNA aptamer toward NF-κB. Nucleic Acids Res. 2004;32:3815–3825. doi: 10.1093/nar/gkh705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoshika S, Minakawa N, Kamiya H, Harashima H, Matsuda A. RNA interference induced by siRNAs modified with 4′-thioribonucleosides in cultured mammalian cells. FEBS Lett. 2005;579:3115–3118. doi: 10.1016/j.febslet.2005.04.073. [DOI] [PubMed] [Google Scholar]
- 25.Hoshika S, Minakawa N, Shionoya A, Imada K, Ogawa N, Matsuda A. Study of modification pattern–RNAi activity relationships by using siRNAs modified with 4′-thioribonucleosides. Chembiochem. 2007;8:2133–2138. doi: 10.1002/cbic.200700342. [DOI] [PubMed] [Google Scholar]
- 26.Kato Y, Minakawa N, Komatsu Y, Kamiya H, Ogawa N, Harashima H, Matsuda A. New NTP analogs: the synthesis of 4′-thioUTP and 4′-thioCTP and their utility for SELEX. Nucleic Acids Res. 2005;33:2942–2951. doi: 10.1093/nar/gki578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minakawa N, Sanji M, Kato Y, Matsuda A. Investigations toward the selection of fully-modified 4′-thioRNA aptamers: optimization of in vitro transcription steps in the presence of 4′-thioNTPs. Bioorg. Med. Chem. 2008;16:9450–9456. doi: 10.1016/j.bmc.2008.09.048. [DOI] [PubMed] [Google Scholar]
- 28.Dande P, Prakash TP, Sioufi N, Gaus H, Jarres R, Berdeja A, Swayze EE, Griffey RH, Bhat B. Improving RNA interference in mammalian cells by 4′-thio-modified small interfering RNA (siRNA): effect on siRNA activity and nuclease stability when used in combination with 2′-O-alkyl modifications. J. Med. Chem. 2006;49:1624–1634. doi: 10.1021/jm050822c. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi M, Nagai C, Hatakeyama H, Minakawa N, Harashima H, Matsuda A. Intracellular stability of 2′-OMe-4′-thioribonucleoside modified siRNA leads to long-term RNAi effect. Nucleic Acids Res. 2012;40:5787–5793. doi: 10.1093/nar/gks204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato Y, Hatakeyama H, Sakurai Y, Hyodo M, Akita H, Harashima H. A pH-sensitive cationic lipid facilitates the delivery of liposomal siRNA and gene silencing activity in vitro and in vivo. J. Control. Release. 2012;163:267–276. doi: 10.1016/j.jconrel.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Sakurai Y, Hatakeyama H, Sato Y, Hyodo M, Akita H, Harashima H. Gene silencing via RNAi and siRNA quantification in tumor tissue using MEND, a liposomal siRNA delivery system. Mol. Ther. 2013;21:1195–1203. doi: 10.1038/mt.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA.J. Cell. Mol. Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi M, Daidouji S, Shiro M, Minakawa N, Matsuda A. Synthesis and crystal structure of 2′-deoxy-2′-fluoro-4′-thioribonucleosides: substrates for the synthesis of novel modified RNAs. Tetrahedron. 2008;64:4313–4324. [Google Scholar]
- 34.Horwich M, Zamore P. Design and delivery of antisense oligonucleotides to block microRNA function in cultured Drosophila and human cells. Nat. Protoc. 2008;3:1537–1549. doi: 10.1038/nprot.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lennox KA, Behlke MA. A direct comparison of anti-microRNA oligonucleotide potency. Pharm. Res. 2010;9:1788–99. doi: 10.1007/s11095-010-0156-0. [DOI] [PubMed] [Google Scholar]
- 36.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 37.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 38.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 39.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis S, Propp S, Freier SM, Jones LE, Serra MJ, Kinberger G, Bhat B, Swayze EE, Bennett CF, Esau C. Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res. 2009;37:70–77. doi: 10.1093/nar/gkn904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjärn M, Hansen JB, Hansen HF, Straarup EM, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freier SM, Altmann KH. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA: RNA duplexes. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stein CA, Subasinghe C, Shinozuka K, Cohen JS. Physicochemical properties of phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. 1988;16:3209–3221. doi: 10.1093/nar/16.8.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


