Abstract
Spirochetes of the genus Borrelia include the tick-transmitted causative agents of Lyme disease and relapsing fever. They possess unusual genomes composed mainly of linear replicons terminated by closed DNA hairpin telomeres. Hairpin telomeres present an uninterrupted DNA chain to the replication machinery overcoming the ‘end-replication problem’ for the linear replicons. Hairpin telomeres are formed from inverted repeat replicated telomere junctions by the telomere resolvase, ResT. ResT uses a reaction mechanism similar to that of the type IB topoisomerases and tyrosine recombinases. We report here that ResT also possesses single-strand annealing activity and a limited ability to promote DNA strand exchange reactions on partial duplex substrates. This combination of activities suggests ResT is a nexus between the seemingly distinct processes of telomere resolution and homologous recombination. Implications for hairpin telomere replication and linear plasmid recombination, including antigenic variation, are discussed.
INTRODUCTION
Spirochetes of the genus Borrelia include important tick-transmitted zoonotic pathogens that cause Lyme disease and relapsing fever maladies (1–3). Borrelia species possess unusual, highly segmented genomes with the majority of the replicons, including the chromosome, being linear double-stranded DNA (dsDNAs) terminated by covalently closed DNA hairpins referred to as hairpin telomeres (4–6).
The prevailing paradigm is that the ‘end-replication problem’ for these linear DNAs is solved by the simple and elegant means of the hairpin telomeres eliminating the discontinuity in the DNA chain that causes the problem. Bidirectional replication from an internal origin would produce, after replication through the hairpin telomeres, replication intermediates with replicated telomere (rTel) junctions with inverted repeat sequence symmetry (7–10). Daughter DNA molecules covalently linked via the rTel junctions must be separated by a specialized DNA breakage and reunion reaction, referred to as telomere resolution that reforms the hairpin telomeres to allow for subsequent segregation. rTel junctions are processed in vivo and in vitro into two hairpin telomeres (9,11). The essential specialized telomere resolvase that performs this reaction for Borrelia is known as ResT (12,13). A similar replication strategy has been demonstrated for the lysogen of the N15 bacteriophage; N15 lysogens exist as extrachromosomal linear molecules terminated by hairpin telomeres (14–16). The telomere resolvases from the characterized phage systems with linear prophage genomes are commonly referred to as protelomerases. The N15 protelomerase (TelN) has been confirmed to be required in lysogen maintenance but has also, unexpectedly, been shown to be required for lytic replication of N15 (17).
ResT and other confirmed telomere resolvases have a catalytic domain that shares the protein fold and most of the catalytic side chains of the catalytic domain of tyrosine recombinases (18,19). Biochemical analyses have confirmed that telomere resolvases and tyrosine recombinases share a common chemical mechanism (9). ResT has a two-domain structure with site-specific telomere recognition and the tyrosine recombinase catalytic domain residing in the C-terminal two-thirds of the protein. The N-terminal domain possesses non-specific DNA binding activity, and near its C-terminal end, it possesses side chains implicated in creating DNA distortions between the scissile phosphates to allow cleavage by the catalytic domain (20,21). A more complete view of the ResT mechanism is found in (10,22). Genomic sequence analysis of multiple Borrelia burgdorferi strains shows that there is frequent inter-plasmid exchange between the linear plasmids that seems especially frequent near the telomeres (23,24). These exchanges produce complex mosaic relationships among the linear plasmids both within and between strains.
It is an intriguing possibility that this recombinogenic property of hairpin telomeres has been harnessed in the service of infection persistence in the mammalian host. Both the relapsing fever and Lyme disease spirochetes undergo gene conversion-driven antigenic variation of the VlsE surface lipoprotein, the expression site for which is located immediately upstream of a hairpin telomere on a linear plasmid (25). For B. burgdorferi, this switching usually involves short patch conversions of the variable portion of the VlsE expression site from unexpressed donor site(s) in a process that is RecA-independent but dependent on the Holliday junction branch migration motor RuvAB (26–28).
In this report, we show that ResT promotes single-strand annealing (SSA) and limited strand exchange between single-stranded DNA (ssDNA) donors and partial duplex target DNA. These activities are reminiscent of the activities of the λ beta protein, the recombinase component of the λ Red recombination system (29). These results point to a, hitherto, unanticipated connection between the processes of telomere resolution and homologous recombination. Possible implications for hairpin telomere replication and linear plasmid recombination, including antigenic variation of VlsE, are discussed.
MATERIALS AND METHODS
Proteins and oligonucleotide substrates
All oligonucleotides were purchased from Integrated DNA Technologies as polyacrylamide gel electrophoresis (PAGE)-purified oligos (see Supplementary Materials and Methods). All reactions using wild-type ResT used N-terminal (6×-His) protein purified as previously reported except with the addition of step elution off of a heparin-Sepharose 6-LB column loaded and washed with buffer HG 0.5 M NaCl [25 mM HEPES (pH = 7.6), 0.2 mM ethylenediaminetetraacetic acid (EDTA; pH = 8.0), 10% (w/v) glycerol and 500 mM NaCl] and eluted with buffer HG 1.5 M NaCl (20). ResT (1–163) and ResT (164–449) were purified as reported in (21). Cre recombinase was purchased from New England Biolabs, and recombinant human FEN-1 was purchased from MyBioSource.
SSA assays
Fourteen-nanomolar unlabeled complementary oligonucleotide was mixed into reaction buffer (25 mM Tris-Cl, pH = 8.5; 1 mM EDTA, pH = 8.0; 100 mM NaCl) and chilled on ice for 2 min to inhibit the rate of spontaneous annealing on addition of 14-nM 5′-32P endlabeled reporter oligonucleotide and 150 nM ResT followed by incubation at 30°C. Timepoints were taken by removing 21 µl from the 120 µl master annealing reactions to tubes with pre-aliquoted sodium dodecyl sulphate (SDS) stop dye containing an excess unlabeled version of the reporter oligo to prevent further annealing of the labeled reporter oligo after reaction termination. The 10× stop dye contains 200 mM EDTA, 32% glycerol, 1% SDS, 0.024% bromophenol blue and 600 nM unlabeled reporter oligonucleotide. Electrophoretic analysis was performed on 20 × 20 cm vertical gels with 7 or 9% PAGE 1× TAE/0.1% SDS as indicated in the legends and electrophoresed at 13 V/cm for the times indicated in the legends followed by gel drying, exposure of the gels to phosphor screens and analysis on a BioRad FX phosphorimaging machine. Reactions were performed, at a minimum, in triplicate and the data were quantitated with BioRad’s Image Lab software according to the manufacturer’s instructions. Reaction curves, bar graphs and statistics were generated with Prism’s GraphPad 5.0.
DNA strand exchange assays
Fifteen-nanomolar unlabeled ssDNA donor oligonucleotide was mixed into reaction buffer with or without 222 nM ResT (25 mM Tris-Cl, pH = 8.5; 1 mM EDTA, pH = 8.0; 100 mM NaCl; final concentrations) and incubated at 30°C for 5 min before addition of 15 nM of the partial duplex target DNA, 5′-32P endlabeled on the top strand to be exchanged, followed by continued incubation at 30°C. Timepoints were taken by removing 21 µl from the 120 µl master strand exchange reactions to tubes with pre-aliquoted SDS stop dye to achieve a 1× final concentration of stop dye. Reaction time courses were loaded onto to 20 × 20 cm, 7% polyacrylamide, 1× TAE/0.1% SDS gels run for 2 h at 13.3 V/cm followed by gel drying, exposure of the gels to phosphor screens and analysis on a BioRad FX phosphorimaging machine.
RESULTS
ResT promotes DNA SSA
The lines of evidence discussed in the Introduction indicating that hairpin telomeres in B. burgdorferi are recombinogenic, prompted us to examine the possibility that ResT might have unanticipated activities that promote homologous recombination. The most fundamental of such activities is the pairing of complementary single strands of DNA, a process known as SSA. This is a core common activity shared by the RecA/Rad51 recombinases that promote homologous recombination via strand invasion and by the SSA proteins (SSAPs) typified by λ beta/Rad52 that drive the simpler SSA mode of homologous recombination (30,31). This issue was examined by running SSA assays with ResT. Briefly, the assay involves mixing a radiolabeled reporter DNA strand with an equimolar amount of cold complementary strand in protein-free reactions or with the addition of ResT under buffer conditions typical of a telomere resolution assay (see Materials and Methods for details). Annealing reactions are terminated by addition of SDS load dye containing an excess unlabeled reporter oligonucleotide to sequester any remaining unlabeled complementary strand, freezing the annealing reaction at the point of termination (32). The time courses documented in Figure 1B–D indicate that under our reaction conditions, ResT affords >100-fold stimulation over the rate of the spontaneous reaction. A wide variety of reporter and complementary strand partner DNAs were tested to make sure the SSA activity we noted was not a peculiarity of the particular strands tested in Figure 1. ResT is able to promote SSA on a wide variety of oligonucleotide substrates (24–106 bp range tested; data not shown). ResT was, however, found to be unable to support contiguous annealing of plasmid-length DNA strands (data not shown). Because ResT is an atypical member of the tyrosine recombinase family of enzymes, we tested whether the prototypical tyrosine recombinase Cre would also promote SSA. Addition of Cre over a wide range of concentrations did not promote SSA (see Supplementary Figure S1). Cre is a basic (theoretical pI of 9.6) two-domain protein that binds DNA as a C-clamp, making contacts via major and minor grooves, as do the telomere resolvases (19,33,34). Therefore, SSA promoted by ResT is not simply due to charge neutralization afforded by the addition of a basic DNA binding protein (ResT has a theoretical isoelectric point (pI) of 9.65).
Figure 1.
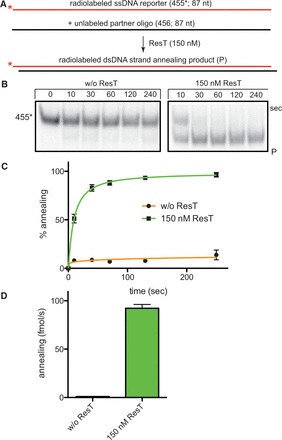
ResT promotes DNA SSA. (A) Schematic representation of the SSA assay. The tested DNA strands anneal into an 83-bp duplex product with 26% GC-content typical of the low GC-content of the B. burgdorferi genome. The 5′-32P endlabeled 455* reporter strand is shown as a shaded or red line, whereas the complementary unlabeled 456 strand is shown in black. Also, represented is the 83-bp heteroduplex product (SSA). The red asterisk represents the 5′-32P endlabel. (B) Nine percent native TAE–SDS PAGE analysis of SSA promoted by ResT. Representative panels of SSA time courses are shown. The migration position on the gel of the labeled reporter strand (455*) is indicated to the left, whereas the migration position of the duplex product of annealing (P) is indicated on the right. Gels were run at 13 V/cm for 150 min. (C) Graphical summary of SSA time courses. The mean and standard deviation of at least three independent experiments are shown; without (w/o) ResT (N = 3) and 150 nM ResT (N = 4). (D) Comparison of the initial rates of spontaneous and ResT-promoted SSA of the 455* and 456 strands detailed in (A); w/o ResT (N = 3) and 150 nM ResT (N = 4).
We tested whether ResT could promote annealing of a model substrate flanked by the unusual GC-rich 17-mer direct repeats that delimit the variable region of VlsE in the expression site and that mark the boundaries between the silent archival variable cassettes (25). The repeats mark the boundary of the gene conversions that drive antigenic variation of the VlsE lipoprotein gene required for infection persistence in mice (35). An SSA assay run with a model substrate flanked by vls 17-mer direct repeats is documented in Figure 2. We labeled the G-rich strand, as we anticipated that this strand would form complex parallel-stranded intermolecular interactions mediated by G-quartet DNA and that this may interfere with normal annealing with its complement (36). This reporter strand forms a complex ladder of bands on our polyacrylamide gel with most of the material retained in the wells. Only the small proportion of the reporter strand not caught up in this web is annealed with the cold complementary strand in protein-free reactions (Figure 2B). Interestingly, addition of ResT to the SSA assay promotes complete annealing of the reporter and cold complementary strands indicating an ability of ResT to disrupt the G-quartet-mediated interactions of the G-rich strand with itself to form a normally annealed duplex DNA with its complementary strand.
Figure 2.
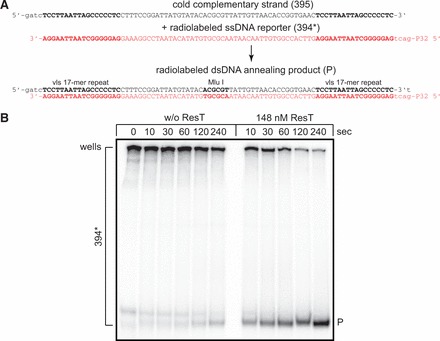
ResT promotes DNA SSA of a model substrate flanked by the 17-bp direct repeats derived from the B. burgdorferi vls locus. (A) Schematic representation of the SSA assay for a model substrate that assembles into an 81-bp duplex flanked by 17-bp direct repeats derived from the boundaries of the variable region of the vls locus that undergoes antigenic variation by gene conversion. The 5′-32P endlabeled reporter strand (394*) is the G-rich strand and is shown in shaded or red script. The complementary unlabeled strand is shown in black (395). Also represented is the 81-bp duplex product of SSA. (B) Nine percent native TAE–SDS PAGE analysis of time courses of spontaneous and ResT-promoted SSA assay detailed in (A). Note that 394* marks the multiple migration positions in the gel of the 394* reporter oligonucleotide, much of which is entrapped in the wells; P marks the migration position of the duplex product of annealing. Electrophoresis was done for 3 h at 13.3 V/cm.
Because SSAPs like λ beta do not use high-energy cofactors, the energy for accelerated annealing of complementary strands of DNA by SSAPs is derived from a high binding energy of these proteins to their heteroduplex products (37). We conclude that ResT also remains bound to its annealed heteroduplex products. This was demonstrated with the vls substrate in an assay based on ResT-binding to the heteroduplex product protecting an Mlu I restriction site in the annealed product (see Supplementary Figure S2).
To determine the minimum length requirements of ResT-mediated SSA and to see whether there is an effect of GC-content on this value, we designed the SSA assay presented in Figure 3. The reporter strand is mixed with a set of cold partner strands that range from perfectly complementary to possessing progressively longer blocks of non-complementary sequence, positioned in such a way as to place the resulting unbasepaired ‘bubble’ asymmetrically within the product, yielding left and right complementary flanks of different lengths and GC-contents (Figure 3A). If both flanks anneal successfully, the heterology in the annealed product forms a bubble product. If only the longer left flank anneals successfully, a frayed-end product is recovered. The gel migration breakpoint between bubble and frayed-end products was determined empirically by comparison of the migration of the SSA products against intentionally designed frayed-end markers. We found that the AT-rich right flank successfully annealed with a flank of only 13 bp, whereas the GC-rich left flank required >19 bp of complementarity to successfully anneal this flank under our reaction conditions (Figure 3B). For the last bubble product formed by ResT (oligo 5), the strand annealed to the reporter strand is only a 63% homology match to the reporter strand.
Figure 3.
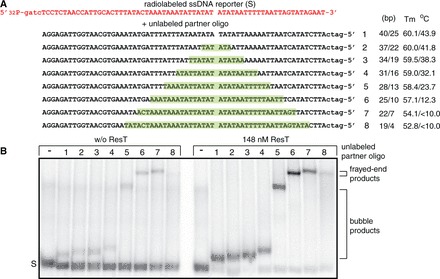
Determination of ssDNA length requirements for ResT-promoted SSA. (A) Schematic representation of the 69-nt 5′-32P endlabeled reporter DNA strand (S) shown in red or shaded script and the set of eight unlabeled partner DNA strands (black) used in SSA assays. Oligo 1 represents a 100% complementary partner that would anneal with the reporter to produce a 65-bp duplex. Each successive partner introduces a 6-nt block of sequence unable to anneal with the reporter (highlighted regions). The block of sequence non-complementarity is asymmetrically positioned so as to give arms of potential annealing of different lengths and melting temperatures (Tm); these values are noted to the right of the partner oligonucleotides. (B) Nine percent native TAE–SDS PAGE analysis of the products of spontaneous and ResT-promoted SSA. SSA assays, performed as indicated in the Materials and Methods section, were incubated for 20 s at 30°C before reaction termination and gel loading. The gel migration breakpoint between bubble and frayed-end products was determined empirically by comparison of the migration of the SSA products against a version of oligo 5 designed to possess only the left flank homology and 41 nt of non-complementary sequence that can only assemble into a frayed-end DNA (Supplementary Methods; OGCB478). S marks the migration position in the gel the 5′-32P endlabeled reporter DNA strand; bubble products are products annealed at both ends but without base pairing in the center; frayed-end products are anchored at the left-end only.
ResT promotes DNA strand exchange
A prokaryotic exemplar of the SSAPs is λ beta. Studies of λ beta indicate that it catalyzes the related reactions of strand assimilation when working in combination with λ exonuclease and of strand exchange on partial duplexes via three-strand branch migration when operating without λ exonouclease (38,39). We tested whether ResT was also able to promote strand exchange on partial duplex substrates. Briefly, the assay consists in pre-incubation of ResT with unlabeled ssDNA donor followed by addition of partial duplex target DNA radiolabeled on the short top strand and continued incubation. Strand exchange was monitored on polyacrylamide gels as the displacement of the labeled strand from the partial duplex target DNA (see the Materials and Methods section for details). For these experiments we used the same substrates tested for beta but provided an equimolar concentration donor ssDNA to target rather than the 25-fold molar excess of donor reported in (39). The results presented in Figure 4 show that ResT also promotes DNA strand exchange. The rapid displacement of the labeled 410* strand from the partial duplex represents bona fide ResT-promoted strand exchange, as spontaneous branch migration was negligible (Figure 4; no ResT + 411 donor panel) and the displacement was dependent on homology of the donor DNA with the target DNA (Figure 4; +ResT and randomized sequence 426 donor). Finally, the possibility that 410* strand displacement was due to contaminating helicase activity was ruled out by the incubation of the partial duplex DNA with ResT without donor addition (Figure 4; +ResT no donor panel). Here the reaction was stopped with SDS stop dye with an excess of unlabeled 410 donor to prevent reannealing of any 410* that may be displaced from the partial duplex by the reaction conditions or possible helicase activity. Strand exchange reactions were also run with radiolabeled donor ssDNA and unlabeled target DNA. This allowed us to verify that the deproteinated heteroduplex products of DNA strand exchange were annealed correctly by treating them with a restriction enzyme (data not shown).
Figure 4.
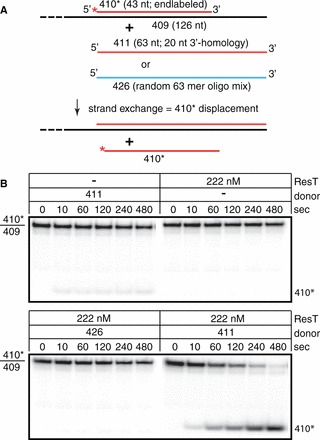
ResT promotes DNA strand exchange. (A) Schematic representation of the DNA strand exchange and control reactions between a 43-bp partial duplex target DNA (410*/409) and a 63-nt ssDNA donor (411) with 20 nt of 3′-flank homology with the bottom strand of the partial duplex target or with 63-nt ssDNA of randomized sequence (426). The top strand is colored red, the bottom strand black and the randomized donor blue; the red asterisk indicates a 5′-32P endlabel. (B) Seven percent native TAE–SDS PAGE analysis of representative time courses of reactions with the combinations of ResT and donor ssDNAs indicated above the gels. The reactions lacking single-stranded donor DNA were stopped with SDS stop dye with an excess of unlabeled 410 oligo to prevent reannealing of any 410* that may be displaced from the partial duplex by the reaction conditions or possible helicase activity. The 410*/409 marks the migration position of the 43-bp partial duplex target labeled on the 410* strand; 410* marks the migration position of the exchanged 410* strand.
The results presented in Supplementary Figure S3 show that ResT is an even more potent strand exchange protein than λ beta being able to promote strand exchange through as many as 63 bp on a partial duplex target, given an equimolar amount of an 83-nt ssDNA donor that can anneal to 20 nt of ssDNA flank next to duplex DNA in the target (Supplementary Figure S3A). λ Beta was unable to strand exchange the equivalent substrate (39). ResT gave no stimulation over spontaneous branch migration in a version of this assay with an 83-bp partial duplex and a 106-nt donor ssDNA, indicating that like beta, ResT has only a limited ability to catalyze strand exchange; neither enzyme is able to use the energy of ATP for their reactions [(37); data not shown].
Strand exchange: the effect of sequence heterology
ResT-promoted strand exchange was shown to be dependent on its SSA activity rather than occurring by strand invasion by use of donor DNAs in the assay with shortened homology to the 3′-flank in the partial duplex target (see Figure 5A). Reducing the length of the flank homology in the donor ssDNA quickly abrogated the strand exchange reaction. Shortening the flank homology to 5 nt or eliminating it abolished strand exchange (Figure 5C). This indicates that ResT is unable to catalyze DNA strand invasion.
Figure 5.
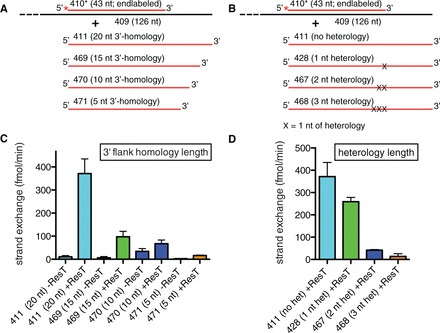
Flank homology dependence and sensitivity to heterology of DNA strand exchange. (A) Schematic representation of the substrate DNAs used in DNA strand exchange reactions between a 43-bp partial duplex target DNA (410*/409) and a series of ssDNA donors with decreasing lengths of 3′-flank homology with the bottom strand of the partial duplex target. The top strand is colored red or shaded grey and the bottom strand black; the red asterisk indicates a 5′-32P endlabel. (B) Schematic representation of the substrate DNAs used in DNA strand exchange reactions between a 43-bp partial duplex target DNA (410*/409) and a series of ssDNA donors with the indicated length of sequence heterology introduced at what would be the initial branch point with the 3′-ss/dsDNA junction of the partial duplex target. (C) Comparison of the initial rates of spontaneous and ResT-promoted DNA strand exchange using the 410*/409 partial duplex substrate and a series of ssDNA donors with the indicated length of 3′ flank homology [detailed in (A)]. Shown is the mean and standard deviation of three independent time courses for each donor DNA and reaction condition. (D) Comparison of the initial rates of ResT-promoted DNA strand exchange using the 410*/409 partial duplex substrate and a series of ssDNA donors with the indicated length of sequence heterology introduced at what would be the initial branch point with the 3′-ss/dsDNA junction of the partial duplex target (detailed in B). Rates of spontaneous branch migration were too slow to be usefully plotted. The mean and standard deviation of three independent time courses for each donor DNA and reaction condition are shown.
We next assessed the effect of sequence changes in the donor ssDNA that correspond to the 3′-ss/dsDNA junction in the target DNA, what would become the initial branch point during strand exchange (Figure 5B). Sequence heterology here presents a strong energetic barrier to spontaneous branch migration and to DNA strand exchange (39). The rate of ResT-promoted strand exchange is strongly reduced by such sequence heterology; >3 nt of heterology completely blocks the strand exchange (Figure 5D). Dispersing 4 nt of heterology throughout the exchanging segment also blocked strand exchange (data not shown). We performed a full set of strand exchange assays with 5′-flank homology in the donor and observed no evidence of a polarity bias for strand exchange (Supplementary Figure S4).
Strand exchange: incomplete strand exchange gives rise to flap complexes
Our strand exchange assays tested the same substrates used to characterize λ beta but differ in that our ‘bottom’ strand is provided by a 126-nt oligonucleotide rather than the M13 genome. This allowed us to use polyacrylamide gels and to visualize ternary complexes formed between the donor ssDNA and the target DNA in strand exchange reactions that have failed to displace the 410* strand owing to the heterology block in the donor ssDNA (see Figure 6 and Supplementary Figure S5). Donor oligonucleotides with increasing sequence heterology decrease the rate of strand exchange but give a corresponding increase in the formation of the slowly migrating ternary complex. A donor ssDNA with four successive nucleotides of sequence heterology at the 3′-ss/dsDNA junction completely blocked strand exchange but yielded a quantitative conversion of the substrate DNAs into ternary complexes, whereas the protein-free reactions gave very little ternary complex (Figure 6B). Similar results were obtained with donors with 5′-flank homology (Supplementary Figure S5).
Figure 6.
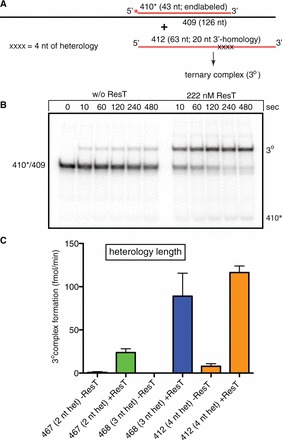
Sequence heterology in the exchanging segment of the single-stranded donor DNA promotes the formation of ternary complexes. (A) Schematic representation of the DNA strand exchange reaction between a 43-bp partial duplex target DNA (410*/409) and a 63-nt ssDNA donor (412) with 20 nt of 3′-flank homology and a contiguous 4-nt block of heterology at the initial branch point with the 3′-ss/dsDNA junction of the partial duplex target. The product is inferred to be a ternary complex (3°) containing the donor DNA and the partial duplex target. The top strand sequence is colored red or shaded grey and the bottom strand black; the asterisk indicates a 5′-32P endlabel; X represents sequence changes in the donor that introduce mismatches with the partial duplex target DNA. (B) Seven percent native TAE–SDS PAGE analysis of representative time courses of spontaneous and ResT-promoted DNA strand exchange reactions. The 410*/409 marks the migration position of the 43-bp partial duplex target labeled on the 410* strand; 410* marks the migration position of displaced 410* strand; 3° marks the migration position of the ternary complex. (C) Comparison of the initial rates of spontaneous and ResT-promoted ternary complex formation using the 410*/409 partial duplex substrate detailed in (A) and a series of ssDNA donors with the indicated length of sequence heterology introduced at what would be the initial branch point with the 3′-ss/dsDNA junction of the partial duplex target. The mean and standard deviation of three independent experiments for each donor DNA and reaction condition are shown.
We investigated the structure of the ternary complexes seen in the experiment in Figure 6 by treating them with the 5′-flap endonuclease, FEN-1 (Figure 7). FEN-1 cleaves 5′-flaps, and its preferred substrate is a double-flap complex (40). FEN-1 treatment of the ternary complexes shows that they are a mix of single- and double-flap complexes. The single-flap complexes are annealed via the 20-nt homology flank only; FEN-1 cleavage releases a 43-nt radiolabeled strand indicating that the 4 nt of heterology has blocked strand exchange at the initial branch point (Figure 7B; lane 4). FEN-1 cleavage also releases a radiolabeled fragment of 15 nt indicating that in the double-flap complexes, strand exchange has progressed 28 nt into the duplex region of the target DNA before stalling (Figure 7B; lane 4). The absence of flap complexes in the protein-free controls suggests that spontaneous branch migration cannot easily progress beyond the 4-nt heterology block and that the 20-nt flank in the target DNA tested is insufficient in the absence of ResT to produce large amounts of gel-stable single-flap complexes annealed to the target solely via the 20-nt single-stranded flank. A similar investigation of the ternary complexes produced with a 4-nt heterology block with donor DNA with a 20-nt 5′-flank homology (Supplementary Figure S5) revealed the presence of only single flap complexes (data not shown). The fact that the 5′-end of the partial duplex target has a much higher GC-content than the 3′-end (5′-CCTG-3′ versus 5′-TACT-3′) likely accounts for this difference.
Figure 7.
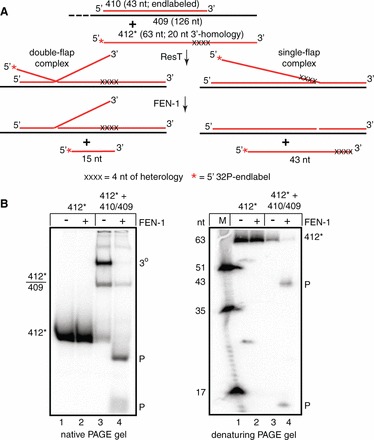
Analysis of the ternary complexes with the 5′-flap endonuclease, FEN-1. (A) Schematic representation of double- or single-flap complex formation by incomplete or blocked strand exchange, respectively, between a 63-nt ssDNA radiolabeled donor (412*) and unlabeled partial duplex target DNA (410/409). The 412* donor oligo has a 4 nt of heterology block at what would be the initial branch point with the partial duplex target DNA (410/409). Treatment with human FEN-1 cleaves off the 5′-flap releasing radiolabeled single-stranded fragments and a smaller unlabeled 3′-single flap complex and a nicked partial duplex product (not visualized on the gels). X represents sequence changes in the donor that introduce mismatches with the target DNA; the asterisk presents the 32P 5′-endlabel. (B) Native and denaturing PAGE analysis of FEN-1 cleavage of deproteinated flap complexes. Strand exchange reactions were set up between 5′-endlabeled 412* donor and unlabeled 410/409 target DNA. Reactions were deproteinated by addition of SDS to a final concentration of 0.1%. The SDS was removed and the buffer was changed by application of the resulting reactions to G-25 Sephadex spin columns for subsequent reaction with 40 nM of human FEN-1 at 37°C for 20 min. The reactions without FEN-1 were treated identically except without the addition of FEN-1 in the final incubation.
The N-terminal domain of ResT is a SSA protein
To determine whether the SSA activity of ResT could be assigned to a particular domain, we compared full-length ResT, ResT (1–163) and ResT (164–449), in SSA and strand exchange assays (Figure 8). ResT (1–163) and ResT (164–449) have been previously defined as domains by partial chymotrypsin cleavage and their independent expression (21). The N-terminal domain possesses non-specific DNA binding activity and acts as an autoinhibition domain for telomere binding/recognition, whereas the C-terminal domain has the tyrosine recombinase catalytic functions and the protein determinants of site-specific telomere binding (21). The SSA and strand exchange assays reveal that ResT (1-163) can support both reactions on its own, although 8-fold more ResT (1–163) is required for half-maximal annealing velocity compared with full-length ResT (Figure 8A). ResT (164–449) was virtually inert in both assays. Approximately 2-fold more ResT (1–163) than ResT is required for a half-maximal strand exchange velocity (Figure 7B). The curves for the strand exchange reaction are more complicated than those for SSA. Reaction rate increases with ResT concentration but begins to drop above a concentration above 250 nM indicating a requirement for a specific stoichiometry of ResT/nucleotides of donor DNA (1 ResT/4 nt DNA) for optimal strand exchange. The curve for ResT (1–163) appears biphasic indicating that the substrates may be present in alternate conformations, one of which requires a higher concentration of ResT (1–163) for optimal strand exchange. ResT (1–163) and ResT (164–449) are both very basic (pI = 9.9 and 9.3, respectively); the segregation of the SSA activity to the N-terminal domain argues for the specificity of our results as opposed to a non-specific effect mediated by charge neutralization (see also Cre’s failure to promote SSA, Supplementary Figure S1). Mixing the two domains together does not reduce the effective concentration of ResT (1–163) required for half-maximal reaction velocity for either SSA or strand exchange (data not shown).
Figure 8.
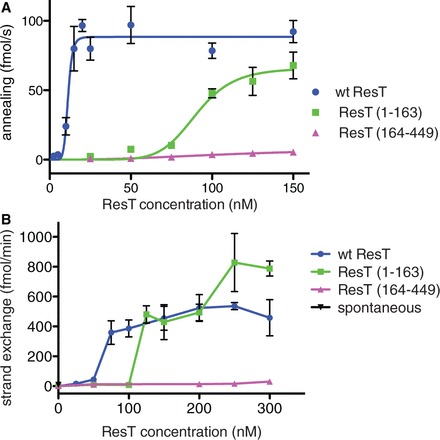
ResT’s N-terminal domain promotes DNA SSA and strand exchange. (A) A plot of the annealing velocity versus ResT concentration is presented comparing full-length ResT and ResT (1–163) and ResT (164–449). The substrate strands used were the 455* + 456 strands used in Figure 1. The mean and standard deviation of at least three independent experiments for each reaction condition are shown. (B) A plot of strand exchange velocity versus ResT concentration is presented comparing full-length ResT and ResT (1–163) and ResT (164–449). The substrates used were the 411 donor ssDNA and the 410/409 partial duplex target used in Figures 4 and 5. The mean and standard deviation of at least three independent experiments for each reaction condition are shown.
DISCUSSION
SSAPs fall into three families typified by Redβ/RecT, ERF and Rad52 (41). Redβ/RecT and ERF families appear to be derived from bacteriophage and the Rad52 family is represented in eukaryotes. SSAPs participate in RecA-dependent and RecA-independent DNA recombination pathways. Despite their different sequence patterns and unrelated predicted structures, representatives of all three families have been shown to form similar ring and filament quaternary superstructures with ssDNA and DNA heteroduplexes, respectively (42,43). In the RecA-independent pathway, the SSAPs act in concert with a 5′-3′ exonuclease to anneal complementary strands created by the exonuclease in a non-conservative DNA recombination event [see Supplementary Figure S6; (38)].
ResT bears no sequence relationship with any of the currently known SSAP families. The N-terminal domain only has similarity to the N-terminal domain of ResT from other Borrelia species. SSA activity has yet to be reported for any other telomere resolvase or tyrosine recombinase or subdomains thereof; the telomere resolvases are considered atypical members of this large family of site-specific recombinases typified by λ integrase (44). There are, however, a growing number of examples of tyrosine recombinases that are active on ssDNA substrates that have folded into hairpin structures that present to the recombinase regions of duplex DNA for site-recognition followed by subsequent DNA cleavage and strand exchange. Examples include integration of the cholera toxin-bearing CTXϕ into the chromosome by host XerC/D in Vibrio cholerae, and the integration of integron gene cassettes into chromosomal sites (45,46). For the integrons that use an arm of an extruded cruciform for integration, it has been suggested that the integrase stabilizes the cruciform conformation that is less energetically favorable than the lineform conformation (47).
What does ResT’s possession of SSA activity mean? We can envision several possibilities: (i) that this activity is implicated in the telomere resolution reaction, (ii) that the SSA activity is required for some unappreciated aspect of linear replicon replication or maintenance and (iii) that the SSA activity is required for other functions in Borrelia.
A role for SSA in telomere resolution?
The telomere resolution reaction involves ResT binding to the rTel junction and DNA cleavage 6 bp apart on opposite strands (9). The formation of the hairpins must involve disruption of the base pairing between the scissile phosphates and reannealing of at least some base pairing to effect strand foldback and strand transfer (19,22,34). It is conceivable that this necessary small-scale annealing activity required for hairpin formation has been elaborated, under selection, into the relatively long-range SSA activity reported here.
Potential implications for hairpin telomere replication
The prevailing model for replication of the linear replicons of Borrelia and the temperate bacteriophages that maintain linear prophage genomes is that bidirectional replication that is initiated internally proceeds around the hairpin telomeres to produce the rTel junctions that are processed into a pair of hairpin telomeres by the telomere resolvase. This model is supported by the fact that both in vivo and in vitro, such rTel junctions are resolved into two hairpin telomeres by the resolvase (9,11). Bidirectional replication from an internal origin of replication has been directly demonstrated for B. burgdorferi’s chromosome and inferred by DNA strand compositional asymmetry mapping for all of B. burgdorferi’s linear plasmids (7,8). This model is further supported by the observation that the hairpin telomeres do not themselves serve as origins of replication as they do in several viral eukaryotic systems with similar telomeres (48,49).
A crucial assumption of this model is that the replisome is able to fully denature the DNA at the hairpin telomere end and that the leading strand polymerase fills any gaps left from lagging strand synthesis. The actual fate of the replisome approaching a hairpin telomere is not currently known. If occasionally the replisome reached the end of the telomere on the leading strand but not the lagging strand, it is conceivable that the ResT SSA and strand exchange activities that we have discovered indicate a need for recombinational repair of a daughter strand gap left on the lagging strand to produce a fully rTel.
Implications for linear plasmid recombination and vls switching
The linear plasmid component of the B. burgdorferi genome is characterized by a fairly uniform complement of genes distributed among the linear plasmids in different strains (24). However, the linear plasmids bear a complex mosaic relationship to each other, indicative of frequent and ongoing recombination (23). The mosaicism is similar to that noted among temperate bacteriophages (50). Among the temperate bacteriophages, it has been demonstrated that λ Red promotes homeologous recombination to produce mosaic phages at a rate far above that produced by the host cell'’s general recombination functions owing to the relative insensitivity of λ beta to sequence mismatches compared with RecA-promoted recombination and the insensitivity of the resulting mismatches to repair by the methyl-directed mismatch repair system (51). Some of the ‘recombination joints’ inferred from the sequencing data indicate recombination by some illegitimate process; we have previously proposed that ResT’s ability to fuse hairpin telomeres together followed by stabilizing deletions could explain this class of linear plasmid recombination and provide the raw material for generating substrates for homologous recombination between different plasmids (10,52). The SSA activity of ResT reported here suggests another way in which ResT could contribute to linear plasmid recombination as detailed in Supplementary Figure S6.
One of the most intriguing examples of recombination at hairpin telomeres is the gene conversion that drives antigenic variation of the vlsE expression site locus. The single expression site for VlsE is located immediately upstream of a linear plasmid telomere. Information in the variable region of the vlsE gene is replaced by information from an array of silent copies of the vls variable region located upstream of the telomeric expression site; the silent vls cassette array is arranged in inverted repeat orientation with respect to the expression site. vls switching usually involves short conversions of the variable portion of the vlsE expression site from unexpressed donor site(s) in a process that is RecA-independent but dependent on the branch migration motor RuvAB, implying the involvement of some variety of RecA-independent strand exchange reaction (26–28). Without data on the initiating events involved in antigen switching, it is difficult to arbitrate between different possible models of switching. Nonetheless, the SSA and strand exchange activity of ResT is consistent with a possible role for ResT in this process. The expression site and silent donors being in inverted orientation indicate that during lagging strand synthesis the single-stranded gaps transiently produced would reveal complementary sequences in the donor vls cassettes and the expression site, creating potential substrates for ResT-mediated DNA SSA or strand exchange.
The discovery of the SSA and strand exchange activities of ResT reported here point to important future studies to examine in more detail the way in which rTel junctions are produced during the Borrelia replication cycle and hint at ways in which hairpin telomere metabolism may contribute to the diversity-generating system represented by the vls locus.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [53].
FUNDING
The Canadian Institutes for Health Research [MOP 79344]; the Saskatchewan Health Research Fund [2570] and the Natural Sciences and Engineering Research Council [RGPIN 326797-2011]. Funding for open access charge: Saskatchewan Health Research Fund [2570].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank George Chaconas for providing constructs for ResT (1-163) and ResT (164-449). They also thank Harold Bull for critical reading of the draft of this report.
REFERENCES
- 1.Barbour AG. Borrelia: a diverse and ubiquitous genus of tick-borne pathogens. In: Scheld MW, Craig WA, Hughes JM, editors. Emerging Infections 5. Washington, DC: American Society for Microbiology; 2001. pp. 153–173. [Google Scholar]
- 2.Dworkin MS, Schwan TG, Anderson DE., Jr Tick-borne relapsing fever in North America. Med. Clin. North Am. 2002;86:417–433, viii–ix. doi: 10.1016/s0025-7125(03)00095-6. [DOI] [PubMed] [Google Scholar]
- 3.Steere AC, Coburn J, Glickstein L. The emergence of Lyme disease. J. Clin. Invest. 2004;113:1093–1101. doi: 10.1172/JCI21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbour AG, Garon CF. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science. 1987;237:409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- 5.Baril C, Richaud C, Baranton G, Saint Girons IS. Linear chromosome of Borrelia burgdorferi. Res. Microbiol. 1989;140:507–516. doi: 10.1016/0923-2508(89)90083-1. [DOI] [PubMed] [Google Scholar]
- 6.Ferdows MS, Barbour AG. Megabase-sized linear DNA in the bacterium Borrelia burgdorferi, the Lyme disease agent. Proc. Natl Acad. Sci. USA. 1989;86:5969–5973. doi: 10.1073/pnas.86.15.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Picardeau M, Lobry JR, Hinnebusch BJ. Physical mapping of an origin of bidirectional replication at the centre of the Borrelia burgdorferi linear chromosome. Mol. Microbiol. 1999;32:437–445. doi: 10.1046/j.1365-2958.1999.01368.x. [DOI] [PubMed] [Google Scholar]
- 8.Picardeau M, Lobry JR, Hinnebusch BJ. Analyzing DNA strand compositional asymmetry to identify candidate replication origins of Borrelia burgdorferi linear and circular plasmids. Genome Res. 2000;10:1594–1604. doi: 10.1101/gr.124000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobryn K, Chaconas G. ResT, a telomere resolvase encoded by the Lyme disease spirochete. Mol. Cell. 2002;9:195–201. doi: 10.1016/s1097-2765(01)00433-6. [DOI] [PubMed] [Google Scholar]
- 10.Chaconas G, Kobryn K. Structure, function, and evolution of linear replicons in Borrelia. Annu. Rev. Microbiol. 2010;64:185–202. doi: 10.1146/annurev.micro.112408.134037. [DOI] [PubMed] [Google Scholar]
- 11.Chaconas G, Stewart PE, Tilly K, Bono JL, Rosa P. Telomere resolution in the Lyme disease spirochete. EMBO J. 2001;20:3229–3237. doi: 10.1093/emboj/20.12.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byram R, Stewart PE, Rosa P. The essential nature of the ubiquitous 26-kilobase circular replicon of Borrelia burgdorferi. J. Bacteriol. 2004;186:3561–3569. doi: 10.1128/JB.186.11.3561-3569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobryn K. The Linear Hairpin Replicons of Borrelia burgdorferi. In: Klassen FMR, editor. Microbial Linear Plasmids. Vol. 7. Heidlberg: Springer-Verlag; 2007. pp. 117–140. [Google Scholar]
- 14.Deneke J, Ziegelin G, Lurz R, Lanka E. The protelomerase of temperate Escherichia coli phage N15 has cleaving-joining activity. Proc. Natl Acad. Sci. USA. 2000;97:7721–7726. doi: 10.1073/pnas.97.14.7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravin NV, Strakhova TS, Kuprianov VV. The protelomerase of the phage-plasmid N15 is responsible for its maintenance in linear form. J. Mol. Biol. 2001;312:899–906. doi: 10.1006/jmbi.2001.5019. [DOI] [PubMed] [Google Scholar]
- 16.Ravin NV, Kuprianov VV, Gilcrease EB, Casjens SR. Bidirectional replication from an internal ori site of the linear N15 plasmid prophage. Nucleic Acids Res. 2003;31:6552–6560. doi: 10.1093/nar/gkg856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mardanov AV, Ravin NV. Conversion of linear DNA with hairpin telomeres into a circular molecule in the course of phage N15 lytic replication. J. Mol. Biol. 2009;391:261–268. doi: 10.1016/j.jmb.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 18.Deneke J, Burgin AB, Wilson SL, Chaconas G. Catalytic residues of the telomere resolvase ResT: a pattern similar to, but distinct from, tyrosine recombinases and type IB topoisomerases. J. Biol. Chem. 2004;279:53699–53706. doi: 10.1074/jbc.M409001200. [DOI] [PubMed] [Google Scholar]
- 19.Aihara H, Huang WM, Ellenberger T. An interlocked dimer of the protelomerase TelK distorts DNA structure for the formation of hairpin telomeres. Mol. Cell. 2007;27:901–913. doi: 10.1016/j.molcel.2007.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bankhead T, Chaconas G. Mixing active site components: a recipe for the unique enzymatic activity of a telomere resolvase. Proc. Natl Acad. Sci. USA. 2004;101:13768–13773. doi: 10.1073/pnas.0405762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tourand Y, Lee L, Chaconas G. Telomere resolution by Borrelia burgdorferi ResT through the collaborative efforts of tethered DNA binding domains. Mol. Microbiol. 2007;64:580–590. doi: 10.1111/j.1365-2958.2007.05691.x. [DOI] [PubMed] [Google Scholar]
- 22.Briffotaux J, Kobryn K. Preventing broken Borrelia telomeres: ResT couples dual hairpin telomere formation with product release. J. Biol. Chem. 2010;285:41010–41018. doi: 10.1074/jbc.M110.150060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casjens S, Palmer N, Van Vugt R, Huang WH, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, et al. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 24.Casjens SR, Mongodin EF, Qiu WG, Luft BJ, Schutzer SE, Gilcrease EB, Huang WM, Vujadinovic M, Aron JK, Vargas LC, et al. Genome stability of Lyme disease spirochetes: comparative genomics of Borrelia burgdorferi plasmids. PLoS One. 2012;7:e33280. doi: 10.1371/journal.pone.0033280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang JR, Hardham JM, Barbour AG, Norris SJ. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- 26.Dresser AR, Hardy PO, Chaconas G. Investigation of the genes involved in antigenic switching at the vlsE locus in Borrelia burgdorferi: an essential role for the RuvAB branch migrase. PLoS Pathog. 2009;5:e1000680. doi: 10.1371/journal.ppat.1000680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin T, Gao L, Edmondson DG, Jacobs MB, Philipp MT, Norris SJ. Central role of the Holliday junction helicase RuvAB in vlsE recombination and infectivity of Borrelia burgdorferi. PLoS Pathog. 2009;5:e1000679. doi: 10.1371/journal.ppat.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liveris D, Mulay V, Sandigursky S, Schwartz I. Borrelia burgdorferi vlsE antigenic variation is not mediated by RecA. Infect. Immun. 2008;76:4009–4018. doi: 10.1128/IAI.00027-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radding CM. The role of exonuclease and beta protein of bacteriophage lambda in genetic recombination. I. Effects of red mutants on protein structure. J. Mol. Biol. 1970;52:491–499. doi: 10.1016/0022-2836(70)90415-8. [DOI] [PubMed] [Google Scholar]
- 30.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 1999;63:751–813, table of contents. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Sneeden J, Heyer WD. In vitro assays for DNA pairing and recombination-associated DNA synthesis. Methods Mol. Biol. 2011;745:363–383. doi: 10.1007/978-1-61779-129-1_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo F, Gopaul DN, van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 34.Shi K, Huang WM, Aihara H. An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation. PLoS Biol. 2013;11:e1001472. doi: 10.1371/journal.pbio.1001472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bankhead T, Chaconas G. The role of VlsE antigenic variation in the Lyme disease spirochete: persistence through a mechanism that differs from other pathogens. Mol. Microbiol. 2007;65:1547–1558. doi: 10.1111/j.1365-2958.2007.05895.x. [DOI] [PubMed] [Google Scholar]
- 36.Walia R, Chaconas G. Suggested role for G4 DNA in recombinational switching at the antigenic variation locus of the Lyme disease spirochete. PLoS One. 2013;8:e57792. doi: 10.1371/journal.pone.0057792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karakousis G, Ye N, Li Z, Chiu SK, Reddy G, Radding CM. The beta protein of phage lambda binds preferentially to an intermediate in DNA renaturation. J. Mol. Biol. 1998;276:721–731. doi: 10.1006/jmbi.1997.1573. [DOI] [PubMed] [Google Scholar]
- 38.Cassuto E, Radding CM. Mechanism for the action of lambda exonuclease in genetic recombination. Nat. New Biol. 1971;229:13–16. doi: 10.1038/newbio229013a0. [DOI] [PubMed] [Google Scholar]
- 39.Li Z, Karakousis G, Chiu SK, Reddy G, Radding CM. The beta protein of phage lambda promotes strand exchange. J. Mol. Biol. 1998;276:733–744. doi: 10.1006/jmbi.1997.1572. [DOI] [PubMed] [Google Scholar]
- 40.Kao HI, Henricksen LA, Liu Y, Bambara RA. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 2002;277:14379–14389. doi: 10.1074/jbc.M110662200. [DOI] [PubMed] [Google Scholar]
- 41.Iyer LM, Koonin EV, Aravind L. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics. 2002;3:8. doi: 10.1186/1471-2164-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Passy SI, Yu X, Li Z, Radding CM, Egelman EH. Rings and filaments of beta protein from bacteriophage lambda suggest a superfamily of recombination proteins. Proc. Natl Acad. Sci. USA. 1999;96:4279–4284. doi: 10.1073/pnas.96.8.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erler A, Wegmann S, Elie-Caille C, Bradshaw CR, Maresca M, Seidel R, Habermann B, Muller DJ, Stewart AF. Conformational adaptability of Redbeta during DNA annealing and implications for its structural relationship with Rad52. J. Mol. Biol. 2009;391:586–598. doi: 10.1016/j.jmb.2009.06.030. [DOI] [PubMed] [Google Scholar]
- 44.Kobryn K, Briffotaux J, Karpov V. Holliday junction formation by the Borrelia burgdorferi telomere resolvase, ResT: implications for the origin of genome linearity. Mol. Microbiol. 2009;71:1117–1130. doi: 10.1111/j.1365-2958.2008.06584.x. [DOI] [PubMed] [Google Scholar]
- 45.Val ME, Bouvier M, Campos J, Sherratt D, Cornet F, Mazel D, Barre FX. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell. 2005;19:559–566. doi: 10.1016/j.molcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 46.MacDonald D, Demarre G, Bouvier M, Mazel D, Gopaul DN. Structural basis for broad DNA-specificity in integron recombination. Nature. 2006;440:1157–1162. doi: 10.1038/nature04643. [DOI] [PubMed] [Google Scholar]
- 47.Loot C, Bikard D, Rachlin A, Mazel D. Cellular pathways controlling integron cassette site folding. EMBO J. 2010;29:2623–2634. doi: 10.1038/emboj.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeLange AM, Reddy M, Scraba D, Upton C, McFadden G. Replication and resolution of cloned poxvirus telomeres in vivo generates linear minichromosomes with intact viral hairpin termini. J. Virol. 1986;59:249–259. doi: 10.1128/jvi.59.2.249-259.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willwand K, Baldauf AQ, Deleu L, Mumtsidu E, Costello E, Beard P, Rommelaere J. The minute virus of mice (MVM) nonstructural protein NS1 induces nicking of MVM DNA at a unique site of the right-end telomere in both hairpin and duplex conformations in vitro. J. Gen. Virol. 1997;78:2647–2655. doi: 10.1099/0022-1317-78-10-2647. [DOI] [PubMed] [Google Scholar]
- 50.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl Acad. Sci. USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinsohn JT, Radman M, Petit MA. The lambda red proteins promote efficient recombination between diverged sequences: implications for bacteriophage genome mosaicism. PLoS Genet. 2008;4:e1000065. doi: 10.1371/journal.pgen.1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobryn K, Chaconas G. Fusion of hairpin telomeres by the B. burgdorferi telomere resolvase ResT implications for shaping a genome in flux. Mol. Cell. 2005;17:783–791. doi: 10.1016/j.molcel.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 53.Gupta RC, Folta-Stogniew E, O'Malley S, Takahashi M, Radding CM. Rapid exchange of A:T base pairs is essential for recognition of DNA homology by human Rad51 recombination protein. Mol. Cell. 1999;4:705–714. doi: 10.1016/s1097-2765(00)80381-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


