Abstract
The highly conserved Paf1 complex (PAF1C) plays critical roles in RNA polymerase II transcription elongation and in the regulation of histone modifications. It has also been implicated in other diverse cellular activities, including posttranscriptional events, embryonic development and cell survival and maintenance of embryonic stem cell identity. Here, we report the structure of the human Paf1/Leo1 subcomplex within PAF1C. The overall structure reveals that the Paf1 and Leo1 subunits form a tightly associated heterodimer through antiparallel beta-sheet interactions. Detailed biochemical experiments indicate that Leo1 binds to PAF1C through Paf1 and that the Ctr9 subunit is the key scaffold protein in assembling PAF1C. Furthermore, we show that the Paf1/Leo1 heterodimer is necessary for its binding to histone H3, the histone octamer, and nucleosome in vitro. Our results shed light on the PAF1C assembly process and substrate recognition during various PAF1C-coordinated histone modifications.
INTRODUCTION
In eukaryotes, most DNA is wrapped around a histone octamer to form a nucleosome. Transcription elongation, mediated by RNA polymerase II (Pol II), occurs in the context of nucleosomal arrays within the chromatin; however, nucleosomes may become an obstacle for Pol II passage. During elongation, various specific Pol II-associated factors may facilitate transcriptional elongation and modulate the elongation complex at multiple stages (1). The Paf1 complex (PAF1C) was originally identified in Saccharomyces cerevisiae as an RNA polymerase II-associated factor (PAF) (2) and contributes to multiple aspects of Pol II transcriptional regulation (3,4).
PAF1C is a multifunctional complex comprising the Paf1, Leo1, Ctr9, Cdc73 and Rtf1 subunits in budding yeast (5,6). Human PAF1C contains an additional subunit, Ski8/Wdr61 (7). PAF1C is associated with the promoter and coding sequences of active genes (8,9) and is recruited to chromatin through both Cdc73 and Rtf1 (10–12). PAF1C has been implicated in transcription elongation through physical and functional interactions with various elongation factors, including Spt16-Pob3/FACT, Spt4-Spt5/DSIF and TFIIS (7,13,14).
Beyond its role in transcription elongation, PAF1C has established roles in H2B monoubiquitylation and downstream histone H3 methylation. The recruitment of the Bre1/Rad6 ubiquitin conjugase-ligase complex by PAF1C is essential for H2B monoubiquitylation (15,16), which is a prerequisite for the subsequent methylation of H3K4 and H3K79 by the Set1/COMPASS and Dot1 methyltransferases, respectively (15,17–20). Moreover, various studies have implicated PAF1C function in other diverse cellular processes, including the processing of mRNA and small nucleolar RNA during posttranscriptional events (12,21–23), mammalian embryonic development and survival in adults (24,25) and embryonic stem cell identity maintenance (26). Additional evidence implicates PAFC1 in immunological responses to disease states and cancer progression (27,28).
To date, structural information on PAFC1 is limited. Thus far, the plus 3 domain of Rtf1 (29) and the Ras-like C-domain of Cdc73 (30,31) have been determined. No structural data on PAF1C formation has been reported. In this study, we determine the crystal structure of the human Paf1/Leo1 subcomplex to elucidate the assembly of PAFC1 and the mechanisms underlying its function.
MATERIALS AND METHODS
Expression and purification
To co-express Paf1 and Leo1, DNA fragments corresponding to human Paf1 (full-length residues 1–531) and Leo1 (full-length residues 1–666) were polymerase chain reaction (PCR)-amplified and cloned into an in-house modified version of the pET32a vector (Novagen). ribosome binding sequence (RBS)-Leo1, which contains an RBS on the N-terminus of Leo1, was PCR-amplified from pET32a-Leo1 and inserted into pET32a-Paf1. The resulting proteins contained a Thioredoxin (Trx)-his6 tag on the N-terminus of both Paf1 and Leo1.
The recombinant proteins were expressed in BL21(DE3) Codon Plus Escherichia coli cells at 16°C for 16–18 h. The cells were then lysed by AH-1500 (ATS Engineering Limited). The Trx-His6-tagged protein complex was purified by Ni-NTA affinity chromatography (QIAGEN) followed by size-exclusion chromatography on a HiLoad 26/60 Superdex 200 (GE Healthcare) in 50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA and 1 mM dithiothreitol (DTT). The Trx-His6-tagged complex proteins were further purified on a Hiprep Q FF 16/10 anion-exchange column (GE Healthcare). The final purification step was size-exclusion chromatography on a HiLoad 26/60 Superdex 200 column in 50 mM Tris, pH 8.0, 200 mM NaCl, 1 mM EDTA and 1 mM DTT. The sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of the recombinant Trx-Paf1/Trx-Leo1 complex is shown in Supplementary Figure S1A.
We used similar co-expression strategies to prepare various truncations of the Paf1/Leo1 complex, including residues 57–392 of Paf1 and residues 277–562 of Leo1 (referred to as Paf1(57−392)/Leo1(277−562), similar nomenclature hereafter), Paf1(161−392)/Leo1(355−562), Paf1(161−392)/Leo1(277–562), Paf1(161–375)/Leo1(277–562), Paf1(161–250)/Leo1(277–562), Paf1(161–250)/Leo1(370–477), Paf1(161–250)/Leo1(277–495), Paf1(161–250)/Leo1(370–495) and Paf1(161–250)/Leo1(370–462). All of the expression and purification processes were similar to the procedures for the full-length complex. The SDS-PAGE analyses of all recombinant protein complexes are shown in Supplementary Figures S1 and S2.
To make a single-chain fusion protein of the Paf1(161–250)/Leo1(370–462) complex (hereafter named Paf1(161–250)-Leo1(370–462)), DNA fragments corresponding to the Paf1 residues 161–250 (Paf1(161–250)) and Leo1 residues 370–462 (Leo1(370–462)) were amplified by PCR and linked with a tobacco etch virus (TEV) protease-cleavable segment (Glu-Asn-Leu-Tyr-Phe-Gln-Ser). Two amino acids (Ser-Gly) were inserted before the TEV protease-cleavable segment. The single open reading frame was cloned into an in-house modified version of the pET32a vector. The resulting protein contained a Trx-His6 tag on its N-terminus.
The recombinant protein was expressed in BL21(DE3) Codon Plus E. coli cells at 16°C for 16–18 h. The Trx-His6-tagged protein was purified by Ni-NTA affinity chromatography, followed by size-exclusion chromatography on a HiLoad 26/60 Superdex 200 column in 50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA and 1 mM DTT. After digestion with PreScission Protease to cleave the N-terminal Trx-His6 tag, the target protein was purified on a Hiprep Q FF 16/10 anion-exchange column. The final purification step was size-exclusion chromatography on a HiLoad 26/60 Superdex 200 column in 20 mM N-2-Hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), pH 7.5, 50 mM NaCl, 1 mM EDTA and 1 mM DTT.
Crystallization and data collection
Wild-type Paf1(161–250)-Leo1(370–462) crystals were grown at 20°C using the sitting-drop vapor-diffusion method. The reservoir solution contained 3.5 M sodium formate, pH 7.0. Initial crystals were needle-shaped and unsuitable for diffraction. We used several additives for further screening and found that AdoMet or cyclic AMP could significantly improve crystal formation. Finally, sitting drops of 3 μl total volume were prepared from 1 μl Paf1(161–250)-Leo1(370–462) protein (30 mg/ml, 20 mM HEPES, pH 7.5, 50 mM NaCl, 1 mM DTT and 1 mM EDTA) mixed with 1 μl of reservoir solution and 1 μl AdoMet (20 mg/ml). Tetragonal crystals appeared within 1 week. Diffraction data were collected on beam line BL17U1 of the Shanghai Synchrotron Radiation Facility (SSRF). The data were processed and scaled to 2.5 Å using the HKL2000 software package (32). The Paf1(161–250)-Leo1(370–462) crystal belongs to space group R32 with unit cell dimensions a = b =115.2 Å, c = 157.4 Å.
Se-Met-substituted protein was expressed in B834 cells using LeMaster medium. The purification and crystallization conditions were the same as those described above, except the reservoir solution was supplemented with 4% 1,4-butanediol. The single anomalous dispersion (SAD) data set for the Se-Met-substituted Paf1(161–250)-Leo1(370–462) crystal was collected at the peak wavelength for Se on beam line BL17U1 of the SSRF. The anomalous signal data set was processed and scaled to 2.8 Å using the HKL2000 software package (32).
Structure determination and refinement
The HKL2MAP program (33) yielded eight out of 10 theoretical Se sites. Initial SAD phases were then calculated using PHENIX software (34). The model was built manually using the COOT program (35). After the initial alanine model was built, iterative refinement was performed against the wild-type data to assign all side chains. After several refinement cycles with the PHENIX program (34) and COOT (35), the orientations of amino acid side chains and bound water molecules were modeled on the basis of 2Fo–Fc and Fo–Fc difference Fourier maps. The final structure had an Rcryst value of 23.2% and an Rfree value of 26.8%. The Ramachandran plot calculated by the PROCHECK program (36) showed that 93.5% of the residues were in their most favored regions, 6.5% of the residues were in additionally allowed regions and no residues were in generously allowed regions or disallowed regions. Detailed data collection and refinement statistics are summarized in Supplementary Table S1.
Glutathione S-transferase pull-down assays
H3(1−28)-glutathione S-transferase (GST) and H3(1–28)(K4A, R17A)-GST fusion proteins were expressed in E. coli BL21(DE3) Codon Plus cells and purified using a glutathione-Sepharose 4B column (GE Healthcare) and a HiLoad 26/60 Superdex 200 column in 50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA and 1 mM DTT. Transfected HEK293T cells were lysed using 1 ml of ice-cold cell lysis buffer [50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 3% glycerol, 0.5% NP40, 0.5% Triton X-100 and 1 mM phenyl-methylsulphonylfluoride (PMSF)] and cleared by centrifugation at 13 000 rpm for 30 min at 4°C. Soluble fractions were incubated with GST-fusion proteins coupled to glutathione-Sepharose 4B beads at 4°C for 2 h. The beads were washed with cell lysis buffer three times and boiled in SDS sample buffer. The prepared samples were then separated by SDS-PAGE and analyzed by western blots.
GST-Paf1/Trx-Leo1 fusion proteins were expressed in E. coli BL21 (DE3) Codon Plus cells and purified using a glutathione-Sepharose 4B column (GE Healthcare) and a HiLoad 26/60 Superdex 200 column in 50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA and 1 mM DTT. Human Histone octamer was prepared according to a modified method (37). GST fusion proteins were incubated with glutathione-Sepharose 4B beads at 4°C for 2 h. The beads were washed three times with the buffer (50 mM Tris–HCl, pH 8.0, 100 mM NaCl and 5% glycerol). Histone octamer was incubated with GST fusion proteins coupled to glutathione-Sepharose 4B beads at 4°C for 2 h. The beads were washed three times with the buffer (50 mM Tris–HCl, pH 8.0, 100 mM NaCl and 5% glycerol). The prepared samples were then separated by SDS-PAGE and the gel was stained with Coomassie blue.
Co-Immunoprecipitation
HEK293T cells were transfected with the indicated combinations of plasmids. At 24 h after transfection, HEK293T cells were lysed using 1 ml of ice-cold cell lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 3% glycerol, 0.5% NP40, 0.5% Triton X-100 and 1 mM PMSF) and cleared by centrifugation at 13 000 rpm for 30 min at 4°C. The supernatants were then incubated with argarose conjugated anti-Myc (clone PL14, Medical & Biological Laboratories) for 2 h at 4°C. The argarose beads were washed three times with cell lysis buffer and eluted with SDS sample buffer. Samples were then subjected to SDS-PAGE and western blot analysis.
Dot-blot overlay assay
Recombinant human histone octamer and nucleosome were expressed, purified and assembled with a 147-bp DNA sequence as described previously [Supplementary Figure S6B; (37)]. The purified Paf1/Leo1, Paf1/Leo1(370−666), Paf1(1–375)/Leo1 and Paf1(1–375)/Leo1(370–666) complex proteins were stored in buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA and 1 mM DTT). Each recombinant protein was first resolved on a 12% SDS-PAGE gel to normalize the sample inputs before the proteins were absorbed onto polyvinylidene difluoride (PVDF) membranes (Millipore). The membranes were subsequently blocked with 10% nonfat milk in TBST (50 mM Tris–HCl, pH 7.4, 150 mM NaCl and 0.1% Tween 20) for 1 h. The membranes were then incubated with 10 μg/ml histone octamer or nucleosome in TBST at room temperature for 1 h. After extensive washing, the membrane-bound histone octamer or nucleosome core particle was immunoblotted using anti-H3 antibody. Finally, the membrane was stained using Coomassie blue to confirm the sample loading.
Analytical gel filtration
Size-exclusion chromatography was performed on an AKTA FPLC system using a Superose 12 10/300 column (GE Healthcare) for the single-chain fusion protein Paf1(161–250)-Leo1(370–462) and a Superdex 200 10/300 column (GE Healthcare) for the other Paf1/Leo1 complex proteins shown in Supplementary Figures S1 and S2. The column was equilibrated in buffer containing 50 mM Tris, pH 8.0, 200 mM NaCl, 1 mM EDTA and 1 mM DTT.
Analytical ultracentrifugation
Sedimentation velocity (SV) experiments were performed in a Beckman Coulter XL-I analytical ultracentrifuge (Beckman Coulter) using double sector centerpieces and sapphirine windows. SV experiments were conducted at 42 000 rpm and 4°C using interference detection for Paf1(161–250)-Leo1(370–462), 40 000 rpm for Paf1(57–392)/Leo1(277–562) and 36 000 rpm for Trx-Paf1/Trx-Leo1. The SV data was analyzed using the SEDFIT program (38,39).
RESULTS
Interaction between Paf1 and Leo1
Previous studies have shown that in human PAF1C, Paf1 and Leo1 form a binary subcomplex (7,19). We first confirmed the Paf1/Leo1 interaction by showing that two Thioredoxin (Trx)-tagged fusion proteins (Trx-Paf1 and Trx-Leo1) form a heteromeric complex with a stoichiometry of 1:1 in the analytical size-exclusion column (Supplementary Figure S1A). Analytical ultracentrifugation further confirmed that the Paf1/Leo1 subcomplex assembles into a heterodimer with a molecular mass of ∼172 kDa (Supplementary Figure S1D). The Paf1/Leo1 subcomplex protein, however, is unstable (SDS-PAGE insert in Supplementary Figure S1A). Using a combination of limited proteolysis followed by size exclusion and mass spectrum fingerprinting, we identified a region in Paf1 (aa 57–392, Paf1(57–392)) and a region in Leo1 (aa 277–562, Leo1(277–562)) that also form a 1:1 stoichiometric heterodimer complex (Supplementary Figures S1B and D). Using a similar truncation-based approach, we mapped the minimal Leo1-binding region of Paf1 to a 90-residue fragment (aa 161–250, Paf1(161−250)) and the minimal Paf1-binding region of Leo1 to a 93-residue fragment (aa 370–462, Leo1(370−462)) (Figure 1A and B, and Supplementary Figure S2I). Given these results, we conclude that the Paf1/Leo1 subcomplex forms a heterodimer through interaction between fragments Paf1(161–250) and Leo1(370–462).
Figure 1.
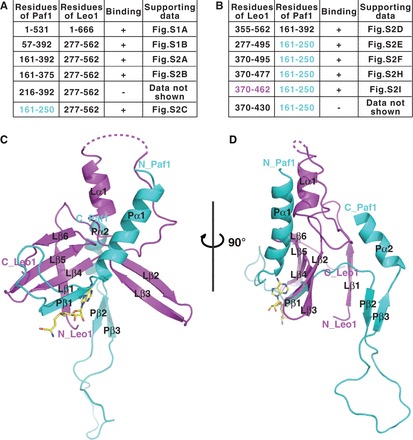
Crystal structure of the human Paf1/Leo1 subcomplex. (A and B) Gel filtration-based test of the binding of Paf1 and Leo1 by the co-expression of target proteins. ‘+’ indicates protein binding, according to the supporting data in Supplementary Figures S1 and S2; ‘−’ indicates no protein binding. The protein fragments of the complex used for structural determination were Paf1(161–250) colored in cyan, and Leo1(370–462) colored in magenta. (C) A cartoon representation of the structure of the Paf1(161–250)-Leo1(370–462) heterodimer complex. Paf1 is shown in cyan, and Leo1 is shown in magenta. The bound AdoMet is shown as a stick model and colored in yellow. (D) A side view of the Paf1(161–250)-Leo1(370–462) heterodimer complex. The N- and C-termini of the two proteins are labeled. For clarity, the β-strand formed by the artificial linker is not shown.
Overall structure of the Paf1/Leo1 heterodimer
To understand how Paf1 and Leo1 bind to each other, we attempted to determine the crystal structure of the Paf1(161–250) and Leo1(370–462) heterodimer using two separate chains; these experiments were unsuccessful. However, we succeeded in obtaining crystals of the single polypeptide created by the fusion of Paf1(161–250) to the N-terminus of Leo1(370−462) with a TEV-cleavable segment. The purified single-chain fusion protein of Paf1(161−250) and Leo1(370−462) (herein named Paf1(161–250)-Leo1(370–462)) eluted as a single peak from an analytical size-exclusion column and assembled into a heterodimer with a molecular mass of ∼25 kDa from the SV experiment (Supplementary Figure S1C and D). The crystal structure of Paf1(161–250)-Leo1(370–462) was determined at a resolution of 2.5 Å and with one molecule per asymmetric unit (Supplementary Table S1). S-adenosyl-L-methionine (AdoMet) was added into the crystallization solution to facilitate the formation of the crystal complex. In the final model, all residues are clearly visible, except residues 399–406 of Leo1(370−462), presumably owing to conformational flexibility (Figure 1C and D). The entire complex was butterfly shaped, with the Leo1 segment encompassed by the Paf1 segment. The Paf1(161−250) segment consists of two α-helices (Pα1-Pα2) and three β-strands (Pβ1-Pβ3), while the Leo1(370−462) segment consists of one α-helix (Lα1) and six β-strands (Lβ1-Lβ6).
Part of an AdoMet molecule is visible and modeled based on the 2Fo-Fc electron density map (Figure 1C and D). A detailed structural analysis shows that one AdoMet molecule has several interactions with two symmetry-related molecules of the Paf1(161−250)/Leo1(370−462) heterodimer in the crystal (Supplementary Figure S3A). The adenine moiety of AdoMet is stabilized by the hydrophobic pocket formed by Paf1 in one molecule, which consists of residues V204, P206 and I190 (Supplementary Figure S3B and C). Moreover, the N6 amino group of the adenine moiety forms a hydrogen bond with the main-chain carboxyl group of K188 and A186 in the same molecule, while the N3 and N9 amines form hydrogen bonds with the main-chain carboxyl group of E428 of Leo1 in the other symmetry-related molecule, and the O2 of the ribose moiety forms a hydrogen bond with the side-chain carboxyl group OE1 of E428.
Paf1 and Leo1 interactions in the heterodimer
In the structure of the Paf1(161−250)-Leo1(370−462) heterodimer, Pβ1, Lβ4, Lβ5 and Lβ6 form a set of antiparallel β-sheets, which are the main interactions between Paf1 and Leo1. The Pβ1 strand from Paf1 lies antiparallel to the Lβ4 strand from Leo1 to form multiple main-chain hydrogen bonds (Figure 2A). Moreover, K442 from Lβ4 forms an ionic bond with the side-chain atom of E203 on Pβ1. It was noted that the TEV-cleavable linker forms another antiparallel β-sheet with the N-terminal Lβ1 of Leo1 (‘TEV linker’-Lβ1antiparallel sheet) (Supplementary Figure S4).
Figure 2.
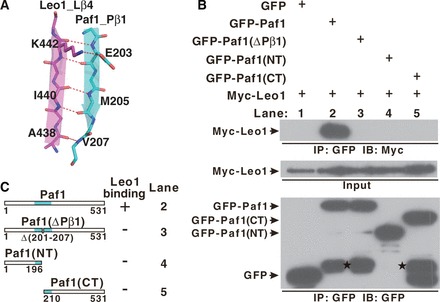
The interaction between Paf1 and Leo1. (A) The interface details in the Paf1(161–250)-Leo1(370–462) heterodimer. The β1 strand of Paf1 (Pβ1) is colored in cyan, and the β4 strand of Leo1 (Lβ4) is colored in magenta. Hydrogen bonds and ionic bonds are shown as dotted red lines. (B) Co-IP experiments of the interaction between Leo1 and various Paf1 constructs. Extracts were prepared from HEK293T cells transfected with various combinations of plasmids as indicated, immunoprecipitated with agarose-conjugated anti-GFP and subsequently immunoblotted with anti-Myc (top panel) or anti-GFP (bottom panel) as indicated. The top panel shows the immunoprecipitation (IP) results. The middle panel represents 2% of the input material Myc-Leo1 for each IP. The bottom panel represents the IP of GFP and various GFP fusion proteins (GFP-Paf1, GFP-Paf1(ΔPβ1), GFP-Paf1(NT), GFP-Paf1(CT)). Bands indicating degradation products in GFP-Paf1, GFP-Paf1(ΔPβ1) and GFP-Paf1(CT) are marked by stars. (C) Summary of results obtained in (B). ‘+’ or ‘−’ indicates binding or no binding, respectively, between Leo1 and Paf1 in the co-IP experiments. A schematic representation of the full length and mutated Paf1 constructs used in the IP experiments is shown.
We performed a series of mutagenesis studies to validate the interactions observed in the structure of the Paf1(161−250)-Leo1(370−462) complex. To probe the Pβ1-Lβ4 antiparallel interaction, we used a co-immunoprecipitation (Co-IP) strategy using two different tags on the various truncations of Paf1 and full-length Leo1. As expected, Myc-Leo1 specifically co-precipitated with green fluorescent protein (GFP)-Paf1, whereas negligible amounts of Myc-Leo1 co-precipitated with any GFP-Paf1 mutants in which the Pβ1 strand was omitted (Figure 2B and C), indicating that the Pβ1-Lβ4 antiparallel sheet is vital for the Paf1/Leo1 subcomplex formation.
Assembly of human Paf1C
The biochemical and structural data above demonstrated that the Paf1 and Leo1 subunits can specifically bind to each other to form a heterodimer. We next wanted to investigate the role of the Paf1 and Leo1 subcomplex formation in the assembly of human PAF1C. We used a Co-IP strategy. In the wild-type PAF1C, both Myc-Paf1 and Myc-Leo1 specifically co-precipitated with other PAF1C subunits lacking Rtf1 (Figure 3A, lane 2, and Figure 3B, lane 2). In the mutant PAF1C, which contained the myc-Paf1(ΔPβ1) mutant, myc-Paf1(ΔPβ1) could also co-precipitate with Ctr9, Cdc73 and Ski8, but not Leo1 or Rtf1 (Figure 3A, lane 3). In contrast, myc-Leo1 did not co-precipitate with any proteins (Figure 3B, lane 3), indicating that Leo1 binds to PAF1C through the Paf1 subunit. In the same experiments, when the Ctr9 subunit was omitted, we found that Myc-Leo1 co-precipitated only with Paf1 in the context of wild-type PAF1C (Figure 3B, lane 4); Myc-Paf1(ΔPβ1) did not co-precipitate with any proteins in the context of mutant PAF1C (Figure 3A, lane 4). These data indicated that the Ctr9 subunit is a key scaffold protein binding to the Cdc73 and Ski8 subunits during PAF1C formation. Together, these results led to the establishment of the interaction network for the assembly of human PAF1C (Figure 3C).
Figure 3.
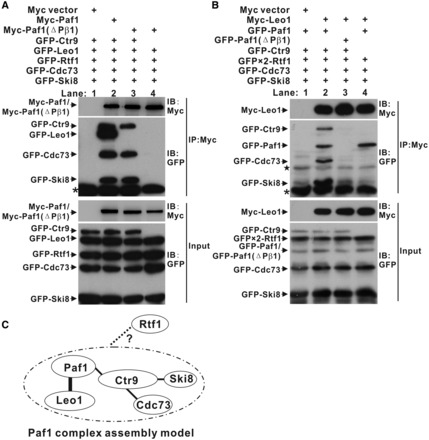
Assembly of human PAF1C. (A and B) Co-IP experiments of PAF1C formation by Paf1 (A) and Leo1 (B). Extracts were prepared from HEK293T cells transfected with various combinations of plasmids as indicated, immunoprecipitated with agarose-conjugated anti-Myc and subsequently immunoblotted with anti-Myc (upper top panels) or anti-GFP (lower bottom panels) as indicated. The top panel shows the IP results. The bottom panel shows 20% of the GFP fusion proteins for each IP. Nonspecific bands and the heavy chain of the antibody are marked by a star and asterisk, respectively. (C) Model of PAF1C assembly. The bold line represents the interaction in the crystal structure of the Paf1(161–250)-Leo1(370–462) heterodimer. The fine lines represent the interaction obtained from the IP results. The dotted line represents interactions that we could not detect in the IP experiments.
The heterodimer of the Paf1/Leo1 subcomplex specifically recognizes histone
Previous studies have demonstrated that PAF1C plays an essential role in transcription-coupled histone modifications (15,17–20,28,40–43) and that the Paf1 subunit may be involved in histone H3 binding (28,43). We speculated that the Paf1/Leo1 heterodimer may bind to histone protein. To test this hypothesis, the first 28 amino acids of the N-terminal histone H3 were fused to the N-terminus of GST (H3(1−28)-GST). In a GST pull-down assay, H3(1−28)-GST could bind to both the Paf1 and Leo1 subunits but not to any other subunits of PAF1C (Figure 4A). Next, we mapped each histone H3 binding domain of Paf1 and Leo1 using GST pull-down assays and found that the C-terminal region of Paf1 (aa 376–531) (Supplementary Figure S5A) and the N-terminal region of Leo1 (aa 1–369) (Supplementary Figure S5B) are essential for histone H3 binding. Importantly, we showed that H3(1−28)-GST binds more strongly to the wild-type Paf1/Leo1 heterodimer than to the Paf1(ΔPβ1)/Leo1 mutant or the Paf1(1−375)/Leo1 and Paf1/Leo1(370−666) complexes (lane 2, compared with lane 3, 4 and 5 in Figure 4B; the results are summarized in Figure 4C). The Paf1(ΔPβ1)/Leo1 mutant is incapable of heterodimer formation (lane 3 in Figure 2B), and the Paf1(1−375)/Leo1 and the Paf1/Leo1(370−666) complexes contained only one histone H3-binding site (cartoon in Figure 4C). These findings indicate that the Paf1/Leo1 heterodimer containing both histone H3-binding domains in Paf1 and Leo1 is important for H3(1−28)-GST binding. Furthermore, we introduced two point mutations K4A and R17A in H3(1−28)-GST (referred to as H3(1−28)(K4A, R17A)-GST) and found that the H3 mutant cannot bind to the Paf1/Leo1 complex (Figure 4D). Together, these data indicate that the Paf1/Leo1 subcomplex specifically recognizes the N-terminus of histone H3.
Figure 4.
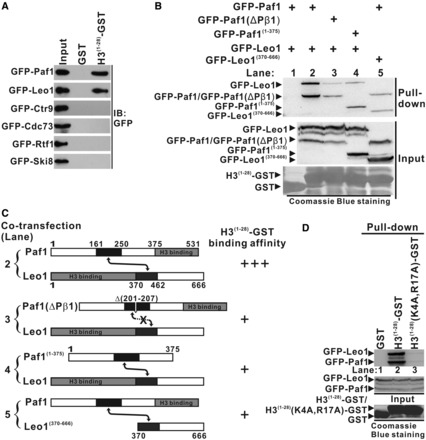
Interaction of the Paf1/Leo1 subcomplex with histone H3. (A) GST pull-down assays of histone H3 with each subunit in PAF1C. H3(1–28)-GST fusion proteins or GST alone were incubated with the extracts of HEK293T cells transfected with each PAF1C subunit. The input lane shows 20% input for the corresponding pull-down. The PVDF membrane was immunoblotted with α-GFP. (B) GST pull-down assays of the Paf1/Leo1 subcomplex with histone H3. H3(1–28)-GST fusion proteins were incubated with the extracts of HEK293T cells transfected with the Pafl/Leo1, the Paf1(ΔPβ1)/Leo1, the Paf1(1–375)/Leo1 and the Paf1/Leo1(370–666) complex, respectively. The PVDF membrane was immunoblotted with α-GFP (top panel) and subsequently stained with Coomassie blue (bottom panel). The middle panel represents 20% of the input material for the corresponding pull-down. (C) Summary of the results obtained in (B). A schematic representation of various combinations of Paf1/Leo1 complex used in the GST pull-down assays is shown. Greater numbers of “+”s correlate with higher H3(1–28)-GST binding affinity with corresponding Paf1/Leo1 complex. (D) GST pull-down assays of the Paf1/Leo1 subcomplex with wild-type and mutant histone H3. H3(1–28)-GST and H3(1–28)(K4A, R17A)-GST fusion proteins were incubated with the extracts of HEK293T cells transfected with the Pafl/Leo1complex. The PVDF membrane was immunoblotted with α-GFP (top panel) and subsequently stained with Coomassie blue (bottom panel). The middle panel represents 20% of the input material for the corresponding pull-down.
The H3(1−28)-GST fusion protein contains two histone H3 tails because the crystal structure of GST shows that the protein forms a dimer (44). Consistent with this observation, histone octamer in the nucleosome also contains two histone H3 units; thus, we hypothesized that PAF1C may specifically bind the nucleosome through the Paf1/Leo1 heterodimer. As expected, the Paf1/Leo1 subcomplex interacted with the reconstituted histone octamer in a GST pull-down assay (Supplementary Figure 6A). Furthermore, we showed that the Paf1/Leo1 subcomplex binds specifically to both histone octamer (Dot No. 1 in Figure 5A) and nucleosome (Dot No. 1 in Figure 5C) in the dot-blot overlay assays, whereas the Paf1/Leo(370−666) and the Paf1(1−375)/Leo1 complexes bind to histone octamer (Figure 5A and B) or nucleosome (Figure 5C and D) with more than half the affinity of the wild-type Paf1/Leo1 complex. Interestingly, the Paf1(1−375)/Leo1(370−666) complex lacking both the histone H3-binding domains of Paf1 and Leo1 binds weakly to both histone octamer and nucleosome (Dot No. 4 in Figure 5A and C), and thereby serves as a negative control. Together, these results indicate that the Paf1/Leo1 heterodimer may play an important role in recognizing substrates during various PAF1C-coordinated histone modifications.
Figure 5.
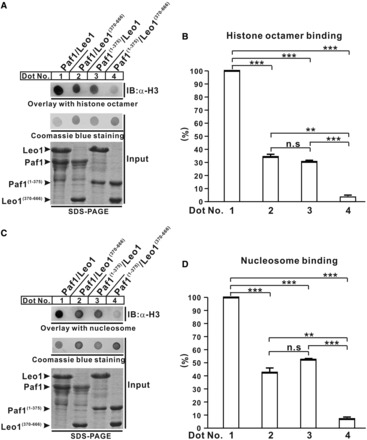
Interaction of the Paf1/Leo1 subcomplex with histone octamer and nucleosome. (A and C) Dot-blot overlay assays of the Paf1/Leo1 complex and histone octamer and nucleosome. The bound histone octamer or nucleosome was detected by immunoblotting with antibody against histone H3 (top panel). The PVDF membrane was stained with Coomassie blue (middle panel). Each recombinant Paf1/Leo1 complex was resolved by SDS-PAGE to normalize the sample inputs (bottom panel). (B and D) Bar graph of the binding histone octamer and nucleosome. The error bars indicate the standard error of the mean (n = 3, separate experiments). n.s., not significant (P > 0.05). **P < 0.01, ***P < 0.001.
DISCUSSION
The crystal structure of the Paf1/Leo1 subcomplex resolved in this study provides structural information regarding the interaction between the Paf1 and Leo1 subunits. A 90-residue fragment (aa 161–250) of human Paf1 binds to a 93-residue fragment (aa 370–462) of human Leo1 primarily through antiparallel sheet interactions (Figures 1 and 2). There is extremely high conservation of the amino acid sequence of Paf1 and Leo1 from zebrafish to human (in particular, the corresponding regions involved in the formation of the Paf1/Leo1 heterodimer, except in fungi; Supplementary Figure S7A and B). Therefore, it is reasonably safe to assume that the structural and biochemical features of the Paf1/Leo1 subcomplex described here are shared by the complexes formed by the orthologs of these two proteins in other metazoan species.
Although the sequence identity is low between human and yeast of Paf1 and Leo1, the predicted secondary structures of the corresponding sequences of yeast Paf1(150−225) and Leo1(165–259) are similar to human Paf1(161–250) and Leo1(370–462), respectively (Supplementary Figure S7A and B). Consistent with this observation, GFP-tagged yeast Paf1 (GFP-yPaf1) co-precipitated with Myc-tagged yeast Leo1 (Myc-yLeo1), whereas the yPaf1(Δ179–187) mutant (lacking residues 179–187 of the predicted β1′-strand in yeast Paf1) co-precipitated negligible amounts of Myc-yLeo1 (lane 3 compared with lane 2 in Supplementary Figure S7C), indicating that the yeast Paf1/Leo1 subcomplex may form a heteromeric complex through the similar antiparallel beta-sheet interactions described here in the human Paf1/Leo1 subcomplex.
In the crystal structure of the Paf1(161–250)-Leo1(370–462) heterodimer, we clearly observed the electron density of the adenine moiety and ribose moiety of AdoMet bound to two symmetry-related molecules of the Paf1(161–250)/Leo1(370–462) heterodimer (Supplementary Figure S3A–C). The AdoMet forms hydrogen bonds with the main-chain carboxyl group of K188 and A186 in Paf1 and E428 in Leo1 to stabilize the corresponding loops of Paf1 (loop between Pα1 and Pβ1) and Leo1 (loop between L′β2 and L′β3), respectively. Not all of the residues involved in AdoMet binding were conserved between fungi and metazoa in both Paf1 and Leo1 (Supplementary Figure S7A and B), and we could not detect any interaction between the Paf1/Leo1 subcomplex and AdoMet in solution using isothermal titration calorimetry (Supplementary Figure S8). Additionally, during the screening with additives, we found that the cyclic AMP molecule, which has a structure similar to AdoMet, can also facilitate crystal packing (Supplementary Figure S3D). Taken together, these results indicate that the introduction of AdoMet facilitates crystal packing rather than performing any specific biological function.
PAF1C in yeast is composed of five subunits, including Paf1, Leo1, Ctr9, Cdc73 and Rtf1 (5,6), and PAF1C in human contains six subunits, including Ski8/Wdr61 (7). However, the Co-IP results shown in Figure 3 indicate that Rtf1 is a less stable subunit in human PAF1C, consistent with several other studies (19,45,46). Furthermore, the histone modification domain (HMD) in Rtf1 can promote histone H3 methylation and H2B ubiquitylation, independent of the other subunits in PAF1C (47). The HMD lacks the region required for interaction with other PAF1C subunits (11), indicating that Rtf1 alone can accomplish some functions.
In our study, we found that both Paf1 and Leo1 can bind to the histone H3 tail (Figure 4A), which is somewhat inconsistent with two recent studies showing that PAF1C interacts with the histone H3 tail only through the Paf1 subunit (28,43). It is possible that we used a longer H3 tail (28 aa) than was used in the other studies. Xu and colleagues showed that the asymmetrically dimethylated histone H3 at R17 (H3R17me2a) binds to the Cdc73/Ctr9/Ski8 subunits of PAF1C (43) and that the unmodified H3 tail binds to the Paf1 subunit. These data, together with the results of this study, indicate that PAF1C may use different subunits to recognize native and/or various posttranslational modifications of histone in the context of temporal and spatial regulation.
Previous studies have established a role for PAF1C in H2B monoubiquitylation (15,16) and histone H3 methylation (15,17–20,40,42,43). Histones H2B and H3 are the substrates for the corresponding histone modification enzymes. In this study, we showed that the heterodimer of Paf1/Leo1 subcomplex is necessary for specifically binding to both histone octamer and nucleosome in vitro (Figures 4 and 5). Taken together, these results indicate that PAF1C may act as a platform or mediator to facilitate the histone modification process. Future studies are required to test this hypothesis.
ACCESSION NUMBERS
The atomic coordinates and structure factors for the structure of the Paf1/Leo1 subcomplex have been deposited in the protein data bank with accession code 4M6T.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
973 Program [2009CB825504 and 2014CB910201 to J.L., 2012CB917201 and 2013CB910400 to Y.S.]; National Natural Science Foundation of China [31270815 to J.L., 31100527 to H.Z. and 31170684 to Y.S.]; Fundamental Research Funds for the Central Universities [65122020 to J.L. and 65011621 to H.Z.]. Funding for open access: The Fundamental Research Funds for the Central Universities [65122020 to J.L.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the staff at the beam line BL17U1 of the Shanghai Synchrotron Radiation Facility (SSRF) and the staff at the beam line NW3A at Photon Factory (Tsukuba, Japan) for excellent technical assistance during data collection.
REFERENCES
- 1.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu. Rev. Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi X, Finkelstein A, Wolf AJ, Wade PA, Burton ZF, Jaehning JA. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol. Cell. Biol. 1996;16:669–676. doi: 10.1128/mcb.16.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaehning JA. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim. Biophys. Acta. 2010;1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomson BN, Arndt KM. The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim. Biophys. Acta. 2013;1829:116–126. doi: 10.1016/j.bbagrm.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wade PA, Werel W, Fentzke RC, Thompson NE, Leykam JF, Burgess RR, Jaehning JA, Burton ZF. A novel collection of accessory factors associated with yeast RNA polymerase II. Protein Expr. Purif. 1996;8:85–90. doi: 10.1006/prep.1996.0077. [DOI] [PubMed] [Google Scholar]
- 6.Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 2004;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 10.Qiu H, Hu C, Gaur NA, Hinnebusch AG. Pol II CTD kinases Bur1 and Kin28 promote Spt5 CTR-independent recruitment of Paf1 complex. EMBO J. 2012;31:3494–3505. doi: 10.1038/emboj.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warner MH, Roinick KL, Arndt KM. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol. Cell. Biol. 2007;27:6103–6115. doi: 10.1128/MCB.00772-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller CL, Porter SE, Hoffman MG, Jaehning JA. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- 13.Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rondon AG, Gallardo M, Garcia-Rubio M, Aguilera A. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep. 2004;5:47–53. doi: 10.1038/sj.embor.7400045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Roeder RG. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J. Biol. Chem. 2009;284:20582–20592. doi: 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng HH, Dole S, Struhl K. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 2003;278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- 18.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 19.Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Guermah M, McGinty RK, Lee J-S, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol. Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 22.Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc. Natl Acad. Sci. USA. 2009;106:755–760. doi: 10.1073/pnas.0812023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheldon KE, Mauger DM, Arndt KM. A Requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol. Cell. 2005;20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosimann C, Hausmann G, Basler K. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with β-catenin/Armadillo. Cell. 2006;125:327–341. doi: 10.1016/j.cell.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 25.Wang P, Bowl MR, Bender S, Peng J, Farber L, Chen J, Ali A, Zhang Z, Alberts AS, Thakker RV, et al. Parafibromin, a component of the human PAF complex, regulates growth factors and is required for embryonic development and survival in adult mice. Mol. Cell. Biol. 2008;28:2930–2940. doi: 10.1128/MCB.00654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Chaudhary K, Deb S, Moniaux N, Ponnusamy MP, Batra SK. Human RNA polymerase II-associated factor complex: dysregulation in cancer. Oncogene. 2007;26:7499–7507. doi: 10.1038/sj.onc.1210582. [DOI] [PubMed] [Google Scholar]
- 28.Marazzi I, Ho JS, Kim J, Manicassamy B, Dewell S, Albrecht RA, Seibert CW, Schaefer U, Jeffrey KL, Prinjha RK, et al. Suppression of the antiviral response by an influenza histone mimic. Nature. 2012;483:428–433. doi: 10.1038/nature10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Jong RN, Truffault V, Diercks T, Ab E, Daniels MA, Kaptein R, Folkers GE. Structure and DNA binding of the human Rtf1 Plus3 domain. Structure. 2008;16:149–159. doi: 10.1016/j.str.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 30.Amrich CG, Davis CP, Rogal WP, Shirra MK, Heroux A, Gardner RG, Arndt KM, VanDemark AP. Cdc73 subunit of Paf1 complex contains C-terminal Ras-like domain that promotes association of Paf1 complex with chromatin. J. Biol. Chem. 2012;287:10863–10875. doi: 10.1074/jbc.M111.325647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H, Shi N, Gao Y, Li X, Teng M, Niu L. Crystallographic analysis of the conserved C-terminal domain of transcription factor Cdc73 from Saccharomyces cerevisiae reveals a GTPase-like fold. Acta Crystallogr. D Biol. Crystallogr. 2012;68:953–959. doi: 10.1107/S0907444912017325. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.Pape T, Schneider TR. HKL2MAP: a graphical user interface for phasing with SHELX programs. J. Appl. Cryst. 2004;37:843–844. [Google Scholar]
- 34.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 37.Hanson B, Alexander C, Harp J, Bunick G. Preparation and crystallization of nucleosome core particle. Methods Enzymol. 2004;375:44–62. doi: 10.1016/s0076-6879(03)75003-4. [DOI] [PubMed] [Google Scholar]
- 38.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 40.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu Y, Simic R, Warner MH, Arndt KM, Prelich G. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 2007;26:4646–4656. doi: 10.1038/sj.emboj.7601887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA, Basrur V, Elenitoba-Johnson KS, Hess JL. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17:609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu J, Xu W. Histone H3R17me2a mark recruits human RNA polymerase-associated factor 1 complex to activate transcription. Proc. Natl Acad. Sci. USA. 2012;109:5675–5680. doi: 10.1073/pnas.1114905109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McTigue MA, Williams DR, Tainer JA. Crystal structures of a schistosomal drug and vaccine target: glutathione S-transferase from Schistosoma japonicaand its complex with the leading antischistomal drug praziquantel. J. Mol. Biol. 1995;246:21–27. doi: 10.1006/jmbi.1994.0061. [DOI] [PubMed] [Google Scholar]
- 45.Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, Resau JH, Meyerson M. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol. Cell. Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yart A, Gstaiger M, Wirbelauer C, Pecnik M, Anastasiou D, Hess D, Krek W. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol. Cell. Biol. 2005;25:5052–5060. doi: 10.1128/MCB.25.12.5052-5060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piro AS, Mayekar MK, Warner MH, Davis CP, Arndt KM. Small region of Rtf1 protein can substitute for complete Paf1 complex in facilitating global histone H2B ubiquitylation in yeast. Proc. Natl Acad. Sci. USA. 2012;109:10837–10842. doi: 10.1073/pnas.1116994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


