Abstract
Single-stranded DNA overhangs at the ends of human telomeric repeats are capable of adopting four-stranded G-quadruplex structures, which could serve as potential anticancer targets. Out of the five reported intramolecular human telomeric G-quadruplex structures, four were formed in the presence of K+ ions and only one in the presence of Na+ ions, leading often to a perception that this structural polymorphism occurs exclusively in the presence of K+ but not Na+. Here we present the structure of a new antiparallel (2+2) G-quadruplex formed by a derivative of a 27-nt human telomeric sequence in Na+ solution, which comprises a novel core arrangement distinct from the known topologies. This structure complements the previously elucidated basket-type human telomeric G-quadruplex to serve as reference structures in Na+-containing environment. These structures, together with the coexistence of other conformations in Na+ solution as observed by nuclear magnetic resonance spectroscopy, establish the polymorphic nature of human telomeric repeats beyond the influence of K+ ions.
INTRODUCTION
Telomeres, the ends of linear eukaryotic chromosomes, perform vital roles in the maintenance of genome integrity and the regulation of cell proliferation (1). Human telomeric DNA consists of thousands of tandem repeats of (TTAGGG)n sequences (2,3), terminating as single-stranded DNA of 100–200 nt at the 3′-ends (4). Such guanine-rich repeats are capable of adopting four-stranded G-quadruplex structures (5–8) under physiological conditions. G-quadruplex formation at the telomeric ends has been shown to inhibit the activity of the enzyme telomerase (9), which is essential for the proliferation of most cancer cells (10,11). Insights from high-resolution G-quadruplex structures adopted by these sequences could thus aid in drug design efforts targeting telomeric DNA for potential anticancer treatment (12,13).
To date, at least five distinct intramolecular G-quadruplex folding topologies (Supplementary Figure S1) have been reported for natural human telomeric repeats (14–23), four of which were observed in the presence of K+ ions. Form 1 (15–17) and Form 2 (18,19), observed in K+ solution, both comprise the (3+1) G-tetrad core, but differ in procession of the different loops; Form 3 (20), also observed in K+ solution, consists of a two-G-tetrad basket-type core with extensive base stacking interactions in the loops. In a K+-containing crystal (21) and under water-depleted conditions (22,24–26), the all-parallel-stranded propeller-type G-quadruplex structure was shown to be the preferred form. On the other hand, the only conformation to have been characterized in Na+ solution consists of a three-layered basket-type G-quadruplex (23). Myriad studies on human telomeric G-quadruplexes have been carried out using a wide variety of techniques (27–41), ranging from ultraviolet (UV) (27), circular dichroism (CD) (28), fluorescence (29–31), Raman (32), electron paramagnetic resonance (33) and nuclear magnetic resonance (NMR) spectroscopy (34), to X-ray crystallography (21), mass spectrometry (35), gel electrophoresis (36), single-molecule Förster resonance energy transfer (37,38), laser-tweezers pulling (39), calorimetry (40) and molecular dynamics (MD) simulations (41), and the results have revealed the polymorphic nature of human telomeric repeats in the presence of K+ ions. While not fully addressed, this structural polymorphism has also been implicated in Na+ solution (29,36,37), yet the basket form has largely been used as the sole reference structure for interpretation of studies under Na+-containing environment. Knowledge on the structural disparities, if any, in the presence of Na+ ions would contribute toward fundamental understanding of the cationic influence on human telomeric repeats under different contexts. Using NMR, here we show that four-repeat human telomeric sequences could adopt multiple G-quadruplex conformations in Na+ solution. We present the solution structure of one of these forms, which exhibits features distinct from the known topologies. This latest addition to the ensemble of telomeric G-quadruplex structures establishes the polymorphic nature of human telomeric repeats beyond the influence of K+ ions, and builds on the available pool of structural motifs for targeting of these important entities.
METHODS
Sample preparation
Unlabeled (Supplementary Table S1) and site-specific labeled (2%-15N-enriched or 2H-labeled; Supplementary Table S2) DNA oligonucleotides were chemically synthesized on an ABI 394 DNA/RNA synthesizer using products from Glen Research, Spectra Gases and Cambridge Isotope Laboratories. The oligonucleotides were de-protected following the manufacturer’s protocols and purified using Poly-Pak™ cartridges. Samples were dialyzed successively against water, ∼25 mM NaCl solution and water again. They were subsequently frozen, lyophilized and suspended in a buffer containing 20 mM sodium phosphate (pH 7.0) and 70 mM NaCl. DNA concentration is expressed in strand molarity using a nearest-neighbor approximation for the absorption coefficients of the unfolded species (42). The same extinction coefficient was used for the natural and BrG-substituted oligonucleotides.
UV spectroscopy
Thermal stability of different DNA oligonucleotides was characterized by recording the UV absorbance at 295 nm (43) as a function of temperature (20–90°C) using a JASCO V-650 spectrophotometer. The heating and cooling rates were 0.2°C/min. Two baselines corresponding to the completely folded (low temperature) and completely unfolded (high temperature) states were manually drawn to determine the fractions of folded and unfolded species during the melting/folding transition. The melting temperature (Tm) is defined as the temperature for which there are equal fractions of folded and unfolded species. Experiments were performed with quartz cuvettes (1-cm pathlength for low DNA concentrations and 0.2-cm pathlength for high DNA concentrations).
CD spectroscopy
CD spectra were recorded at 20°C on a JASCO-815 spectropolarimeter over the range of 220–320 nm using a 1-cm pathlength quartz cuvette with a reaction volume of 600 µl. For each sample, an average of three scans was taken, the spectrum of the buffer was subtracted and the data were zero-corrected at 320 nm.
NMR spectroscopy
Strand concentration of the NMR samples was typically 0.5–2.0 mM. NMR experiments were performed on Bruker AVANCE 600- and 700-MHz spectrometers at 25°C, unless otherwise specified. Resonances for guanine residues were assigned unambiguously using site-specific low-enrichment 15N labeling (44), site-specific 2H labeling (45) and through-bond correlations at natural abundance (46,47). Spectral assignments were assisted by NOESY, COSY, TOCSY and {1H-13C}-HSQC experiments. The spectra were processed with the software TopSpin and analyzed using the program FELIX (Felix NMR, Inc.).
Structure calculation
Inter-proton distances for the d[(TTAGGG)3TTA(BrG)GGTTA] (htel27[Br22]) G-quadruplex were deduced from NOESY experiments performed in H2O (mixing time, 200 ms) and D2O (mixing times, 100 and 350 ms). Structures were calculated based on distance-restrained molecular dynamics refinement following distance geometry simulated annealing using the program XPLOR-NIH (48). Hydrogen bond restraints, inter-proton distance restraints, dihedral restraints, planarity restraints and repulsive restraints were imposed during structure calculations. Structures were displayed using the program PyMOL (49). Detailed procedures for structure calculation are described in Supplementary Text.
Data deposition
The coordinates for the d[(TTAGGG)3TTA(BrG)GGTTA] (htel27[Br22]) G-quadruplex have been deposited in the Protein Data Bank (accession code 2MBJ).
RESULTS AND DISCUSSION
Four-repeat human telomeric sequences adopt diverse G-quadruplex conformations in Na+ solution
NMR spectra of four-repeat natural human telomeric sequences with different flanking ends (Supplementary Table S1) in Na+ solution indicated the adoption of multiple conformations (Supplementary Figure S2). In agreement with the previous study (23), the sequence d[AGGG(TTAGGG)3] (denoted as htel22; Table 1) displayed 12 major imino proton peaks at ∼10.6–12.0 ppm (Figure 1A), corresponding to the establishment of the basket-type G-quadruplex (Supplementary Figure S1A). In addition, two sequences with the same 5′-flanking end as htel22 showed a major species with a highly similar spread of imino proton peaks (Supplementary Figure S2F and G), suggesting their adoption of the same topology as the prevailing form. The other sequences mostly displayed the coexistence of two or more major species, judging from the number and intensity of imino proton peaks. CD spectra of all 16 sequences displayed two positive maxima at ∼250 and ∼295 nm, and a trough at ∼260–270 nm (Supplementary Figure S3). The 295-nm peak is characteristic of opposite-polarity stacking of G-tetrads (50), suggesting that these sequences largely conform to antiparallel G-quadruplexes in Na+ solution. Note that htel22 produced a sharp negative trough at ∼260 nm (Figure 1B), which was less intense in the other sequences.
Table 1.
Representative natural and modified human telomeric sequences used in this studya
| Name | Sequence |
|---|---|
| htel22 | A GGG TTA GGG TTA GGG TTA GGG |
| htel27 | TTA GGG TTA GGG TTA GGG TTA GGG TTA |
| htel27[Br22] | TTA GGG TTA GGG TTA GGG TTA BrGGG TTA |
aThe modified 8-bromoguanine (BrG) residue is in boldface.
Figure 1.
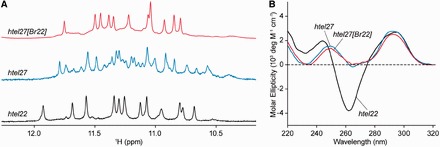
(A) NMR imino proton spectra and (B) CD spectra of human telomeric sequences: htel22, htel27 and htel27[Br22] in ∼100 mM Na+.
Formation of a G-quadruplex with an alternative conformation distinct from htel22
We chose to probe the structures present in the sequence d[(TTAGGG)4TTA] (denoted as htel27; Table 1) (Figure 1A), which has the largest number of flanking nucleotides for potential interactions. Previously, 8-bromoguanine (BrG)-for-G substitution has been used to drive the folding of an oligonucleotide toward a single major G-quadruplex structure by favoring the syn glycosidic conformation at the substituted residue (17,18,20,51). Here, single BrG incorporation at position G22 of htel27, giving rise to the sequence d[(TTAGGG)3TTA(BrG)GGTTA] (denoted as htel27[Br22]; Table 1), led to the emergence of a predominant species (>85%) amenable for detailed structural characterization (Figure 1A). Htel27[Br22] showed 12 major imino proton peaks at ∼10.8–11.8 ppm, indicating the adoption of a three-layered G-quadruplex. The CD spectra of htel27[Br22] and htel27 were highly similar (Figure 1B), suggesting that htel27[Br22] could correspond to the preexisting major conformation(s) in htel27, or that the core topologies of htel27[Br22] and htel27 were closely related. Their CD spectra were substantially different from that of htel22, notably the absence of the sharp negative trough at ∼260 nm, pointing to the adoption of an alternative folding topology. Melting analyses of the three sequences were carried out by monitoring the UV absorbance at 295 nm (43), and the melting temperatures (Tm) of htel22, htel27 and htel27[Br22] were found to be 57.6, 44.5 and 50.8°C, respectively, in ∼100 mM Na+ (Supplementary Figure S4). The BrG-substituted oligonucleotide displayed a Tm ∼6°C higher than that of the natural counterpart, consistent with observations from previous studies (20,51).
NMR spectral assignments
To proceed with the structural elucidation of htel27[Br22], guanine imino (Figure 2A) and adenine/guanine H8 (Figure 2B) protons were unambiguously assigned using site-specific low-enrichment 15N labeling (44) and site-specific 2H labeling (45), respectively (Supplementary Table S2). The guanine imino and H8 proton assignments were further corroborated by through-bond correlations at natural abundance (46) (Figure 2C). Assignments of thymine residues were supplemented by T-to-U substitutions (Supplementary Table S2). The H8/H6-H1′ NOE sequential connectivity of htel27[Br22] (Supplementary Figure S5) was completed with the assistance of other through-bond correlation experiments (COSY, TOCSY and {1H-13C}-HSQC; data not shown) (47). The strong intensity of intraresidue H8-H1′ NOE cross-peaks for G4, G5, G10, G16 and G23 (Supplementary Figure S6) indicated that these guanine residues, together with BrG22, adopt the syn glycosidic conformation. All other guanine residues adopt the anti glycosidic conformation.
Figure 2.
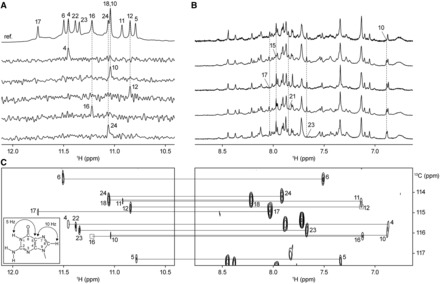
NMR spectral assignments of htel27[Br22] in Na+ solution. (A) Guanine imino protons were assigned based on15N-filtered spectra of samples, 2% 15N-labeled at the indicated positions. The reference spectrum (ref.) with the respective guanine imino proton assignments is shown at the top. (B) Examples of adenine and guanine H8 proton assignments through site-specific 2H labeling at the indicated positions. (C) Through-bond correlations between guanine imino and H8 protons via 13C5 at natural abundance, using long-range J-couplings shown in the inset. Missing/weak correlations are framed in boxes.
Htel27[Br22] adopts a new antiparallel (2+2) folding topology in Na+ solution
Having determined the guanine imino and H8 proton assignments for htel27[Br22], the alignment of the tetrads could then be deduced based on characteristic cyclic imino–H8 NOE connectivity patterns within each tetrad (Figure 3A and B). The top G-tetrad (G6•G24•G16•G10) is oriented in the opposite hydrogen-bond directionality with respect to the other two G-tetrads, (G5•G11•G17•G23) and (G4•G12•G18•BrG22), and the glycosidic conformations of guanines are anti•anti•syn•syn for all three tetrads. The central positioning of (G5•G11•G17•G23) in the tetrad core agrees with solvent exchange data showing that the imino protons from this G-tetrad were the most protected (Supplementary Figure S7). Connecting the sequential guanine residues and joining the corners of the G-tetrad core with the linking TTA loops, we derived an antiparallel G-quadruplex scaffold consisting of two adjacent strands pointing up and two remaining strands pointing down (up–up–down–down) or a (2+2) G-quadruplex scaffold (14) with a successive loop arrangement edgewise–propeller–edgewise (Figure 3C).
Figure 3.
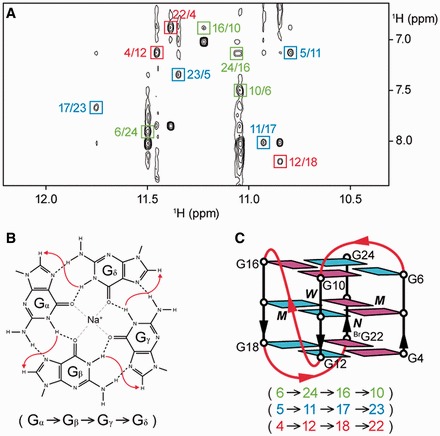
Determination of the folding topology of htel27[Br22]. (A) NOESY spectrum (mixing time, 200 ms) showing the imino–H8 connectivity of htel27[Br22]. Cross-peaks that identify the arrangement of the G-tetrads are framed and labeled with the residue number of imino protons in the first position and that of H8 protons in the second position. (B) Characteristic guanine imino–H8 NOE connectivity patterns around a Gα•Gβ•Gγ•Gδ tetrad as indicated with arrows (connectivity between Gδ and Gα implied). (C) Schematic structure of htel27[Br22]. Anti guanines are colored in cyan, while syn guanines and 8-bromoguanine are colored in magenta. The backbones of the core and loops are colored in black and red, respectively. W, M and N represent wide, medium and narrow grooves, respectively.
Na+ solution structure of the htel27[Br22] G-quadruplex
The structure of htel27[Br22] G-quadruplex in Na+ solution (Figure 4) was computed on the basis of NMR restraints (Table 2). There are one wide, one narrow and two medium grooves. The first (edgewise) loop crosses the top of the narrow groove, the second (propeller) loop traverses across one of the medium grooves, while the third (edgewise) loop completes a turn over the bottom of the wide groove. The 5′- and 3′-flanking TTA nucleotides take part in hydrogen-bond and stacking interactions at the bottom and the top of the structure, respectively. There is a Watson–Crick A9•T25 base pair capping the top of the tetrad core. A15 from the propeller loop also stacks over the top G-tetrad, potentially establishing interactions with the loop elements at the top. T14 from the same loop projects into the medium groove, whereas T13 flips out of the propeller loop. At the bottom of the structure, A3 and T20 establishes a Watson–Crick base pair, with A21 completing the formation of a triple platform. These stabilizing interactions of the loop and terminal residues would agree with the observation of additional broad imino proton peaks at ∼12.2–14.0 ppm (Supplementary Figure S8). The establishment of base pair stacking interactions across the ends of the tetrad core could have provided considerable contribution toward the adoption of the major conformation of htel27[Br22], consistent with previous observations regarding other human telomeric G-quadruplex structures formed in K+ solution (15–20). In a longer sequence context, the various structural forms could exist in a state of equilibrium, with possible interconversion among them.
Figure 4.
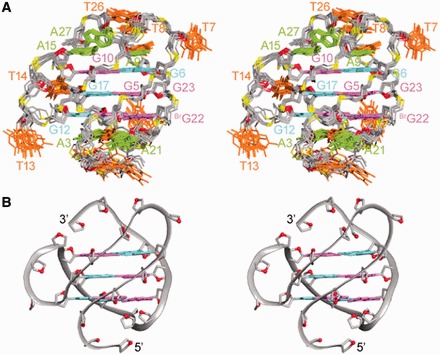
Stereoviews of the htel27[Br22] G-quadruplex structure in Na+ solution. (A) Ten superimposed refined structures. (B) Ribbon view of a representative structure. Anti guanines are colored in cyan; syn guanines and 8-bromoguanine, magenta; adenines, green; thymines, orange; backbone and sugars, gray; O4′ atoms, red; phosphorus atoms, yellow.
Table 2.
Statistics of the computed structures of the 27-nt BrG-modified human telomeric sequence d[(TTAGGG)3TTA(BrG)GGTTA] (htel27[Br22])a
| A. NMR restraints | ||
|---|---|---|
| Distance restraints | D2O | H2O |
| Intraresidue distance restraints | 446 | 0 |
| Sequential (i, i + 1) distance restraints | 197 | 26 |
| Long-range (i, ≥ i + 2) distance restraints | 79 | 32 |
| Other restraints | ||
| Hydrogen bond restraints | 48 | |
| Dihedral restraints | 12 | |
| Planarity restraints | 3 | |
| Repulsive restraintsb | 2 | |
| B. Structure statistics for 10 molecules following distance-restrained MD refinement | ||
| NOE violations | ||
| Number (>0.2 Å) | 0.800 ± 0.872 | |
| Maximum violation (Å) | 0.195 ± 0.050 | |
| RMSD of violations (Å) | 0.022 ± 0.002 | |
| Deviations from the ideal covalent geometry | ||
| Bond lengths (Å) | 0.003 ± 0.000 | |
| Bond angles (°) | 0.694 ± 0.007 | |
| Impropers (°) | 0.367 ± 0.004 | |
| Pairwise all heavy atom RMSD values (Å) | ||
| All heavy atoms except T1, T2 | 1.04 ± 0.12 | |
| All heavy atoms | 1.49 ± 0.35 | |
aPDB ID: 2MBJ.
bDistance restraints between pairs of protons that do not exhibit NOE cross-peaks.
Novel features of the htel27[Br22] G-quadruplex
We have shown that in Na+ solution, htel27[Br22] assumes a novel antiparallel (2+2) folding topology (Figure 3C). Although the tetrad cores of htel27[Br22] (this work) and htel22 (23) possess the same relative strand orientations, they differ in the hydrogen-bond directionalities of the tetrads: anticlockwise–clockwise–clockwise for htel27[Br22] versus clockwise–anticlockwise–clockwise for htel22 (Supplementary Figure S9). Loop arrangement of htel27[Br22] (edgewise–propeller–edgewise) is also different from that of htel22 (edgewise–diagonal–edgewise). To date, antiparallel (2+2) G-quadruplexes that have been investigated largely consist of edgewise and/or diagonal loops (20,23,52–54), while quadruplexes containing propeller loops mostly belong to the all-parallel-stranded (21,22) or (3+1) (15–19,55–57) core topology. Antiparallel (2+2) G-quadruplexes with a propeller loop (58), such as that formed by htel27[Br22], have not been observed in the context of human telomeric repeats.
CONCLUSION
The antiparallel (2+2) G-quadruplex scaffold adopted by htel27[Br22] (and presumably a subpopulation of htel27), as well as the observation of the coexistence of multiple conformations in the series of four-repeat natural human telomeric sequences in Na+ solution, point to additional diversity than had been previously assumed. Even though intracellular Na+ content is ∼10 times lower than that of K+, these structures observed in Na+ solution could potentially take part in the interconversion between various quadruplex forms.
ACCESSION NUMBER
PDB: accession code 2MBJ.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore Ministry of Education and Nanyang Technological University (to A.T.P.). Funding for open access charge: Singapore Ministry of Education.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr Sreelatha Sarangapani for her participation at the early stage of the project.
REFERENCES
- 1.Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allshire RC, Dempster M, Hastie ND. Human telomeres contain at least three types of G-rich repeat distributed non-randomly. Nucleic Acids Res. 1989;17:4611–4627. doi: 10.1093/nar/17.12.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 5.Simonsson T. G-quadruplex DNA structures—Variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 6.Davis JT. G-quartets 40 years later: From 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 7.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 10.Counter CM, Avilion AA, Lefeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PLC, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 12.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 13.De Cian A, Lacroix L, Douarre C, Temime-Smaali N, Trentesaux C, Riou JF, Mergny JL. Targeting telomeres and telomerase. Biochimie. 2008;90:131–155. doi: 10.1016/j.biochi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Phan AT. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 15.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: An intramolecular (3+1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai JX, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang DZ. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: A novel adenine triple formation. Nucleic Acids Res. 2007;35:2440–2450. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsugami A, Xu Y, Noguchi Y, Sugiyama H, Katahira M. Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions, as determined by NMR in a K+ solution. FEBS J. 2007;274:3545–3556. doi: 10.1111/j.1742-4658.2007.05881.x. [DOI] [PubMed] [Google Scholar]
- 18.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai JX, Carver M, Punchihewa C, Jones RA, Yang DZ. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: Insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim KW, Amrane S, Bouaziz S, Xu WX, Mu YG, Patel DJ, Luu KN, Phan AT. Structure of the human telomere in K+ solution: A stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 22.Heddi B, Phan AT. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011;133:9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 24.Xue Y, Kan ZY, Wang Q, Yao Y, Liu J, Hao YH, Tan Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- 25.Lannan FM, Mamajanov I, Hud NV. Human telomere sequence DNA in water-free and high-viscosity solvents: G-quadruplex folding governed by Kramers rate theory. J. Am. Chem. Soc. 2012;134:15324–15330. doi: 10.1021/ja303499m. [DOI] [PubMed] [Google Scholar]
- 26.Yu HQ, Gu XB, Nakano S, Miyoshi D, Sugimoto N. Beads-on-a-string structure of long telomeric DNAs under molecular crowding conditions. J. Am. Chem. Soc. 2012;134:20060–20069. doi: 10.1021/ja305384c. [DOI] [PubMed] [Google Scholar]
- 27.Tran PLT, Mergny JL, Alberti P. Stability of telomeric G-quadruplexes. Nucleic Acids Res. 2011;39:3282–3294. doi: 10.1093/nar/gkq1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renčiuk D, Kejnovská I, Školáková P, Bednářová K, Motlová J, Vorlíčková M. Arrangements of human telomere DNA quadruplex in physiologically relevant K+ solutions. Nucleic Acids Res. 2009;37:6625–6634. doi: 10.1093/nar/gkp701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Risitano A, Fox KR. Inosine substitutions demonstrate that intramolecular DNA quadruplexes adopt different conformations in the presence of sodium and potassium. Bioorg. Med. Chem. Lett. 2005;15:2047–2050. doi: 10.1016/j.bmcl.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 30.Mendez MA, Szalai VA. Fluorescence of unmodified oligonucleotides: A tool to probe G-quadruplex DNA structure. Biopolymers. 2009;91:841–850. doi: 10.1002/bip.21268. [DOI] [PubMed] [Google Scholar]
- 31.Dao NT, Haselsberger R, Michel-Beyerle ME, Phan AT. Following G-quadruplex formation by its intrinsic fluorescence. FEBS Lett. 2011;585:3969–3977. doi: 10.1016/j.febslet.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Palacký J, Vorlíčková M, Kejnovská I, Mojzeš P. Polymorphism of human telomeric quadruplex structure controlled by DNA concentration: A Raman study. Nucleic Acids Res. 2013;41:1005–1016. doi: 10.1093/nar/gks1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh V, Azarkh M, Exner TE, Hartig JS, Drescher M. Human telomeric quadruplex conformations studied by pulsed EPR. Angew. Chem. Int. Ed. 2009;48:9728–9730. doi: 10.1002/anie.200902146. [DOI] [PubMed] [Google Scholar]
- 34.Adrian M, Heddi B, Phan AT. NMR spectroscopy of G-quadruplexes. Methods. 2012;57:11–24. doi: 10.1016/j.ymeth.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 35.Smargiasso N, Rosu F, Hsia W, Colson P, Baker ES, Bowers MT, De Pauw E, Gabelica V. G-quadruplex DNA assemblies: Loop length, cation identity, and multimer formation. J. Am. Chem. Soc. 2008;130:10208–10216. doi: 10.1021/ja801535e. [DOI] [PubMed] [Google Scholar]
- 36.Gaynutdinov TI, Neumann RD, Panyutin IG. Structural polymorphism of intramolecular quadruplex of human telomeric DNA: Effect of cations, quadruplex-binding drugs and flanking sequences. Nucleic Acids Res. 2008;36:4079–4087. doi: 10.1093/nar/gkn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ying LM, Green JJ, Li HT, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JY, Okumus B, Kim DS, Ha TJ. Extreme conformational diversity in human telomeric DNA. Proc. Natl Acad. Sci. USA. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koirala D, Ghimire C, Bohrer C, Sannohe Y, Sugiyama H, Mao HB. Long-loop G-quadruplexes are misfolded population minorities with fast transition kinetics in human telomeric sequences. J. Am. Chem. Soc. 2013;135:2235–2241. doi: 10.1021/ja309668t. [DOI] [PubMed] [Google Scholar]
- 40.Petraccone L, Spink C, Trent JO, Garbett NC, Mekmaysy CS, Giancola C, Chaires JB. Structure and stability of higher-order human telomeric quadruplexes. J. Am. Chem. Soc. 2011;133:20951–20961. doi: 10.1021/ja209192a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cang XH, Šponer J, Cheatham TE. Insight into G-DNA structural polymorphism and folding from sequence and loop connectivity through free energy analysis. J. Am. Chem. Soc. 2011;133:14270–14279. doi: 10.1021/ja107805r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. III. Circular dichroism studies of conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 43.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 44.Phan AT, Patel DJ. A site-specific low-enrichment 15N,13C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 2002;124:1160–1161. doi: 10.1021/ja011977m. [DOI] [PubMed] [Google Scholar]
- 45.Huang XN, Yu PL, LeProust E, Gao XL. An efficient and economic site-specific deuteration strategy for NMR studies of homologous oligonucleotide repeat sequences. Nucleic Acids Res. 1997;25:4758–4763. doi: 10.1093/nar/25.23.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phan AT. Long-range imino proton-13C J-couplings and the through-bond correlation of imino and non-exchangeable protons in unlabeled DNA.J. Biomol. NMR. 2000;16:175–178. doi: 10.1023/a:1008355231085. [DOI] [PubMed] [Google Scholar]
- 47.Phan AT, Guéron M, Leroy JL. Investigation of unusual DNA motifs. Methods Enzymol. 2002;338:341–371. doi: 10.1016/s0076-6879(02)38228-4. [DOI] [PubMed] [Google Scholar]
- 48.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 49.DeLano WL, Bromberg S. PyMOL User's Guide. San Carlos, California, USA: DeLano Scientific LLC; 2004. [Google Scholar]
- 50.Gray DM, Wen JD, Gray CW, Repges R, Repges C, Raabe G, Fleischhauer J. Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality. 2008;20:431–440. doi: 10.1002/chir.20455. [DOI] [PubMed] [Google Scholar]
- 51.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorgan Med Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 52.Smith FW, Lau FW, Feigon J. d(G3T4G3) forms an asymmetric diagonally looped dimeric quadruplex with guanosine 5′-syn-syn-anti and 5′-syn-anti-anti N-glycosidic conformations. Proc. Natl Acad. Sci. USA. 1994;91:10546–10550. doi: 10.1073/pnas.91.22.10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strahan GD, Shafer RH, Keniry MA. Structural properties of the [d(G3T4G3)]2 quadruplex: Evidence for sequential syn-syn deoxyguanosines. Nucleic Acids Res. 1994;22:5447–5455. doi: 10.1093/nar/22.24.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Črnugelj M, Hud NV, Plavec J. The solution structure of d(G4T4G3)2: A bimolecular G-quadruplex with a novel fold. J. Mol. Biol. 2002;320:911–924. doi: 10.1016/s0022-2836(02)00569-7. [DOI] [PubMed] [Google Scholar]
- 55.Črnugelj M, Sket P, Plavec J. Small change in a G-rich sequence, a dramatic change in topology: New dimeric G-quadruplex folding motif with unique loop orientations. J. Am. Chem. Soc. 2003;125:7866–7871. doi: 10.1021/ja0348694. [DOI] [PubMed] [Google Scholar]
- 56.da Silva MW, Trajkovski M, Sannohe Y, Hessari NM, Sugiyama H, Plavec J. Design of a G-quadruplex topology through glycosidic bond angles. Angew. Chem. Int. Ed. 2009;48:9167–9170. doi: 10.1002/anie.200902454. [DOI] [PubMed] [Google Scholar]
- 57.Marušič M, Šket P, Bauer L, Viglasky V, Plavec J. Solution-state structure of an intramolecular G-quadruplex with propeller, diagonal and edgewise loops. Nucleic Acids Res. 2012;40:6946–6956. doi: 10.1093/nar/gks329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuryavyi V, Majumdar A, Shallop A, Chernichenko N, Skripkin E, Jones R, Patel DJ. A double chain reversal loop and two diagonal loops define the architecture of a unimolecular DNA quadruplex containing a pair of stacked G(syn)•G(syn)•G(anti)•G(anti) tetrads flanked by a G•(T-T) triad and a T•T•T triple. J. Mol. Biol. 2001;310:181–194. doi: 10.1006/jmbi.2001.4759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


