Abstract
Riboswitches are cis-acting regulatory elements broadly distributed in bacterial mRNAs that control a wide range of critical metabolic activities. Expression is governed by two distinct domains within the mRNA leader: a sensory ‘aptamer domain’ and a regulatory ‘expression platform’. Riboswitches have also received considerable attention as important tools in synthetic biology because of their conceptually simple structure and the ability to obtain aptamers that bind almost any conceivable small molecule using in vitro selection (referred to as SELEX). In the design of artificial riboswitches, a significant hurdle has been to couple the two domains enabling their efficient communication. We previously demonstrated that biological transcriptional ‘OFF’ expression platforms are easily coupled to diverse aptamers, both biological and SELEX-derived, using simple design rules. Here, we present two modular transcriptional ‘ON’ riboswitch expression platforms that are also capable of hosting foreign aptamers. We demonstrate that these biological parts can be used to facilely generate artificial chimeric riboswitches capable of robustly regulating transcription both in vitro and in vivo. We expect that these modular expression platforms will be of great utility for various synthetic biological applications that use RNA-based biosensors.
INTRODUCTION
RNA-based sensory devices are increasingly viewed as important elements for a range of synthetic biology applications (1–4). One type of these biosensors is the ‘riboswitch’, an RNA element found in bacterial mRNA leaders that directly bind metabolites in the cellular environment to regulate expression of the message (5,6). Small molecule binding to the sensory domain guides folding of a downstream secondary structure switch (also known as the expression platform) that directs the transcriptional and/or translational machinery. Because RNA aptamers can be generated by in vitro selection and evolution methods (7,8), it is theoretically possible to create regulatory devices that respond to almost any conceivable small molecule or protein. Further, riboswitches are broadly distributed across bacteria controlling a multitude of crucial metabolic processes (9,10), underscoring the enormous potential for designed riboswitches to robustly function in the cellular context. The utility of small molecule sensing RNAs has already been demonstrated through applications such as elucidating biosynthetic pathways (11), controlling cellular behavior (12) and in directed protein evolution (13).
While a number of artificial riboswitches that control gene expression at the translational level (12,14–18) or through activation of a ribozyme (19,20) have been developed, only a few artificial regulatory devices modulating transcription have been reported (21–23). A significant motivation for developing transcriptional devices is that they can be used not only to control expression of proteins but also regulatory RNAs to create complex regulatory networks (24,25). Artificial transcriptional regulatory switches have been primarily developed using computational and rational design approaches. Isambert and coworkers designed a set of RNA switches that activate or repress transcription of a message through association of an antisense RNA (22). This approach has been exploited to create a series of orthogonal transcriptional regulators that have the potential to create genetic logic circuits and transcriptional cascades in Escherichia coli (26,27). More recently, Mörl and coworkers combined rational design and computational approaches to create riboswitches that respond to theophylline and promote read through transcription both in vitro and in vivo (23). While in silico approaches are sufficient to model RNA secondary structure, accurate calculation of energies associated with ligand binding and non–Watson-Crick base paired structures within the aptamer is not currently feasible, complicating design of small molecule-responsive devices.
We have taken a different approach to this problem by developing secondary structure switch modules derived from biological sources that can host a variety of aptamers in a ‘mix-and-match’ fashion. Previously, three modular ‘OFF’ expression platforms derived from natural riboswitches were developed (21). These modules can be fused to a diverse array of aptamers using simple design strategies based solely on consideration of secondary structure to create chimeras that function both in vitro and in E. coli. Natural RNA switches have key advantages over those computationally designed. First, the aptamer and the secondary structural switch are directly coupled through a shared secondary structure, the P1 helix. This feature eliminates additional selection or design of a sequence or structure—a communication module—that functionally couples the aptamer to the switch (28–30). Natural expression platforms also contain inconspicuous features such as transcriptional pause sites or sequences that facilitate RNA folding (30,31) important for efficient cellular function but difficult to design. Biological switches have benefited from extensive natural selection to optimize their cellular performance. However, one drawback of the previously engineered switch modules is that they are all ‘OFF’ switches, whereas regulators capable of activation of gene expression by a specific small molecule are highly desirable in synthetic biological applications.
In this work, we have addressed this deficiency by developing two modular and transcriptional ‘ON’ switches. Adapting these modules from biological riboswitches presented novel design challenges including decoupling overlapping sequence requirements of the two domains and secondary structural reorganization that were not encountered in the previous development of chimeric ‘OFF’ switches. In addition, new design strategies had to be explored to optimize the in vitro and in vivo regulatory performance of resultant chimeras. Together, these data yield a robust set of strategies for rapid and facile creation of novel RNA devices that act at the transcriptional level.
MATERIALS AND METHODS
In vitro transcription template construction
Transcription templates encoding riboswitches were synthesized by standard polymerase chain reaction amplification (32) from Bacillus subtilis genomic DNA for wild-type sequences or recursive polymerase chain reaction (33) with overlapping oligonucleotides (Integrated DNA Technologies) for chimeras. The strong T7A1 bacterial RNA polymerase (RNAP) promoter (34) was placed immediately upstream of the predicted transcription start site in all chimeric riboswitch transcription templates. Complete sequences of all riboswitches used in this study are given in Supplementary Table S1. DNA templates were sequence verified prior to use in transcription assays.
In vitro transcription assays
Effector-dependent activity of wild type and chimeric riboswitches was determined using an in vitro single turnover transcription assay (35,36). Before initiation of transcription, E. coli RNA polymerase holoenzyme (0.25 U) (Epicentre Biotechnologies) was incubated with 50 ng of DNA at 37°C for 10 min in 12.5 µl of 2× transcription buffer (140 mM Tris–HCl, pH 8.0, 140 mM NaCl, 0.2 mM EDTA, 28 mM β-mercaptoethanol and 70 mg/ml bovine serum albumin), 2.5 µl of 25 mM MgCl2, 0.5 mCi of α-32P-ATP in a final volume of 17.5 µl. Transcription was initiated by addition of 7.5 µl of ribonucleotide triphosphate (rNTP) mix (165 µM each rNTP), 0.2 mg/ml heparin and a variable amount of the appropriate effector ligand. Reactions were incubated for 10 min at 37°C, quenched by addition of 25 µl of stop mix (8 M urea, containing 0.01% bromophenol blue/xylene cyanol) and incubated for 2 min at 65°C. RNA transcripts were separated by electrophoresing through a denaturing 8% 29:1 acrylamide:bisacrylamide gel and visualized by exposing the dried gel to a phosphor screen. The intensity of each RNA band was quantified using ImageQuant software (Molecular Biosystems) and data fit to a standard two-state model by nonlinear least squares analysis (Kaliedagraph).
Isothermal titration calorimetry
Affinity of the B. subtilis pbuE and Pseudomonas aeruginosa sahH aptamer domains for their effector ligands used in this study [2-aminopurine (2AP) and S-adenosylhomocysteine (SAH), respectively] were measured under transcription buffer and temperature conditions using isothermal titration calorimetry using previously described techniques (37,38). Sequences and secondary structures of the aptamers used are given in Supplementary Figure S1 and Supplementary Table S1. Purified RNA was dialyzed overnight at 4°C against buffer containing 70 mM potassium 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (K-HEPES) or Tris–HCl, pH 8.0, 70 mM NaCl, 2.5 mM MgCl2 and 1 mM β-mercaptoethanol. Ligands were dissolved in the same buffer used for RNA dialysis from solid stocks (Sigma-Aldrich). For the adenine-sensing pbuE aptamer the concentration of RNA in the cell was ∼150 µM yielding a c-value of ∼30 (39,40); the syringe contained 1.5 mM 2AP. For the SAH-sensing metH aptamer, the cell concentration was ∼50 µM yielding a c-value of ∼85; the syringe contained 0.5 mM SAH. Titrations were performed at 37°C using a MicroCal iTC200 (GE Healthcare). Integration and fitting of data were performed using MicroCal Origin 7.0.
In vivo assay
Activity of riboswitches in E. coli was monitored using a reporter system described previously (21). Chimeric riboswitches were cloned upstream of a gfpuv reporter gene under control of a strong tac promoter (41) into pRR1 (21) and sequence verified. Escherichia coli strain BW25113 (Δnep) [Keio collection, (42)] was transformed with the resultant vectors. Single colonies were grown overnight in 3 ml of CSB rich defined media (21) with 100 µg/ml ampicillin. This saturated culture was used to inoculate 100 ml fresh media and allowed to grow to early exponential phase (OD600 = 0.1–0.5). Aliquots were taken in triplicate (3 ml each sample), ligand was added at varying concentration to the media at the concentrations indicated in the titration graphs and the cells were allowed to grow for 6 (pbuE chimeras) or 10 (metH chimeras) h at 37°C.
Optical density (OD600) and fluorescence intensity of each culture was measured using a Tecan Infinite M200 Pro plate reader. Fluorescence measurements were taken at an excitation wavelength of 395 nm, and the average fluorescence was measured between 513 and 515 nm at the maximum emission for a variant of green fluorescent protein that is optimized for maximal fluorescence when irradiated with ultraviolet light (GFPuv). Optical density normalized fluorescent values plotted as a function of the ligand concentration were fit to a standard two-state equation to determine EC50, defined as the concentration of ligand in the media required to elicit a half-maximal expression response. Background fluorescence was measured by titrating ligand into cells carrying the parental plasmid pBR322 and calculating the OD600 normalized fluorescence. An average of the background was subtracted to the values of the cultures containing the plasmids with the riboswitch constructs.
RESULTS
Consideration of biological ‘ON’ riboswitches
To find suitable candidate riboswitches for developing modular expression platforms, a series of biological riboswitches were examined. The vast majority of riboswitches downregulate gene expression in response to a ligand because they primarily control transcriptional units containing genes related to their biosynthesis or transport (43). However, two families of riboswitches are composed of regulatory elements that primarily activate gene expression in the presence of their cognate ligand: SAH (44) and cyclic di-GMP-II (45,46). Additionally, isolated examples of ‘ON’ switches exist in other families including the lysine, glycine and purine families (43), the most characterized of these riboswitches being the B. subtilis pbuE adenine riboswitch (47). Only a subset of these RNAs regulating gene expression at the transcriptional level was further considered.
There are several differences between the ‘OFF’ riboswitches previously used to create modular expression platforms (21) and candidate ‘ON’ switches considered in this study. The alternative secondary structural switch of transcriptional ‘OFF’ riboswitches generally comprises the first helix of the sensory aptamer domain (called P1) and an antiterminator helix (P-AT); the rho-independent terminator (P-T) is merely a readout of the P1/P-AT outcome. However, the ‘ON’ switches present a different architecture where the competing secondary structures are P1 and P-T (Figure 1a). Thus, the P1 helix stabilized by ligand bound to the aptamer domain is the anti-terminator (equivalent to P-AT in ‘OFF’ switches). Further, in contrast to the ‘OFF’ switches previously used, the ‘ON’ switches almost invariably have significant sequence requirement overlap between the two domains, making their separation difficult. For example, in the B. subtilis pbuE riboswitch, a 16-nt sequence is required for both aptamer formation and for the competing terminator structure (yellow, Figure 1a).
Figure 1.
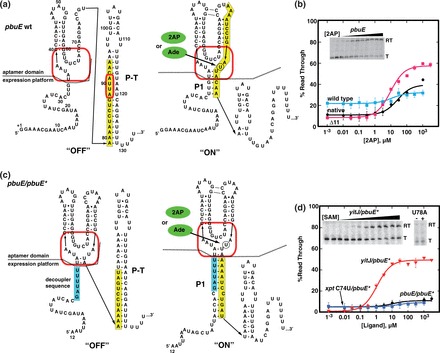
Reengineering the B. subtilis pbuE adenine riboswitch. (a) Sequence and secondary structure of the wild-type pbuE riboswitch in the ‘OFF’ and ‘ON’ states. The red box denotes the nucleotides comprising the ligand-binding pocket, and the yellow box denotes sequence predicted to adopt adenine-dependent alternative secondary structures. Numbering is consistent with the experimental determination of the transcription start site of this riboswitch (48). (b) In vitro assay of effector-dependent transcriptional anti-termination using 2AP for the native B. subtilis pbuE riboswitch transcriptional unit (‘native’), the wild-type pbuE riboswitch under control of the T7A1 promoter (‘wild type’) and the pbuE(Δ1–11) mutant (‘Δ11’). A representative denaturing gel showing 32P-labeled transcription products of the Δ11 mutant as a function of 2AP concentration is shown in the inset; ‘RT’ denotes the anti-terminated read through transcription product, and ‘T’ denotes the terminated product. Data plotted are the average of three independent measurements and the variation represented by the error bars. (c) Sequence and secondary structure of the pbuE ‘decoupled’ riboswitch capable of accommodating different aptamers. The cyan box denotes the insertion sequence that prevents the secondary structural switch from invading the aptamer domain. (d) In vitro transcription assay of the decoupled pbuE/pbuE* riboswitch along with two other aptamer chimeras. The inset demonstrates that the yitJ/pbuE* riboswitch regulates a transcriptional variation of the RNA and that a mutation (U78A) in the yitJ aptamer prevents SAM binding by the aptamer.
Design of a modular ‘ON’ switch from the pbuE adenine riboswitch
Our first effort to create a modular ‘ON’ switch focused on the B. subtilis pbuE riboswitch because it is one of the best-characterized riboswitches to date (47–51). This RNA responds to adenine and its analog 2AP in a single round in vitro transcription assay (47,48). Under our transcription conditions, a T50 (concentration of ligand required to elicit a half-maximal regulatory response in vitro) of 27 ± 6 µM 2AP was observed (‘native’, Figure 1b), which is ∼10-fold higher than that of adenine in a previous report (48). This difference could be in part due to lower Mg2+ concentration in our buffer (2.5 versus 14 mM MgCl2). The observed T50 is also ∼5-fold higher than the KD of the isolated aptamer domain for 2AP (sequence and secondary structure is given in Supplementary Figure S1) under nearly identical solution and temperature conditions (5.6 ± 0.1 µM). Further, the U89A mutation (Figure 1a) that specifically disrupts adenine binding eliminates 2AP-dependent regulation. Placing the pbuE riboswitch downstream of the strong T7A1 promoter for improved transcription yielded a riboswitch with a similar T50 but substantially reduced dynamic range (DR) (12 ± 3 µM and 10%, respectively) despite presumably initiating transcription at the same nucleotide (‘wild type’, Figure 1b). The DR was dramatically improved through deleting the first 11 nt (Δ1–11) from this riboswitch (‘Δ11’, Table 1 and Figure 1b). Notably, the level of transcriptional read through in the absence or low concentrations of ligand (referred to as the RTmin) was lower in the Δ11 variant than either the ‘native’ or ‘wild type’ pbuE riboswitches, an important property for a modular ‘ON’ switch.
Table 1.
Regulatory activity of chimeric ‘ON’ riboswitches
| Ligand | Aptamer | KD, µM | Expression platform | T50 (µM) | DR (%) | RTmin (%) |
|---|---|---|---|---|---|---|
| pbuE expression platform derived module | ||||||
| SAM | yitJ | 1.5 ± 0.3a | pbuE* | 1.2 ± 0.2 | 49 | 1 |
| pbuE*(ΔU126) | 1.6 ± 0.1 | 76 | 1 | |||
| pbuE*(ΔU126–127) | 2.1 ± 0.1 | 86 | 1 | |||
| pbuE*(ΔU126–128) | 2.9 ± 0.7 | 31 | 19 | |||
| 2AP | xpt(C74U) | 8.8 ± 0.8a | pbuE* | 22 ± 10 | 4 | 5 |
| pbuE*(ΔU126) | 12 ± 3 | 11 | 4 | |||
| pbuE*(ΔU126–127) | 18 ± 3 | 25 | 5 | |||
| pbuE*(ΔU126–128) | 28 ± 2 | 47 | 8 | |||
| 2AP | pbuE | 5.6 ± 0.1b | pbuE* | 9.9 ± 1 | 49 | 8 |
| pbuE*(ΔU126) | 90 ± 32 | 20 | 5 | |||
| pbuE*(ΔU126–127) | 24 ± 7 | 36 | 6 | |||
| pbuE*(ΔU126–128) | 20 ± 4 | 48 | 10 | |||
| Guanine | xpt | 0.024 ± 0.003a | pbuE*(ΔU126) | 0.039 ± 0.001 | 58 | 1 |
| pbuE*(ΔU126–127) | 0.041 ± 0.003 | 70 | 1 | |||
| FMN | ribD | 0.91 ± 0.1a | pbuE*(ΔU126) | 0.91 ± 0.06 | 50 | 1 |
| pbuE*(ΔU126–127) | 1.2 ± 0.1 | 55 | 3 | |||
| Lysine | lysC | 18 ± 2a | pbuE*( | 64 ± 20 | 10 | 16 |
| pbuE*(ΔU126) | 37 ± 8 | 17 | 17 | |||
| pbuE*(ΔU126–127) | 26 ± 4 | 25 | 15 | |||
| pbuE*(ΔU126–128) | 36 ± 3 | 26 | 32 | |||
| Theophylline | SELEX | 20 ± 6a | pbuE*(ΔU126) | 1.2 ± 0.1 | 50 | 12 |
| pbuE*(ΔU126–127) | 7.5 ± 2.0 | 63 | 9 | |||
| pbuE*(ΔU126) | 1.2 ± 0.1 | 50 | 12 | |||
| metH expression platform derived module | ||||||
| SAH | meth | 0.57 ± 0.01b | metH | 0.44 ± 0.1 | 23 | 32 |
| SAH | sahH | —c | metH | 0.21 ± 0.06 | 37 | 49 |
| SAH | sahH | — | metH* | 0.10 ± 0.01 | 64 | 25 |
| SAM | yitJ | 1.5 ± 0.3 | metH‡ | n.d.d | n.d. | n.d. |
| 2AP | xpt(C74U) | 8.8 ± 0.8 | metH‡ | 20 ± 2 | 45 | 11 |
| Guanine | xpt | 0.024 ± 0.003 | metH‡ | 0.020 ± 0.001 | 54 | 12 |
| FMN | ribD | 0.91 ± 0.1 | metH‡ | n.d. | n.d. | n.d. |
| Lysine | lysC | 18 ± 2 | metH‡ | n.d. | n.d. | n.d. |
| Tetracycline | SELEX | n.d. | metH‡ | 0.26 ± 0.02 | 41 | 40 |
aThis value is taken from (19).
bThese values are for measurements taken in 70 mM K-HEPES to be consistent with previous measurements; in 70 mM Tris–HCl, the values are 4.2 ± 0.1 and 0.52 ± 0.04 µM for pbuE and metH, respectively.
c—, not determined.
dn.d.; not detectable.
To decouple the two domains of pbuE, a short sequence was introduced into the RNA on the 5′-side of the aptamer domain. U32-G34, which is part of the P1 helix but whose identity is not required for ligand binding to the aptamer domain, was replaced by a 6-nt sequence (5′-GAUUUU) complementary to the extreme 5′-side of the predicted terminator hairpin (Figure 1c, cyan). This additional sequence blocks the rho-independent terminator element from invading into the aptamer domain. This design is conceptually similar to the modular ‘OFF’ switches in which sequence elements of the P1 helix required for ligand binding to the aptamer domain are nonoverlapping with the sequences that participate in the regulatory switch (21). For simplicity, we refer to this modified expression platform that incorporates both the Δ11 mutation and the decoupler sequence as pbuE*.
To test the modularity of the redesigned pbuE* expression platform, we examined ligand-dependent regulatory activity of chimeric riboswitches that incorporate biological or synthetic aptamers (Table 1). These aptamers were chosen because they were previously shown to function within chimeric ‘OFF’ riboswitches (21). For each aptamer, the boundary defining the minimal sequence is based on structural, biochemical and phylogenetic considerations of nucleotide identities in each that are required for function (21). The boundary between each aptamer and the pbuE* expression platform is shown by a dashed line in Figure 1c.
Evaluation of the performance of chimeras with the pbuE* switch by in vitro transcription revealed only one aptamer capable of directing the regulatory switch. The B. subtilis yitJ SAM-sensing aptamer achieved ∼50% read through at saturating SAM concentrations with a T50 of 1.2 ± 0.2 µM (Figure 1d). The observed T50 of the yitJ/pbuE* chimera (artificial riboswitches are named as the aptamer domain/expression platform combination) is consistent with the wild-type yitJ riboswitch and chimeras between the yitJ aptamer and three different ‘OFF’ expression platforms (21,52). Further, a single point mutation in the SAM-binding pocket of the SAM-I riboswitch aptamer [U78A (53,54)] that abrogates binding of SAM eliminates ligand-dependent regulation (inset, Figure 1d). Unfortunately, all other aptamers performed much more poorly. The pbuE and xpt(C74U) purine aptamers yielded a barely detectable increase in read through transcript (∼5% increase; Figure 1d), while others (ribD and xpt) displayed no effector-dependent regulation (data not shown).
All of the inactive chimeras almost completely terminate transcription regardless of effector concentration indicating that the rho-independent terminator is constitutively formed. The relative thermodynamic stability of alternative secondary structures (P1 and P-T in pbuE*) is a central feature of the encoded co-transcriptional folding mechanism governing the switching behavior of the modular ‘OFF’ switches (21,55). Altering P1 helix stability proved highly effective in adjusting the performance of natural and chimeric ‘OFF’ riboswitches (21). Stabilization of the P1 helix by converting a C-C mismatch located within this element to a Watson–Crick pair (P1’, Figure 2a) improved the DR of several chimeras, but at the expense of increased background read through in the absence of ligand (RTmin) from 1 to 5% with the wild-type P1 helix to 10–45% with the P1’ composition for some chimeras (Figure 2b). Thus, stabilizing the P1 helix may not be a productive route to a switching module that has both low read through transcription in the absence of ligand and a large DR.
Figure 2.
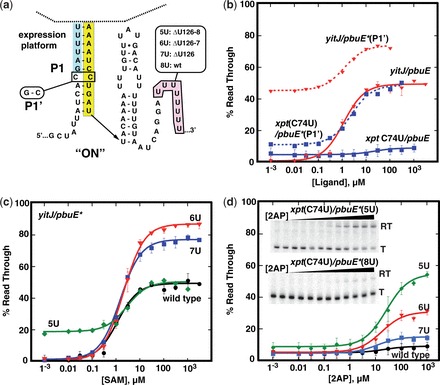
Optimizing performance of the pbuE* expression platform. (a) Secondary structure of the core switching region of the pbuE* expression platform with sites of mutations introduced. (b) Graphical representation of quantified data from in vitro transcription assays of the C31G mutation for yitJ and xpt(C74U) aptamers fused to pbuE* expression platform. (c) Quantified data from in vitro transcription as a function of SAM concentration of yitJ/pbuE* chimeras incorporating deletions of 0, 1, 2 or 3 uridines from the poly-uridine tract. (d) Quantified percentage read through transcription of xpt(C74U)/pbuE* chimeras as a function of 2AP concentration. The insets show two representative denature gels of transcription of chimeras with the wild-type expression platform (bottom) and the deletion of three uridines (top). The data plotted is the average of three independent experiments, and the error bars represent the uncertainty in the measurement.
An alternative strategy explored was to reduce the termination efficiency of the rho-independent terminator by weakening the effect of the uridine-rich region on the 3′-side of P-T (56). Deletion of uridines from this tract (Figure 2a) has the effect of reducing the number of A-U pairs at the base of P-T and/or reducing intrinsic pausing of the polymerase at this site and therefore termination efficiency (57). Strikingly, deletion of one or two uridines (ΔU126 or ‘7U’ and ΔU126-127 or ‘6U’, Figure 2c) resulted in a greatly improved response of all chimeric riboswitches. The yitJ/pbuE* chimera's response improved to 76 and 86% DR from the original 50% change with the deletion of one and two uridines, respectively. Furthermore, read through levels of transcription in the absence of ligand remains unchanged. The T50s of these chimeras are not altered significantly, revealing that this region of the riboswitch only influences the DR properties of the regulatory element. Removal of an additional uridine (ΔU126–128 or ‘5U’) did not yield a greater DR in part owing to increased read through transcript in the absence of ligand.
Alteration of the uridine-rich tract improved the performance of all tested chimeric riboswitches. In the context of the xpt(C74U)/pbuE* and pbuE/pbuE* chimeras, the ΔU126–128 mutation yielded a stronger increase in the read through product by 46 and 49%, respectively, at saturating 2AP concentrations (Table 1, Figure 2d and Supplementary Figure S2). The observed T50s were 20 ± 4 µM for xpt(C74U)/pbuE*, consistent with other chimeras incorporating this aptamer (21), and 28 ± 2 µM for pbuE/pbuE*. As noted above, the ΔU126–128 mutation increases the level of read through in the absence of ligand from 1% of the wild-type terminator to 10%, which could negatively impact the uses of these devices for some applications that require low signal in the absence of the appropriate effector. Other chimeras (xpt/pbuE*, ribD/pbuE* and theo/pbuE*) display similar improvements in performance in the context of the ΔU126 and ΔU126–127 mutations (Table 1 and Figure 3). Together, these data demonstrate that the pbuE* expression platform can host a variety of aptamers to create novel regulatory ‘ON’ devices in a mix-and-match fashion.
Figure 3.
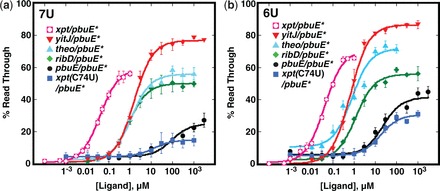
Activity of chimeric riboswitches with the pbuE* expression platform module. (a) In vitro transcription of various chimeric riboswitches as a function of their cognate effector ligand. Each chimera incorporates the pbuE* module with the ΔU126 (7U) mutation that affects termination efficiency. The data plotted are the averages of three independent experiments, and the standard deviation of these measurements are represented by the error bars. (b) Titrations of the same set of chimeras but with the ΔU126–127 (6U) mutation in the pbuE* expression platform. Fitted values for T50 and %RTmin are given in Table 1.
Another highly desirable property of a composable regulatory switch is the ability to host synthetic aptamers derived from in vitro selection (SELEX). Chimeras incorporating the theophylline aptamer (58) efficiently promote read through transcription in the presence of theophylline with comparable performance to the best of the natural aptamers (Table 1 and Figure 3). Further, the observed T50s of the ΔU126 and Δ126–127 variants represent an improvement over those of the theophylline aptamer in ‘OFF’ chimeras by one to two orders of magnitude [1.2 µM for the best ‘ON’ switch versus 13 µM for the best ‘OFF’ switch (21)].
Redesigning the SAH riboswitch
Toward the goal of developing a robust toolbox of riboswitch modules and further demonstrate the generality of this approach, a second ‘ON’ regulatory switch derived from the Dechloromonas aromatica SAH riboswitch expression platform was designed. Along with the challenge of overlapping sequence requirements of the aptamer domain and the expression platform, the helix at the domain boundary is involved in a pseudoknot (P4, Figure 4a). This configuration hinders the central design strategy of splitting the two domains within a single shared P1 helix. Thus, the secondary structure of the expression platform needed to be reconfigured to eliminate the pseudoknot by converting P4 into a conventional P1-type helix (sites of the break denoted by arrowheads in Figure 4a). Of the several SAH riboswitches tested by in vitro transcription, only the wild-type D. aromatica metH riboswitch exhibits a small SAH-dependent activation of read through transcription (metH, Figure 4b). A hybrid SAH riboswitch comprising the aptamer domain sequence from P. aeruginosa PAO1 sahH fused to the D. aromatica expression platform (sahH/metH) yielded an improved DR at elevated SAH concentrations (DR = 37%). However, this chimera is not capable of terminating transcription robustly in the absence of SAH (RTmin = 49%).
Figure 4.
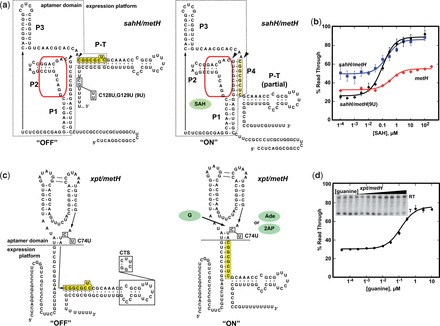
Reengineering the SAH-dependent metH riboswitch. (a) Sequence and secondary structure of the ‘OFF’ and ‘ON’ states of sahH/metH riboswitch. Poly-uridine tract mutations made in this study are denoted in the structure on the left; the yellow box denotes sequence overlap between the aptamer domain and expression platform. The arrowheads denote the break points for splicing in foreign aptamers (see panel c). (b) Activity of the native D. aromatica metH riboswitch (metH, red), the sahH/metH chimeric riboswitch (blue) and the sahH/metH* chimera incorporating the G128U,C129U mutation (black). (c) Sequence and secondary structure of the chimeric xpt/metH* riboswitch in the ‘OFF’ and ‘ON’ states. The dashed line denotes the boundary between the two domains that is used for making all metH chimeras. The C74U mutation that changes the aptamers selectivity from guanine to adenine and 2AP is denoted along with the ‘CTS’ mutation used for in vivo analysis of this riboswitch (Figure 6) is denoted. (d) Quantified activity assay of the xpt/metH* riboswitch with representative gel of transcription products shown in the inset.
To improve the performance of the metH expression platform, we sought to reduce the amount of read through transcription in the absence of effector. It was observed that the uridine-rich region of the terminator element is interrupted by two nonuridine nucleotides (C170, G171; Figure 4a) that may reduce pause time efficiency of the RNAP and thus proper switching (57,59). A mutation creating a nine-uridine (‘9U’) tract resulted in a chimera that showed increased termination efficiency in the absence of SAH (RTmin = 25%) and improved DR (64%) (Figure 4b). This expression platform containing the C170U, G171U mutation is referred to as the metH* module.
To be able to fuse alternative aptamer domains to the metH switch, the secondary structure of the riboswitch needed to be reorganized. Instead of using the aptamer-expression platform boundary as defined by the minimal SAH-binding sequence (60,61), the sequence was split on the 3′-side of the P4 helix that is part of the pseudoknot motif along with P1 (arrowheads, Figure 4a). This converts P4 into the equivalent of the P1 helix in pbuE. The guanine-responsive xpt aptamer was fused to the expression platform on the 3'-side of the P4 helix, resulting in a ‘P1’ helix consisting of 8 bp (Figure 4c) that competes with formation of the terminator element (P-T). This chimera promotes read through transcription in the presence of increasing concentrations of guanine (Figure 4d), but still suffers from substantial read through in the absence of ligand.
To suppress read through transcription at low ligand concentrations, a series of mutations on the 5′-side of the modified P1 helix in the context of the xpt/metH* chimera were constructed and tested (Figure 5). These mutations systematically shorten the P1 helix from eight to five Watson–Crick base pairs (Figure 5a). Reducing the length of P1 yielded substantially increased termination in the absence of guanine (Figure 5b). These data are consistent with observations in modular ‘OFF’ switches in which variation of the length of the P1 helix converted chimeras that exhibited strong constitutive termination in the absence of ligand to robust ligand-dependent transcriptional regulators. These observations reinforce the idea that the stability of the P1 helix is the central feature of the riboswitch (21). The ‘P1(-3)’ mutant was incorporated into a new expression platform module, metH‡, used for all further studies.
Figure 5.
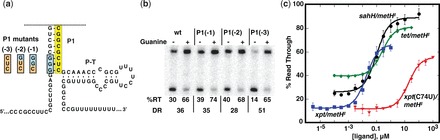
Optimization of metH expression platform performance. (a) Mutations introduced into the metH* platform to repress read through transcription at low effector concentration. Wild-type sequence is highlighted in cyan and mutations [P1(−1), P1(−2) and P1(−3)] in orange. The yellow box denotes the switching sequence. (b) In vitro transcription of xpt/metH* and P1 mutants. The percentage read through product (%RT) in the absence and presence of 1 µM guanine are shown along with the DR. (c) Full titrations of four chimeras of the metH‡ expression platform [xpt, xpt(C74U), tetracycline, sahH] titrated with their cognate effector ligands. The data points represent the average of three independent titrations.
To further interrogate the composability of the metH‡ switch, a series of chimeras was constructed using the same set of aptamers tested for the pbuE* and ‘OFF’ chimeras (21). For those riboswitches exhibiting ligand-dependent transcriptional regulation, the T50s are similar to the values obtained using other switch modules. However, a high level of read through transcription was observed in the absence of the cognate ligand for some aptamer pairings with the metH‡ switch, indicating that further increasing the strength of the terminator may be required to optimize performance. Also, some of the aptamers that worked in the context of other expression platforms failed to regulate when coupled to metH‡: yitJ, ribD and lysC. The reason for these failures is unknown, but does indicate the metH‡ module is less composable and/or reliable than the others. The only commonality between the aptamers that successfully regulated the metH‡ module is that they are three-way junction motifs where the ligand binds to the junction. Nonetheless, these data illustrate that even riboswitches that appear recalcitrant to reengineering can yield a modular RNA switch.
Modular ‘ON’ chimeras function in vivo
While the pbuE* and metH‡ switch modules function under control of a variety of aptamers in vitro, this does not guarantee that they operate in a cellular context. To test the ability of select chimeras to regulate gene expression in E. coli, we used the same GFPuv reporter assay used previously to validate in vivo function of chimeric ‘OFF’ switches (21). Chimeric riboswitches incorporating the 2AP-responsive xpt(C74U) aptamer were incorporated into the 5’-leader sequence of a gfpuv gene and their activity was monitored by measuring changes in GFPuv expression as a function of 2AP concentration in a defined rich medium.
To directly compare the in vivo performance of chimeras against that of a wild-type riboswitch, a series of pbuE/pbuE* riboswitches were first tested. For all in vivo experiments, the Δ1–11 mutation in the pbuE expression platform was used because it results in a significantly increased DR (Supplementary Figure S3). The wild-type pbuE riboswitch carrying the Δ11 mutation yields an observed EC50 = 38 ± 7 µM 2AP (note that the EC50 reflects the concentration of 2AP added to the media; the intracellular concentration of 2AP may be different owing to bioconcentration, degradation and/or efflux) and an induction factor greater than nine (Figure 6a). Further, the ΔU126 mutation (7U, Figure 6a) that weakens the terminator and significantly improves in vitro performance (Figure 2) has little effect on its in vivo regulatory properties. In contrast, the pbuE/pbuE* switch chimera is moderately functional, with a smaller DR (8U, Figure 6b and Supplementary Figure S4). However, decreasing the uridine tract by one and two nucleotides (7U and 6U, Figure 6b) improved the response, while deletion of three uridines (5U, Figure 6b) led to nearly constitutively activated GFPuv expression. Notably, the best performing pbuE/pbuE* chimera in vivo has an observed EC50 and DR that rival the wild-type pbuE riboswitch, indicating that reengineering only had a small effect on the sensitivity of the riboswitch for 2AP. Almost identical trends were seen with xpt(C74U)/pbuE* chimeras (Figure 6c and Supplementary Figure S5). For each chimera, a single point mutation in the ligand-binding pocket abrogated 2AP-dependent GFPuv expression, indicating that the observed responses are the result of specific binding of 2AP to the aptamer domain. These data reinforce that the strategies used for optimization of riboswitch performance in vitro can also be applied in vivo.
Figure 6.
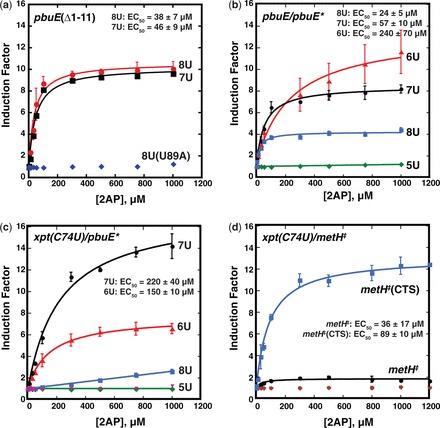
In vivo characterization of pbuE* and metH‡ chimeric riboswitches. (a) Quantification of induction of GFPuv expression as a function of 2AP concentration in a defined medium. The induction factor represents the fold increase in normalized GFP expression of the reporter under control of a riboswitch over the same riboswitch under no ligand conditions. Data was fit to a two-state model and plotted as a function of 2AP to yield the EC50. The U89A mutation (blue diamonds) is a mutant that is defective in 2AP binding as a negative control. (b) Induction of GFP expression by the pbuE/pbuE* chimera (‘8U’, blue) and uridine-rich tract mutations (5U, 6U and 7U). (c) The same series of experiments performed with xpt(C74U)/pbuE* chimeras. The pink triangles represent a U51C mutant of xpt(C74U)/pbuE* that is deficient in ligand binding. (d) Activity of the xpt(C74U)/metH chimera (black) and the ‘CTS’ mutant (blue). Nonbinding mutants are denoted in green and pink as negative controls. The data presented is the average of at least three independent experiments and the standard deviation of the average value is represented as the error bars.
The xpt(C74U)/metH‡ chimera revealed another path for improvement of chimera performance. This chimera yielded only a moderate 2-fold increase in GFPuv expression at high 2AP concentrations. However, encouragingly, the EC50 of this riboswitch is 36 ± 17 µM 2AP, significantly lower than this aptamer in pbuE* or the metE ‘OFF’ switch (730 µM) (21). In these data, we noted that the riboswitch showed substantial expression in the absence of 2AP (Supplementary Figure S6). We hypothesized that by altering the stability and/or folding mechanism of the terminator, this leaky expression could be suppressed. To improve folding of P-T, the capped terminator stem (‘CTS’) mutation (Figure 4c) was introduced into the region of the hairpin that does not participate in the secondary structural switch. This mutation caps the terminator hairpin with a stable UUCG tetraloop and yielded a chimera with a ∼10-fold induction of GFP expression in response to 2AP (EC50 = 89 ± 10 µM, Figure 6d). This mutation improved the performance of this riboswitch by both diminishing expression in the absence of ligand by ∼2-fold and increasing expression in the presence of 2AP by ∼3-fold (Supplementary Figure S6). Thus, similar to pbuE*, performance of chimeras with the metH‡ switch module can be readily improved with rational and directed changes to the structure of the rho-independent terminator.
DISCUSSION
Artificial riboswitches are increasingly viewed as important tools in the synthetic biologist’s toolbox (1,4). In large part, this is owing to the ability to select for RNAs capable of binding small molecules with high affinity and selectivity such as the tetracycline (62) and theophylline (58) aptamers used in this study. However, a significant hurdle faced in engineering practical RNA devices from SELEX-derived aptamers is their coupling to a functional readout (63). In previous work, we demonstrated that three expression platforms from transcriptional ‘OFF’ riboswitches can serve as modular switching domains by using simple design rules that circumvent the need to engineer a communication module (21). In the current study, we have developed two modular transcriptional ‘ON’ switches. To achieve this, a series of alterations were required in the wild-type expression platforms including sequence insertions to decouple the aptamer and expression platform, deletions in the poly-uridine tract to weaken the effect of the rho-independent terminator and alterations to the length of the P1 helix. The resulting ‘ON’ expression platform modules are able to host diverse aptamers using a simple mix-and-match strategy as outlined for the ‘OFF’ chimeras (21). In particular, these data clearly demonstrate that transcriptional riboswitches do not require overlap between the aptamer and the readout domains and that it is possible to fuse terminator elements to aptamers to yield functional riboswitches, in contrast to prior claims (23).
In this study, we illustrate two new extensions of our design strategy for repurposing biological RNA switches. First, we have demonstrated that riboswitches whose sequence requirements for small molecule binding to the aptamer domain and the secondary structural switch overlap can be effectively decoupled. This was achieved in two different secondary structural contexts through introduction of sequences that disallow invasion of the terminator hairpin into the aptamer domain. This enabled the prior ‘mix-and-match’ strategy of appending the minimal P1 helix of the aptamer domain onto the minimized P1 helix of the expression platform (denoted as the dashed line boundary in Figures 1 and 4). The ability to decouple the aptamer domain from the expression platform is important because many natural riboswitches contain substantial sequence overlap between the two domains, as exemplified by the natural pbuE riboswitch. This strategy significantly expands the potential number of biological expression platforms that can be harnessed as modular switches.
The second extension to our strategy relates to optimization of the newly created chimera. As previously observed, the first implementation of the receptor-readout modules can achieve near-optimal performance. However, in some examples, poor performance can readily be corrected; in the ‘OFF’ switches this was primarily achieved by mutations to the 5′-side of the P1 helix to alter the thermodynamic balance between the P1 helix and antiterminator (P-AT) stability (21). Manipulation of the intrinsic terminator strength was not effective because in these switches, formation of P-T is dictated by the P1/P-AT folding outcome. For transcriptional ‘ON’ regulators, because the secondary structural switch comprises the P1 helix and P-T terminator helices, we can manipulate the terminator by either adjusting the stability of P-T or the length of the poly-uridine tract to optimize performance. Thus, our work has presented a simple set of strategies for the creation of a large and diverse set of modular readout domains based on biological riboswitches. Given that only a few expression platforms have been explored, it is highly likely that other expression platforms may be superior, especially given the limitations of the metH‡ module.
From our construction of >50 artificial aptamer–expression platform pairings, several important considerations are emerging. First, most chimeras exhibit T50s close to the KD of the isolated aptamer for the cognate ligand. All five chimeras incorporating the xpt(C74U) aptamer exhibit a T50 of ∼20 µM for 2AP, close to the isolated aptamer’s affinity for 2AP in the same buffer conditions (8.8 ± 0.8 µM). However, this is a property of the in vitro transcription conditions chosen; in vivo the EC50 varies significantly and is always greater than the measured T50. This reflects ‘kinetic control’ of the riboswitch such that RNA folding, effector binding and transcription kinetics considerably influence the EC50 (31,37,49,50,64), although other factors such as influx of ligand into the cell and its metabolic turnover affect this value as well. In contrast, the performance properties of the riboswitch with regard to the minimal and maximal fraction of read through transcription (and the associated DR) are largely controlled by the expression platform. Mutations either to the P1/P-T switch that influences their relative stability or folding kinetics or to the poly-uridine tract modulate these values. For both the ‘OFF’ and ‘ON’ modules, only a limited set of mutations were explored, however. It is highly likely that a comprehensive mutagenic analysis of in vivo activity or computational approaches such as those used to design synthetic riboswitches may yield further improvements to these modules.
The modular transcriptional switches that have been developed represent only a small fraction of the potential of these RNAs. In E. coli and other proteobacteria, most riboswitches control gene expression at the translational level (43). Exploration of these switches may yield even more robust modules with improved performance for these bacteria. Further, development of modules based on riboswitches found in plants and fungi (65–67) would pave the way for the application of these RNA devices in yeast and potentially even mammalian systems. The vast number and diversity of riboswitches and their associated expression platforms that have been identified provides a substantial reservoir of potential candidates for even more robust regulatory switches than the few we have characterized to date.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01 GM073850] and the National Science Foundation [NSF 1150834 to R.T.B.] and through a Creative Training in Molecular Biology [T32 GM07135 to J.J.T.]. Funding for open access charge: National Institutes of Health and National Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the members of the Batey laboratory and Dr. Andrew Garst for providing useful advice and feedback throughout this project and on the manuscript. We would also like to acknowledge the assistance of Christopher G. Bennett in the performance of some of the transcription assays and Joan G. Marcano-Velázquez with cell-based assays.
REFERENCES
- 1.Keasling JD. Synthetic biology and the development of tools for metabolic engineering. Metab. Eng. 2012;14:189–195. doi: 10.1016/j.ymben.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Benenson Y. Synthetic biology with RNA: progress report. Curr. Opin. Chem. Biol. 2012;16:278–284. doi: 10.1016/j.cbpa.2012.05.192. [DOI] [PubMed] [Google Scholar]
- 3.Wieland M, Fussenegger M. Ligand-dependent regulatory RNA parts for synthetic biology in eukaryotes. Curr. Opin. Chem. Biol. 2010;21:760–765. doi: 10.1016/j.copbio.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat. Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 5.Wittmann A, Suess B. Engineered riboswitches: expanding researchers' toolbox with synthetic RNA regulators. FEBS Lett. 2012;586:2076–2083. doi: 10.1016/j.febslet.2012.02.038. [DOI] [PubMed] [Google Scholar]
- 6.Topp S, Gallivan JP. Emerging applications of riboswitches in chemical biology. ACS Chem. Biol. 2010;5:139–148. doi: 10.1021/cb900278x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 8.Joyce GF. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 9.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 2012;4:a003566. doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garst AD, Edwards AL, Batey RT. Riboswitches: structures and mechanisms. Cold Spring Harb. Perspect. Biol. 2011;3:a003533. doi: 10.1101/cshperspect.a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowler CC, Brown ED, Li Y. Using a riboswitch sensor to examine coenzyme B(12) metabolism and transport in E. coli. Chem. Biol. 2010;17:756–765. doi: 10.1016/j.chembiol.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 12.Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat. Chem. Biol. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Michener JK, Smolke CD. High-throughput enzyme evolution in Saccharomyces cerevisiae using a synthetic RNA switch. Metab. Eng. 2012;14:306–316. doi: 10.1016/j.ymben.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Topp S, Reynoso CM, Seeliger JC, Goldlust IS, Desai SK, Murat D, Shen A, Puri AW, Komeili A, Bertozzi CR, et al. Synthetic riboswitches that induce gene expression in diverse bacterial species. Appl. Environ. Microbiol. 2010;76:7881–7884. doi: 10.1128/AEM.01537-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muranaka N, Abe K, Yokobayashi Y. Mechanism-guided library design and dual genetic selection of synthetic OFF riboswitches. Chembiochem. 2009;10:2375–2381. doi: 10.1002/cbic.200900313. [DOI] [PubMed] [Google Scholar]
- 16.Suess B, Fink B, Berens C, Stentz R, Hillen W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desai SK, Gallivan JP. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J. Am. Chem. Soc. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- 18.Hanson S, Berthelot K, Fink B, McCarthy JE, Suess B. Tetracycline-aptamer-mediated translational regulation in yeast. Mol. Microbiol. 2003;49:1627–1637. doi: 10.1046/j.1365-2958.2003.03656.x. [DOI] [PubMed] [Google Scholar]
- 19.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl Acad. Sci. USA. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ceres P, Garst AD, Marcano-Velázquez JG, Batey RT. Modularity of select riboswitch expression platform enables facile engineering of novel genetic regulatory devices. ACS Synth. Biol. 2013;2:463–472. doi: 10.1021/sb4000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dawid A, Cayrol B, Isambert H. RNA synthetic biology inspired from bacteria: construction of transcription attenuators under antisense regulation. Phys. Biol. 2009;6:025007. doi: 10.1088/1478-3975/6/2/025007. [DOI] [PubMed] [Google Scholar]
- 23.Wachsmuth M, Findeiss S, Weissheimer N, Stadler PF, Morl M. De novo design of a synthetic riboswitch that regulates transcription termination. Nucleic Acids Res. 2013;41:2541–2551. doi: 10.1093/nar/gks1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 25.Qi L, Lucks JB, Liu CC, Mutalik VK, Arkin AP. Engineering naturally occurring trans-acting non-coding RNAs to sense molecular signals. Nucleic Acids Res. 2012;40:5775–5786. doi: 10.1093/nar/gks168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucks JB, Qi L, Mutalik VK, Wang D, Arkin AP. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc. Natl Acad. Sci. USA. 2011;108:8617–8622. doi: 10.1073/pnas.1015741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi M, Lucks JB. A modular strategy for engineering orthogonal chimeric RNA transcription regulators. Nucleic Acids Res. 2013;41:7577–7588. doi: 10.1093/nar/gkt452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinha J, Topp S, Gallivan JP. From SELEX to cell dual selections for synthetic riboswitches. Methods Enzymol. 2011;497:207–220. doi: 10.1016/B978-0-12-385075-1.00009-3. [DOI] [PubMed] [Google Scholar]
- 29.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wittmann A, Suess B. Selection of tetracycline inducible self-cleaving ribozymes as synthetic devices for gene regulation in yeast. Mol. Biosyst. 2011;7:2419–2427. doi: 10.1039/c1mb05070b. [DOI] [PubMed] [Google Scholar]
- 31.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 33.Prodromou C, Pearl LH. Recursive PCR: a novel technique for total gene synthesis. Protein Eng. 1992;5:827–829. doi: 10.1093/protein/5.8.827. [DOI] [PubMed] [Google Scholar]
- 34.Kadesch TR, Rosenberg S, Chamberlin MJ. Binding of Escherichia coli RNA polymerase holoenzyme to bacteriophage T7 DNA. Measurements of binding at bacteriophage T7 promoter A1 using a template competition assay. J. Mol. Biol. 1982;155:1–29. doi: 10.1016/0022-2836(82)90489-2. [DOI] [PubMed] [Google Scholar]
- 35.Artsimovitch I, Henkin TM. In vitro approaches to analysis of transcription termination. Methods. 2009;47:37–43. doi: 10.1016/j.ymeth.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dann CE, Wakeman CA, Sieling CL, Baker SB, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 37.Garst AD, Porter EB, Batey RT. Insights into the regulatory landscape of the lysine riboswitch. J. Mol. Biol. 2012;423:17–33. doi: 10.1016/j.jmb.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilbert SD, Batey RT. Monitoring RNA-ligand interactions using isothermal titration calorimetry. Methods Mol. Biol. 2009;540:97–114. doi: 10.1007/978-1-59745-558-9_8. [DOI] [PubMed] [Google Scholar]
- 39.Broecker J, Vargas C, Keller S. Revisiting the optimal c value for isothermal titration calorimetry. Anal. Biochemi. 2011;418:307–309. doi: 10.1016/j.ab.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 40.Turnbull WB, Daranas AH. On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J. Am. Chem. Soc. 2003;125:14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- 41.de Boer HA, Comstock LJ, Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc. Natl Acad. Sci. USA. 1983;80:21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang JX, Breaker RR. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem. Cell. Biol. 2008;86:157–168. doi: 10.1139/O08-008. [DOI] [PubMed] [Google Scholar]
- 45.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen AG, Sudarsan N, Breaker RR. Mechanism for gene control by a natural allosteric group I ribozyme. RNA. 2011;17:1967–1972. doi: 10.1261/rna.2757311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 48.Lemay JF, Desnoyers G, Blouin S, Heppell B, Bastet L, St-Pierre P, Masse E, Lafontaine DA. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 50.Frieda KL, Block SM. Direct observation of cotranscriptional folding in an adenine riboswitch. Science. 2012;338:397–400. doi: 10.1126/science.1225722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 52.Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. J. Bacteriol. 2008;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu C, Ding F, Chowdhury A, Pradhan V, Tomsic J, Holmes WM, Henkin TM, Ke A. SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch. J. Mol. Biol. 2010;404:803–818. doi: 10.1016/j.jmb.2010.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 55.Xayaphoummine A, Viasnoff V, Harlepp S, Isambert H. Encoding folding paths of RNA switches. Nucleic Acids Res. 2007;35:614–622. doi: 10.1093/nar/gkl1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol. Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 57.Nudler E, Gottesman ME. Transcription termination and anti-termination in E. coli. Genes Cells. 2002;7:755–768. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- 58.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 59.Nudler E, Gusarov I. Analysis of the intrinsic transcription termination mechanism and its control. Methods Enzymol. 2003;371:369–382. doi: 10.1016/S0076-6879(03)71028-3. [DOI] [PubMed] [Google Scholar]
- 60.Edwards AL, Reyes FE, Heroux A, Batey RT. Structural basis for recognition of S-adenosylhomocysteine by riboswitches. RNA. 2010;16:2144–2155. doi: 10.1261/rna.2341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang JX, Lee ER, Morales DR, Lim J, Breaker RR. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol. Cell. 2008;29:691–702. doi: 10.1016/j.molcel.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berens C, Thain A, Schroeder R. A tetracycline-binding RNA aptamer. Bioorg. Med. Chem. 2001;9:2549–2556. doi: 10.1016/s0968-0896(01)00063-3. [DOI] [PubMed] [Google Scholar]
- 63.Suess B, Weigand JE. Engineered riboswitches: overview, problems and trends. RNA Biol. 2008;5:24–29. doi: 10.4161/rna.5.1.5955. [DOI] [PubMed] [Google Scholar]
- 64.Garst AD, Batey RT. A switch in time: detailing the life of a riboswitch. Biochim. Biophys. Acta. 2009;1789:584–591. doi: 10.1016/j.bbagrm.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 66.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


