Abstract
The successful synthesis of a transcript by RNA polymerase II (RNAPII) is a multistage process with distinct rate-limiting steps that can vary depending on the particular gene. A growing number of genes in a variety of organisms are regulated at steps after the recruitment of RNAPII. The best-characterized Saccharomyces cerevisiae gene regulated in this manner is CYC1. This gene has high occupancy of RNAPII under non-inducing conditions, defining it as a poised gene. Here, we find that subunits of the head module of Mediator, Med18 and Med20, and Med19 are required for activation of transcription at the CYC1 promoter in response to environmental cues. These subunits of Mediator are required at the preloaded promoter for normal levels of recruitment and activity of the general transcription factor TFIIH. Strikingly, these Mediator components are dispensable for activation by the same activator at a different gene, which lacks a preloaded polymerase in the promoter region. Based on these results and other studies, we speculate that Mediator plays an essential role in triggering an inactive polymerase at CYC1 into a productively elongating form.
INTRODUCTION
Control of gene expression via the regulation of events after the recruitment of RNA polymerase II (RNAPII) is now a well-recognized feature of transcriptional regulation (1–4). At genes regulated in this manner, events after the recruitment of polymerase are rate limiting for successful production of transcripts (5–7). Consequently, the promoter regions of these genes contain key members of the transcription machinery (including RNAPII) in the absence of transcript production and exhibit increased expression via the stimulation of inactive complexes. Here, we refer to genes with general transcription factor and RNAPII promoter occupancy in the absence of transcription as poised, or preloaded (8). Poised promoters are found across the evolutionary spectrum; poising has been observed in bacteria, yeast, worms, flies and humans (5–7,9–13). Despite their prevalence, how poised genes are triggered in response to diverse biological signals remains unclear.
The yeast CYC1 gene is preloaded and postrecruitment regulated: in the uninduced (non-activated) state, the CYC1 promoter region contains RNAPII, TATA binding protein, Spt-Ada-Gcn5-acetyltransferase (SAGA) and Spn1 occupancy, as well as detectable Ser5 phosphorylation on the Rpb1 C-terminal domain (CTD) (7–9,14,15). We previously found that the Mediator coactivator complex is required for activating the preloaded CYC1 gene during growth in non-fermentable carbon sources (8). Mediator is a large multi-subunit coactivator that is conserved from yeast to humans with diverse and complex roles in transcriptional processes (4,16–18). The core complex consists of three modules termed the head, middle and tail (19,20). Mediator integrates the transcription process by linking upstream signals from the activator with the general transcription machinery (8,21–26), participating in postrecruitment (27–33) and termination steps of transcription (34), as well as in silencing and chromatin structure (35–38). Here, we investigate the activation of the preloaded CYC1 gene during the response to oxidative stress. We find that activation of the poised promoter by oxidative stress occurs via an independent activator from the response to non-fermentable carbon source (8) and displays vastly different activation kinetics. Despite the differences in these responses, both pathways are dependent on similar Mediator subunits to upregulate CYC1 gene expression. We also show that a specific activator (Yap1) can stimulate expression of postrecruitment and recruitment regulated genes. Importantly, the investigation of these two gene classes, which are coordinately regulated in the response to oxidative stress, revealed that specific Mediator subunits are essential for activation of the poised promoter and dispensable for activation of the recruitment regulated promoter. Taken together, our results provide important new insights on regulation of poised promoter gene expression and the functions of Mediator in transitioning genes to an activated state.
MATERIALS AND METHODS
Yeast strains and culturing conditions
Strains used in this study are listed in Table 1. The parent BY4741 (MATa his3Δ1 ura3Δ0 leu2Δ0 met15Δ0) strain and deletion strains were purchased from Research Genetics, except the med2Δ strain, which was generated using common protocols (8). We generated all tagged strains from the BY4741 parent strain using established protocols (42). Media used for routine culture of yeast is as described (43).
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research genetics |
| med2Δ | BY4741 med2Δ::URA3 | (8) |
| yap1Δa | BY4741 yap1Δ::kanMX | Research genetics |
| YAP1-myc | BY4741 YAP1-myc13::HIS3 | This work |
| YAP1-myc, med20Δ | BY4741 YAP1-myc13::HIS3, med20Δ::kanMX | This work |
| MED14-HA | BY4741 MED14-HA3::HIS3 | This work |
| MED15-myc | BY4741 MED15-myc13::HIS3 | (39) |
| MED15-myc, med20Δ | BY4741 MED15-myc13::HIS3, med20Δ::kanMX | (39) |
| MED18-myc | BY4741 MED18-myc13::HIS3 | (40) |
| SRB4 wt | srb4Δ::HIS3 {RY2844 SRB4+} | (41) |
| srb4 ts | srb4Δ::HIS3 {RY2882 srb4-138} | (41) |
| RAD3-HA | BY4741 RAD3-HA3::HIS3 | This work |
| RAD3-HA, med20Δ | BY4741 RAD3-HA3::HIS3, med20Δ::kanMX | This work |
| KIN28-myc | BY4741 KIN28-myc13::HIS3 | This work |
| KIN28-myc, med20Δ | BY4741 KIN28-myc13::HIS3, med20Δ::kanMX | This work |
aAll other deletion strains were purchased from Research Genetics and have the same markers.
For phenotypic analysis, yeast cultures were grown overnight in rich media. The next morning, cultures were diluted and allowed to undergo two cell doublings to an OD600 of 0.7–0.9. Cells were collected and diluted in water to an OD600 of 0.1. Serial dilutions (10-fold) were plated on the indicated plates and incubated at 30° for 2–5 days (depending on the media). Yeast extract-peptone-dextrose (YPD) plates with hydrogen peroxide were made by supplementing cooled YPD with hydrogen peroxide (Sigma) to a final concentration between 2.5 and 4.5 mM. Yeast extract-peptone-ethanol-glycerol (YPEG) plates were made by supplementing YP with ethanol (3%) and glycerol (3%).
For oxidative stress induction, cultures were grown overnight in rich media, diluted and allowed to undergo two cell doublings. When cultures reached an OD600 of 0.7–0.8, cells were treated with hydrogen peroxide to a final concentration of 0.3 mM. Samples were taken at various time points after induction. For oxidative stress induction of the MED17 wild-type and temperature sensitive strains, cells were grown at 30°C for the permissive temperature. For the non-permissive temperature, cells were shifted to 37°C for 45 min. At this point, the uninduced sample was removed, and H2O2 was added to a final concentration of 0.3 mM. For inducing CYC1 via growth in non-fermentable carbon sources, yeast cultures were grown overnight in rich media, then diluted and allowed to undergo two cell doublings in rich media. Cells were washed with YP three times and diluted into YP medium containing 3% ethanol as the sole carbon source and were cultured at 30°C for various times (30 min to 6 h, as indicated). For uninduced samples, cells were grown in YPD media for 6 h at 30°C.
RNA abundance
S1 nuclease assays were conducted as described (44). Yeast cells were harvested, and total RNA was extracted by the hot-phenol extraction method. Thirty micrograms of total RNA was hybridized with excess 32P labeled probe in a 55°C water bath overnight. S1 nuclease digestion was performed on hybridized samples for 30 min at 37°C. Band intensity was normalized to the intensity of the tRNAw band.
Chromatin immunoprecipitation experiments
Chromatin immunoprecipitation (ChIP) was performed as previously described (9) with few modifications. Cultures were induced as described earlier in the text. At the indicated time, cultures were cross-linked with 1% formaldehyde for 15 min, and then glycine (125 mM) was added to stop cross-linking. Sheared chromatin material (500 µl, corresponding to 25 ml of the initial culture) was incubated with 10 µl of anti-myc (Upstate), anti-HA (Santa Cruz), anti-RNAPII (Cat#: 8WG16, Covance) or anti-CTD Ser5 (Cat#: 39750, Active Motif) antibodies, rotating overnight at 4°C. Protein–DNA cross-links were reversed by incubation at 65°C, and the DNA was purified by phenol-chloroform extraction and used for quantitative PCR analysis. For all quantitative PCR samples, occupancy at the GAL10 promoter region was subtracted from the occupancy of the promoter of interest from the identical replicate. Values equal to or below background levels after GAL10 subtraction were set to 0. Primer locations are as follows: CYC1 promoter (−149 to +36 relative to the ATG), GTT2 promoter (−230 to +110 relative to the ATG), GAL10 promoter (−139 to +142 relative to the ATG). ChIPs were performed in biological duplicate or triplicate, as indicated in the figure legend, RT-PCR of individual samples was performed in duplicate or triplicate. Unpaired t-tests were used to compare the means of two groups, when appropriate (www.graphpad.com). Two-tailed P-values are indicated in parentheses in the text.
Immunoblotting
Yeast cells were grown to log phase (OD600 of 0.8) in rich media. Cells were harvested, washed and resuspended in 200 µl of lysis buffer [0.5 M phosphate buffer (pH 7.5)]. Whole-cell extracts were prepared by vigorous bead beating. Cellular debris was removed by spinning the extracts at 3000 rpm at 4°C for 15 min. An equal amount of total proteins from whole-cell extract was separated on a 10% SDS–PAGE gel and then transferred to a nitrocellulose membrane. The following antibodies were used at the given dilutions: anti-myc (Upstate, 1:500) and polyclonal anti-TATA binding protein (1:5000), Alexa Fluor goat anti-mouse (LI-COR Biosciences, 1:15 000) and goat anti-rabbit (LI-COR Biosciences 1:15 000). Proteins were detected with the Odyssey Infrared Imaging System (LI-COR).
RESULTS
Yap1 controls CYC1 gene expression during oxidative stress
We previously discovered that the CYC1 gene is poised with a preloaded RNAPII complex before activation, and that this complex is activated by growth in ethanol (8,9,45). Despite the constitutive presence of general transcription factors and RNAPII at the CYC1 promoter, induction in response to ethanol takes several hours (4–5 h). This time scale seems inconsistent with the idea that poising potentiates genes for rapid activation (12,46–48). Given the connections between poising and speed of the transcriptional response, we were curious whether CYC1 responds to other environmental changes with faster kinetics than the response to ethanol. Results from whole genome transcriptional profiling suggested that oxidative stress also induces CYC1 expression (49). Indeed, we confirmed that sub-lethal amounts of hydrogen peroxide (H2O2) activate the CYC1 gene quickly and transcript levels peak within 10 min after the addition of H2O2 (Figure 1A). This is in stark contrast to the 4 h required for growth in ethanol to elicit a similar transcriptional response at CYC1. After oxidative stress, transcript levels decline, reaching pre-induction levels 50 min after the treatment with H2O2. Yap1 is a transcriptional activator protein that is important during oxidative stress in yeast (50). Using a strain deleted for the Yap1 activator (yap1Δ), we found that the expression of CYC1 during oxidative stress is dependent on this factor (Figure 1A). Yap1 is essential for cellular survival during growth in oxidative stress-inducing conditions, as cells lacking the YAP1 gene fail to grow on plates containing hydrogen peroxide (51) (Figure 1B).
Figure 1.
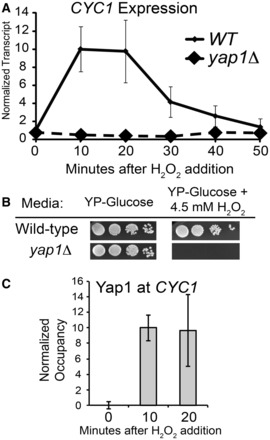
Yap1 directly regulates CYC1 expression during oxidative stress. (A) CYC1 transcript levels are rapidly induced in response to oxidative stress. An S1 nuclease protection assay was performed with RNA isolated from wild-type and yap1Δ cells before exposure to H2O2 and in 10-min intervals after adding H2O2 (0.3 mM). A probe for tRNAw was used as a loading control, and the transcript level in the wild-type strain 10 min after H2O2 exposure was set to 10. Results represent the average of at least three biological replicates ±SD. (B) Serial spot dilutions of the wild-type and yap1Δ strains on YP-Glucose and on YP-Glucose containing H2O2 (4.5 mM). Plates were incubated at 30°C for 3 days before photographing. (C) Yap1-myc occupies the CYC1 promoter during oxidative stress. Occupancy was determined with a ChIP using a Yap1-myc strain. Formaldehyde was added to cross-link proteins and DNA at the time point indicated on the x-axis. Occupancy at the GAL10 promoter region was subtracted from the occupancy at CYC1, and the 10-min time point was set to 10. Bars represent the average ±SD of three biological replicates.
We next determined whether the transcriptional dependence on Yap1 is a direct result of promoter occupancy by this activator. A time course ChIP assay was performed using a strain containing a tagged version of Yap1. We found no significant occupancy of Yap1 at the CYC1 promoter before induction with H2O2, whereas occupancy increases greatly after induction (Figure 1C). This occupancy pattern is consistent with the pattern of CYC1 transcript production. Therefore, Yap1 is directly involved in the rapid activation of the poised CYC1 promoter during the response to oxidative stress in yeast.
Yap1 is not required for preloading the CYC1 promoter or for the response to changing carbon sources
During normal conditions, Yap1 is maintained primarily in the cytoplasm via interaction with the nuclear exporter Crm1 (52). During oxidative stress, this interaction is masked owing to oxidant-induced changes in the structure of Yap1, and Yap1 accumulates in the nucleus (53,54). As previously shown, the CYC1 promoter contains preloaded RNAPII (7,8,15). Thus, the RNAPII enzyme occupies the promoter region prior to gene activity. Given that the Yap1 protein shuttles into and out of the nucleus, it is possible that Yap1 molecules that escape the export machinery are responsible for recruiting the inactive RNAPII at the CYC1 promoter during uninduced conditions. To test this, we analyzed RNAPII occupancy in the wild-type and yap1Δ strain under normal conditions and found that RNAPII still occupies this promoter in the yap1Δ strain (Supplementary Figure S1). This indicates that Yap1 is not responsible for loading the promoter with RNAPII before induction. To determine whether the response to oxidative stress is independent from the response to changing carbon source (the traditional way to induce CYC1 expression), we tested the impact of deleting the gene for Yap1 on the activation profile for ethanol activation (Figure 2A). We also tested the converse, in that we assayed whether the response to oxidative stress was impacted in a strain defective for the response to ethanol induction. Carbon-source dependent activation requires the evolutionarily conserved Hap2, Hap3, Hap4 and Hap5 complex of proteins (55–57). The Hap4 protein provides the transcriptional activation function of the complex (58), and expression of the HAP4 gene is induced during growth in non-fermentable carbon sources (58). Therefore, we tested this deletion strain for the response to oxidative stress (Figure 2B). We found that the Yap1 and Hap4 activators are specific to their respective inducing conditions, and there is no evidence for connectivity between these two pathways upstream of the poised RNAPII complex.
Figure 2.
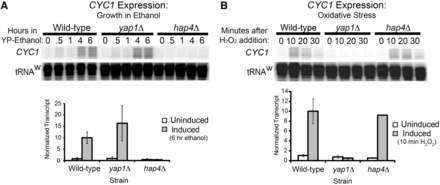
Activator dependence at CYC1. (A) CYC1 activation during growth in non-fermentable carbon sources is Hap4-dependent. An S1 nuclease protection assay was used to analyze RNA prepared from the wild-type, yap1Δ and hap4Δ strains after cells were transferred to media containing 3% ethanol as the carbon source. A tRNAw probe was used as a loading control. Representative gel is shown with quantification of transcript levels 6 h after transfer to a non-fermentable carbon source. Levels were analyzed in biological triplicate. (B) CYC1 activation during oxidative stress is Yap1-dependent. Transcript levels were analyzed as described earlier in the text with RNA collected 10, 20 and 30 min after the addition of H2O2 to growth media. A tRNAw probe was used as a loading control in the S1 assays. Representative gel is shown with quantification of CYC1 transcript levels in each strain 10 min after the addition of H2O2. Levels were analyzed in biological triplicate.
Yap1-dependent Mediator requirements at CYC1
Given our finding that two different activators stimulate the preloaded CYC1 gene, we next wondered whether the co-activator requirements overlap or are distinct. We previously found that CYC1 is Mediator-dependent during growth in non-fermentable carbon sources like ethanol (8). Therefore, we explored the role of Mediator at CYC1 during oxidative stress. We first tested a classical temperature-sensitive strain of MED17 (srb4-138) and found that this strain is defective for CYC1 expression under the non-permissive conditions in response to oxidative stress (Figure 3A). We next tested for specific Mediator subunit requirements by assaying deletion strains of the non-essential Mediator subunits (Figure 3B). We found that CYC1 transcript levels during oxidative stress are statistically significantly diminished in strains containing deletions of the MED18, MED19 and MED20 genes (P < 0.015 using a t-test). Transcript levels were not statistically significantly changed in the uninduced condition in any of the strains. MED18 and MED20 encode proteins that form a heterodimer in the head module of the complex (59). They are the only non-essential proteins within this module. The precise location of the Med19 protein within Mediator is currently unknown, as it is not included in any high-resolution models of the complex (23,60). Although Med19 was previously assigned to the head module (61), more recent studies suggest this subunit is better viewed as a connector between the head and middle modules (23,62–64). The transcriptional defect of CYC1 is largely specific for these head-related non-essential subunits, as transcript levels are not statistically significantly affected in strains containing deletions of the middle or tail module subunits of Mediator. Thus, activation of the poised complex during oxidative stress requires specific subunits of Mediator, which are similar to the requirements observed for activation during ethanol induction (8).
Figure 3.
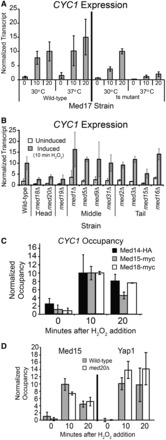
CYC1 expression is Mediator dependent during oxidative stress. (A) CYC1 expression in strains with wild-type Med17 and a temperature sensitive mutant of Med17 (srb4-138). Transcript levels are shown at the permissive (30°C) and non-permissive temperature (37°C) in the uninduced condition, and 10 and 20 min after the addition of H2O2, as indicated. CYC1 transcript levels were normalized to a tRNAw probe, expression level in the wild-type strain 20 min after activation was set to 10. The average ±SD of three biological replicates is shown. (B) Certain non-essential subunits of the Mediator complex are required for CYC1 activation during oxidative stress. Transcript levels were analyzed with RNA prepared from the wild-type strain and strains containing deletions of the indicated Mediator subunit in both uninduced (YP-Glucose, unfilled bars) and induced (10 min H2O2 treatment, gray bars) conditions. CYC1 transcript level was normalized as described earlier in the text. Expression in the wild-type strain at 10 min after activation was set to 10. The average ±SD of three biological replicates is shown for all strains except the tail module deletions, which is shown in biological duplicate. (C) Mediator subunits are recruited to CYC1 during oxidative stress. Occupancy of Med14-HA, Med15-myc and Med18-myc was determined with a time course ChIP. Occupancy at the GAL10 promoter region was subtracted from the occupancy at CYC1, and the 10-min time point was set to 10. Bars represent the average ±SD of two or three biological replicates. (D) Mediator and Yap1 occupancy at the CYC1 promoter is independent of Med20, as occupancy of Med15-myc and Yap1-myc increases on gene activation in both the wild-type (gray bars) and med20Δ strain (white bars). Occupancy of Med15-myc and Yap1-myc was determined with a time course ChIP. Occupancy at the GAL10 promoter region was subtracted from the occupancy at CYC1, and the 10-min time point was set to 10. Bars represent the average ±SD of three biological replicates.
We also performed a ChIP assay to determine whether Mediator occupies the CYC1 promoter during oxidative stress. We tested occupancy of three tagged subunits of Mediator, representing the head module (Med18), the middle module (Med14) and the tail module (Med15). We observed that occupancy of each subunit is not detectable before activation and increases significantly on gene induction with oxidative stress (Figure 3C). This demonstrates that Mediator does not occupy the promoter in the uninduced condition but is recruited to CYC1 during activation.
Since activator proteins interact with Mediator, activator occupancy could be affected by a change in Mediator composition (65). To test this, we performed a ChIP for Yap1 in the med20Δ strain. We found that Yap1 occupancy is not compromised on deletion of Med20 but is recruited normally in the med20Δ background (Figure 3D and Supplementary Figure S2). Therefore, lack of Med20 does not affect activator occupancy at CYC1. We also tested whether Mediator recruitment is changed in the absence of Med20, by examining occupancy of the Med15 subunit in the med20Δ strain (Figure 3D and Supplementary Figure S2). There is no statistically significant difference in Med15 occupancy when Med20 is missing; therefore, Yap1 and Mediator are present even when transcription is compromised by deletion of this head module subunit.
Mediator subunits are necessary during oxidative stress and growth on non-fermentable carbon sources
We found that Med18, Med19 and Med20 are critical for controlling transcription of the preloaded CYC1 gene; yet, these subunits are non-essential for cell viability under normal conditions. To test the physiological importance of these proteins, we analyzed phenotypes of 11 Mediator deletion strains through serial spot analysis on media supplemented with H2O2 or on media containing a non-fermentable carbon source (ethanol). We found that strains lacking MED18, MED19 and MED20 show diminished cell growth on both types of plates compared with growth on glucose (Figure 4). This indicates the head-related proteins are only non-essential during optimal conditions and are important for expression of genes that help the cell survive during growth on both oxidative stress inducing conditions and non-fermentable carbon sources. Thus, MED18, MED19 and MED20 are essential for induction of the preloaded CYC1 gene and for cell viability under the activating conditions.
Figure 4.
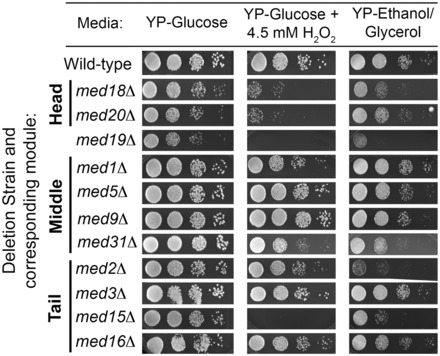
MED18, MED19 and MED20 are required during oxidative stress and growth on non-fermentable carbon sources. Serial spot dilutions of the wild-type strain and strains containing deletions of 11 Mediator subunits on YP-Glucose, YP-Glucose supplemented with H2O2 and YP-Ethanol/Glycerol. Plates were incubated at 30°C for 2–5 days before photographing.
GTT2 is a Yap1-dependent gene induced by oxidative stress but is not poised
We next wanted to test whether other genes induced by H2O2 were preloaded with RNAPII and poised for activation. Using genomic transcriptional profiling data (49,66), we selected a group of H2O2-responsive genes: YSR3, FAD1, AIM13, TRX2, GLR1 and GTT2. We found that YSR3, FAD1 and AIM13 were not H2O2-responsive in our strain background, and although TRX2 and GLR1 were induced in response to H2O2, these genes have high levels of constitutive transcription before induction (data not shown). This basal transcription confounds our ability to clearly distinguish between poised and non-poised promoters; therefore, we did not follow-up with these genes. However, we did find that GTT2 is H2O2-responsive, Yap1-dependent and has low transcript levels in the uninduced condition (Figure 5A). Yap1 binds to the promoter only during activation (Figure 5B), and RNAPII does not occupy this promoter before activation, but is recruited shortly after (Figure 5C). Therefore, unlike CYC1, GTT2 is a bona fide recruitment-regulated gene, as RNAPII is only detectable at the promoter after gene activation.
Figure 5.
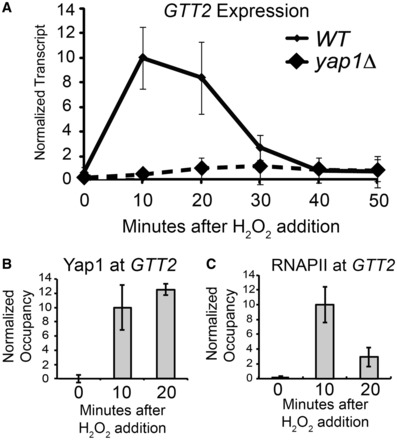
GTT2 is a Yap1-dependent gene without preloaded RNAPII. (A) GTT2 transcript levels are rapidly induced in response to oxidative stress. An S1 nuclease protection assay was performed with RNA isolated from wild-type and yap1Δ cells before exposure to H2O2 and in 10-min intervals after adding H2O2 (0.3 mM). A probe for tRNAw was used as a loading control, and the transcript level in the wild-type strain 10 min after H2O2 exposure was set to 10. Points represent the average ±SD of at least three biological replicates for the wild-type, and in biological duplicate for the yap1Δ strain. (B) Yap1 directly stimulates GTT2 transcription, as Yap1-myc occupies this promoter during oxidative stress. Occupancy was determined with a ChIP of a Yap1-myc fusion protein. Occupancy at the GAL10 promoter region was subtracted from the occupancy at GTT2, and the 10-min time point was set to 10. Bars represent the average ±SD of three biological replicates. (C) RNAPII does not occupy the GTT2 promoter before gene activation but is recruited on activation. Occupancy at the GAL10 promoter region was subtracted from the occupancy at GTT2, and the 10-min time point was set to 10. Bars represent the average ±SD of three biological replicates.
Mediator requirements at GTT2
To determine whether the Mediator-dependency profile of CYC1 is shared with GTT2, we next tested the requirement for this co-activator during activation of GTT2. Similar to CYC1, we found the temperature-sensitive strain of MED17 (srb4-138) was defective in its ability to activate the GTT2 gene in response to oxidative stress at the non-permissive temperature (Figure 6A). This indicates that Mediator is required for normal GTT2 activation. Given the finding that Med18, Med19 and Med20 are required for stimulating the preloaded CYC1 gene, we next tested the effect of removing these proteins on GTT2 expression. Although there is a slight reduction in GTT2 transcript levels in the med18Δ and med20Δ strains (Figure 6B), the difference from wild-type is not statistically significant, whereas the reduction is significant and severe for CYC1 expression (Figure 3B). To confirm Mediator occupancy at the GTT2 promoter, we performed ChIP assays for head, middle and tail subunits before and during treatment with H2O2. We found Mediator does not occupy this promoter in the uninduced condition, but occupancy of each module increases significantly on activation (Figure 6C). Furthermore, absence of Med20 does not disrupt recruitment of Med15 or Yap1 (Figure 6D). Taken together, these results indicate that Mediator is required for GTT2 activation, but the Med18, Med19 and Med20 subunits are dispensable.
Figure 6.
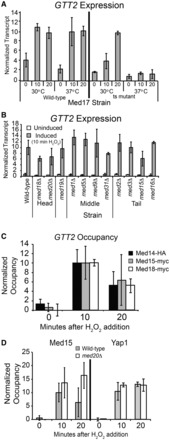
GTT2 activation is Mediator dependent, but does not rely on the Med18, Med20 and Med20 subunits for expression. (A) GTT2 expression in strains with wild-type Med17 and a temperature sensitive mutant of Med17 (srb4-138). Transcript levels are shown at the permissive (30°C) and non-permissive temperature (37°C) in the uninduced condition, and 10 and 20 min after the addition of H2O2, as indicated. Transcript levels were analyzed as described in Figure 3. The average ±SD of three biological replicates is shown. (B) GTT2 activation is largely independent of the Med18, Med19 and Med20 proteins. RNA levels from indicated strains were analyzed as described in Figure 3. The average ±SD of three biological replicates is shown for all strains except the tail module deletions, which is shown in biological duplicate. (C) Mediator subunits are recruited to GTT2 during oxidative stress. Occupancy of Med14-HA, Med15-myc and Med18-myc were determined with a time course ChIP and analyzed as described in Figure 3. (D). Mediator and Yap1 occupancy at the GTT2 promoter is independent of Med20, as occupancy of Med15-myc and Yap1-myc increases on gene activation in both the wild-type (gray bars) and med20Δ strain (white bars). Occupancy of Med15-myc and Yap1-myc was determined with a time course ChIP and analyzed as described in Figure 3.
Mediator promotes postrecruitment events at CYC1
Mediator is well-characterized for its functional role in recruiting RNAPII to promoters upon activation (25,67). However, this function is unnecessary for CYC1 expression, as RNAPII is present at the poised promoter prior to gene activation (7–9,14,15), and this occupancy does not depend on Med20, as it is maintained in the med20Δ strain (Figure 7A). While this promoter is classified as poised (i.e. possesses RNAPII occupancy before induction), we do observe an ∼3-fold increase in RNAPII occupancy after induction with H2O2 (Figure 7A). This is not observed when CYC1 is induced by ethanol (8), which may be due to the much more rapid induction observed during oxidative stress. This increase in occupancy is still observed in the med20Δ strain, although it is not correlated with a corresponding increase in productive transcripts (to the contrary; Figure 3B) and may depend on other subunits of Mediator or other unknown components. Like CYC1, the Drosophila Hsp70 gene, which is the best-characterized postrecruitment regulated gene, also shows an increase in RNAPII occupancy during induction (68).
Figure 7.
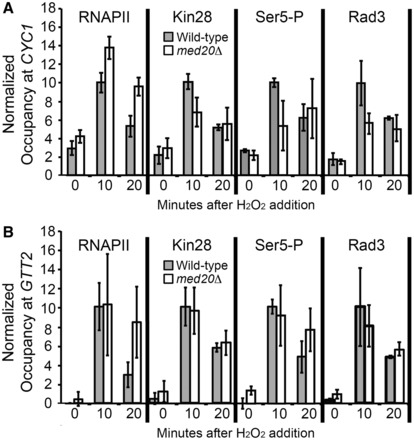
Med20 is required for normal TFIIH recruitment and Ser5 phosphorylation at the promoter of CYC1 but not at GTT2. (A) Occupancy of RNAPII, Kin28-myc, Ser5-P and Rad3-HA in the wild-type (gray bars) and med20Δ (white bars) strains was determined at the CYC1 promoter with a time course ChIP. Occupancy at the GAL10 promoter region was subtracted from the occupancy at CYC1, and the 10-min time point in the wild-type strain was set to 10. Bars represent the average ±SD of three biological replicates. (B) Occupancy of RNAPII, Kin28-myc, Ser5-P and Rad3-HA in the wild-type (gray bars) and med20Δ (white bars) strains was determined at the GTT2 promoter as described in (A).
Mediator is also known to stimulate TFIIH-dependent phosphorylation of the CTD (25,69). Kin28, the cyclin-dependent kinase activity of TFIIH, phosphorylates the CTD of RNAPII (41,70–72). Therefore, we tested whether Kin28 occupancy at the CYC1 and GTT2 promoters is dependent on the Med20 subunit of the Mediator complex. We tagged Kin28 in a wild-type and med20Δ strain (Supplementary Figure S2) and performed a ChIP assay. In wild-type cells, we found that Kin28 is preloaded at the CYC1 promoter, but the occupancy increases upon activation by oxidative stress (Figure 7A). At the GTT2 promoter Kin28 is recruited only during activation (Figure 7B). H2O2-induced Kin28 recruitment is significantly diminished at the CYC1 promoter region upon deletion of the Med20 protein (P < 0.05 at the 10-min time point). In contrast, recruitment of Kin28 is unaffected at the GTT2 promoter. Consistent with the kinase occupancy observation, we also observe statistically significantly diminished Ser5 phosphorylation of the CTD of RNAPII at CYC1 upon induction in the med20Δ strain (P < 0.05 at the 10-min time point) but found no impact at the GTT2 gene (Figure 7A and B). Mediator also interacts with one of the helicase subunits of TFIIH, Rad3 (25). Recent structural studies of TFIIH and new models of the preinitiation complex reveal Rad3, along with the kinase submodule, is positioned near the proposed location of the Mediator head (64,73,74). Therefore, we also tagged Rad3 (Supplementary Figure S2) and tested Rad3 occupancy in the presence and absence of Med20. We found that H2O2-induced Rad3 recruitment is statistically significantly diminished at CYC1 in the absence of Med20 (P < 0.05 at the 10-min time point), but the deletion does not affect recruitment at GTT2 (Figure 7A and B). These findings reveal that the head module of Mediator, specifically the Med20 protein, is required for normal levels and activity of the Kin28 kinase subunit of TFIIH, and for normal recruitment of the Rad3 helicase at the preloaded CYC1 gene. Although Med20 is required for wild-type levels and activity of TFIIH, there is still a considerable increase in recruitment of these factors in the med20Δ strain (at the 20-min time point, the defect seen at 10 min is abolished). At the GTT2 gene, which does not contain a preloaded polymerase, Kin28 recruitment and activity and Rad3 recruitment are completely independent of the Med20 subunit (Figure 7B).
DISCUSSION
Here, we investigated the response of the preloaded CYC1 gene to different biological stimuli. We find that this preloaded promoter responds to two environmental conditions via two different activators: the Hap complex and Yap1. The Hap complex controls CYC1 transcription during growth in non-fermentable carbon sources such as ethanol. Yap1 controls CYC1 transcription during oxidative stress. Both activators stimulate this gene via Mediator. Interestingly, our investigation of CYC1 expression under both conditions reveals that three non-essential subunits (Med18, Med19 and Med20) are required for gene activation. Therefore, two different activator proteins stimulate the poised CYC1 promoter via Mediator, and the same subunits of this large complex are required for each activation scheme.
Our study of the regulation of GTT2 transcription revealed differences at this promoter compared with CYC1. In the uninduced condition, the GTT2 promoter does not contain polymerase, whereas polymerase is present at CYC1 (Figure 7). On induction with H2O2, Yap1 is rapidly recruited to both genes (Figures 1 and 5), followed by activation of gene expression of both CYC1 and GTT2. These findings highlight the versatility of activator proteins, as Yap1 can operate at two fundamentally different promoter settings in vivo (one preloaded and one lacking polymerase before stimulation). Further, Med18, Med19 and Med20 appear to play a more important role at CYC1 than at GTT2. These proteins are not generally required for transcriptional processes, as these subunits are non-essential under rich growth conditions. In addition, genome-wide studies show the expression of only a small set of genes is impacted by the deletion of MED18 or MED20 when yeast are grown under standard conditions (59). However, we demonstrate that these proteins are critical for cell growth in ethanol or during oxidative stress (Figure 4). These conditions induce the CYC1 gene, indicating they form an accessory domain that is essential in certain contexts. A study of the genes induced by the Met4 activator demonstrated that Med18 and Med20 are not required for activation of recruitment-regulated Met4 target genes (75). Our study of CYC1 and GTT2 transcription also suggests that this accessory domain is more important at poised genes than recruitment-regulated genes.
At CYC1, we found that Kin28 occupancy is diminished 10 min after activation when Med20 is deleted. Although the defect was slight (albeit statistically significant), we observed a similar pattern for two uniquely tagged subunits of TFIIH (Kin28-myc and Rad3-HA), and for the phospho-Ser5 mark on the CTD of RNAPII, providing support for the trend. The diminished occupancy and activity we observed could result from a loss of direct interactions between Mediator and TFIIH, as TFIIH interacts with several subunits of the Mediator complex, including the head region (24,25,76). Further, genetic interaction with the CTD was used in the original SRB screen that discovered many Mediator components, including head module subunits (77). Although we observed a statistically significant decrease in Kin28, Ser5-phosphorylation and Rad3 occupancy at CYC1 in the med20Δ strain, there is still some Kin28 and Rad3 present and active in the absence of this Mediator subunit (Figure 7A). Therefore, TFIIH does not exclusively rely on Med20 for normal occupancy at CYC1. Furthermore, it seems unlikely that failure to fully recruit or activate Kin28 and Rad3 is the only defect in the med20Δ strain, given the significant decrease in transcript level observed in the absence of this protein.
What other function could these Mediator subunits play at the preloaded CYC1 promoter? We propose two (not mutually exclusive) possibilities. First, it is possible that these subunits are required for recruitment or stability of an additional factor(s) at the CYC1 promoter. As we found that MED18, MED19 and MED20 are not required for GTT2 transcription, these subunits do not perform this proposed function at GTT2. A second possibility is that these Mediator subunits are critical for altering the conformation of the preloaded RNAPII at CYC1 into an actively transcribing state. Activation of stalled human RNAPII is correlated with conformational changes induced by activator contact with the essential Med17 (Srb4) subunit in the Head (21). Biophysical evidence supports the idea that structural shifts occur in both Mediator and RNAPII on interaction in human and yeast systems (23,64,79–82). Recent structural studies of Mediator reveal that Med18 and Med20 are in a prime location to affect conformational changes in RNAPII. This heterodimer is at the distal end of a movable jaw in the head module (23,63). The Rpb4/7 heterodimer of RNAPII is between the fixed and movable jaw of the head module (63). Rpb4/7 can dissociate from the polymerase and appears to control the conformation of the clamp domain of the enzyme (64,83). Given our findings at CYC1 and GTT2, together with the biophysical evidence, we favor the second possibility. Here, Mediator, containing Med18, Med19 and Med20, is required for affecting a conformational change in RNAPII at the poised promoter, shifting it to a more active conformation that is compatible with full Kin28 and Rad3 binding and activity (Supplementary Figure S3). In this model, the impact we observed on TFIIH at CYC1 is a marker for the essential Med20-dependent conformational change that occurs in the poised complex to transition into a TFIIH-stably associated and actively transcribing state.
Identification and characterization of additional poised genes will allow further study into the specific role of Mediator at genes with preloaded RNAPII. With these tools, we can test our model more rigorously. Additionally, biophysical studies can probe the Mediator-polymerase interaction more fully to determine the role of the accessory domain. Furthermore, we found that a single gene (CYC1) requires the same Mediator subunits for activation in response to growth in non-fermentable carbon sources or oxidative stress. Activation in response to these stimuli occurs via two distinct activators: the Hap complex and Yap1. As poising, Mediator and TFIIH are conserved features of transcriptional regulation, these findings provide important new insight on the regulation of gene expression at preloaded promoters.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [64,73,74].
FUNDING
National Institutes of Health (NIH) [P01GM088409 to L.A.S]; Welch Foundation [R-0021 to S.K.L.]. Funding for open access charge: NIH; and the Chemistry and Biochemistry Department at Abilene Christian University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Alan Hinnebusch for providing the Med15-myc and Med18-myc strains, Julie Fischbeck for generating the Kin28-myc med20Δ strain and the Rad3-HA med20Δ strain, and Tyler Fara for help with RNA preparations.
REFERENCES
- 1.Nechaev S, Adelman K. Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine M. Paused RNA Polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selth LA, Sigurdsson S, Svejstrup JQ. Transcript elongation by RNA polymerase II. Annu. Rev. Biochem. 2010;79:271–293. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- 4.Conaway RC, Conaway JW. Origins and activity of the Mediator complex. Semin. Cell Dev. Biol. 2011;22:729–734. doi: 10.1016/j.semcdb.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reppas NB, Wade JT, Church GM, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol. Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 6.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 8.Lee SK, Fletcher AG, Zhang L, Chen X, Fischbeck JA, Stargell LA. Activation of a poised RNAPII-dependent promoter requires both SAGA and Mediator. Genetics. 2010;184:659–672. doi: 10.1534/genetics.109.113464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Fletcher A, Cheung V, Winston F, Stargell LA. Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Mol. Cell. Biol. 2008;28:1393–1403. doi: 10.1128/MCB.01733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kininis M, Isaacs GD, Core LJ, Hah N, Kraus WL. Postrecruitment regulation of RNA polymerase II directs rapid signaling responses at the promoters of estrogen target genes. Mol. Cell. Biol. 2009;29:1123–1133. doi: 10.1128/MCB.00841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- 14.Martens C, Krett B, Laybourn P. RNA polymerase II and TBP occupy the repressed CYC1 promoter. Mol. Microbiol. 2001;40:1009–1019. doi: 10.1046/j.1365-2958.2001.02445.x. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Ding M, Pederson DS. Binding of TFIID to the CYC1 TATA boxes in yeast occurs independently of upstream activating sequences. Proc. Natl Acad. Sci. USA. 1994;91:11909–11913. doi: 10.1073/pnas.91.25.11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional Mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjorklund S, Gustafsson CM. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 2005;30:240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD. Conserved structures of Mediator and RNA polymerase II holoenzyme. Science. 1999;283:985–987. doi: 10.1126/science.283.5404.985. [DOI] [PubMed] [Google Scholar]
- 20.Dotson MR, Yuan CX, Roeder RG, Myers LC, Gustafsson CM, Jiang YW, Li Y, Kornberg RD, Asturias FJ. Structural organization of yeast and mammalian Mediator complexes. Proc Natl Acad Sci USA. 2000;97:14307–14310. doi: 10.1073/pnas.260489497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer KD, Lin SC, Bernecky C, Gao Y, Taatjes DJ. p53 activates transcription by directing structural shifts in Mediator. Nat. Struct. Mol. Biol. 2010;17:753–760. doi: 10.1038/nsmb.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis JA, Takagi Y, Kornberg RD, Asturias FA. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 23.Cai G, Imasaki T, Yamada K, Cardelli F, Takagi Y, Asturias FJ. Mediator head module structure and functional interactions. Nat. Struct. Mol. Biol. 2010;17:273–279. doi: 10.1038/nsmb.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakurai H, Fukasawa T. Functional connections between Mediator components and general transcription factors of Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:37251–37256. doi: 10.1074/jbc.M004364200. [DOI] [PubMed] [Google Scholar]
- 25.Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol. Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 26.Soutourina J, Wydau S, Ambroise Y, Boschiero C, Werner M. Direct interaction of RNA polymerase II and Mediator required for transcription in vivo. Science. 2011;331:1451–1454. doi: 10.1126/science.1200188. [DOI] [PubMed] [Google Scholar]
- 27.Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Malik S, Barrero MJ, Jones T. Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. Proc Natl Acad Sci USA. 2007;104:6182–6187. doi: 10.1073/pnas.0608717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guglielmi B, Soutourina J, Esnault C, Werner M. TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc Natl Acad Sci USA. 2007;104:16062–16067. doi: 10.1073/pnas.0704534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balamotis MA, Pennella MA, Stevens JL, Wasylyk B, Belmont AS, Berk AJ. Complexity in transcription control at the activation domain-Mediator interface. Sci. Signal. 2009;2:ra20. doi: 10.1126/scisignal.1164302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al. Human Mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conaway RC, Conaway JW. The Mediator Complex and transcription elongation. Biochim. Biophys. Acta. 2012;1829:69–75. doi: 10.1016/j.bbagrm.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukundan B, Ansari A. Novel role for Mediator complex subunit Srb5/Med18 in termination of transcription. J. Biol. Chem. 2011;286:37053–37057. doi: 10.1074/jbc.C111.295915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding N, Zhou H, Esteve PO, Chin HG, Kim S, Xu X, Joseph SM, Friez MJ, Schwartz CE, Pradhan S, et al. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol. Cell. 2008;31:347–359. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol. Cell. 2011;42:536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kremer SB, Kim S, Jeon JO, Moustafa YW, Chen A, Zhao J, Gross DS. Role of Mediator in regulating Pol II elongation and nucleosome displacement in Saccharomyces cerevisiae. Genetics. 2012;191:95–106. doi: 10.1534/genetics.111.135806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlsten JO, Szilagyi Z, Liu B, Lopez MD, Szaszi E, Djupedal I, Nystrom T, Ekwall K, Gustafsson CM, Zhu X. Mediator promotes CENP-A incorporation at fission yeast centromeres. Mol. Cell. Biol. 2012;32:4035–4043. doi: 10.1128/MCB.00374-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu H, Hu C, Zhang F, Hwang GJ, Swanson MJ, Boonchird C, Hinnebusch AG. Interdependent recruitment of SAGA and Srb Mediator by transcriptional activator Gcn4p. Mol. Cell. Biol. 2005;25:3461–3474. doi: 10.1128/MCB.25.9.3461-3474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang F, Sumibcay L, Hinnebusch AG, Swanson MJ. A triad of subunits from the Gal11/tail domain of Srb Mediator is an in vivo target of transcriptional activator Gcn4p. Mol. Cell. Biol. 2004;24:6871–6886. doi: 10.1128/MCB.24.15.6871-6886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holstege FC, Jennings EG, Wyrick CJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 42.Longtine M, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 43.Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 44.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yearling MN, Radebaugh CA, Stargell LA. The Transition of poised RNA Polymerase II to an actively elongating state is a “complex” affair. Genet. Res. Int. 2011;2011:206290. doi: 10.4061/2011/206290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boettiger AN, Ralph PL, Evans SN. Transcriptional regulation: effects of promoter proximal pausing on speed, synchrony and reliability. PLoS Comput. Biol. 2011;7:e1001136. doi: 10.1371/journal.pcbi.1001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saha RN, Wissink EM, Bailey ER, Zhao M, Fargo DC, Hwang JY, Daigle KR, Fenn JD, Adelman K, Dudek SM. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat. Neurosci. 2011;14:848–856. doi: 10.1038/nn.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 2009;324:92–94. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- 49.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Cell. Biol. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu A, Wemmie JA, Edgington NP, Goebl M, Guevara JL, Moye-Rowley WS. Yeast bZip proteins mediate pleiotropic drug and metal resistance. J. Biol. Chem. 1993;268:18850–18858. [PubMed] [Google Scholar]
- 51.Coleman ST, Epping EA, Steggerda S, Moye-Rowley WS. Yap1p activates gene transcription in an oxidant-specific fashion. Mol. Cell. Biol. 1999;12:8302–8313. doi: 10.1128/mcb.19.12.8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuge S, Toda T, Iizuka N, Nomoto A. Crm1 (Xpo1)dependent nuclear export of the budding yeast transcription factor yAP-1 is sensitive to oxidative stress. Genes Cells. 1998;3:521–532. doi: 10.1046/j.1365-2443.1998.00209.x. [DOI] [PubMed] [Google Scholar]
- 53.Wood MJ, Storz G, Tjandra N. Structural basis for redox regulation of Yap1 transcription factor localization. Nature. 2004;430:917–921. doi: 10.1038/nature02790. [DOI] [PubMed] [Google Scholar]
- 54.Kuge S, Jones N, Nomoto A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997;16:1710–1720. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forsburg SL, Guarente L. Mutational analysis of upstream activation sequence 2 of the CYC1 gene of Saccharomyces cerevisiae: a HAP2-HAP3-responsive site. Mol. Cell. Biol. 1988;8:647–654. doi: 10.1128/mcb.8.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olesen J, Hahn S, Guarente L. Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell. 1987;51:953–961. doi: 10.1016/0092-8674(87)90582-4. [DOI] [PubMed] [Google Scholar]
- 57.Schuller HJ. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 2003;43:139–160. doi: 10.1007/s00294-003-0381-8. [DOI] [PubMed] [Google Scholar]
- 58.Forsburg SL, Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- 59.Lariviere L, Geiger S, Hoeppner S, Rother S, Strasser K, Cramer P. Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat. Struct. Mol. Biol. 2006;13:895–901. doi: 10.1038/nsmb1143. [DOI] [PubMed] [Google Scholar]
- 60.Imasaki T, Calero G, Cai G, Tsai KL, Yamada K, Cardelli F, Erdjument-Bromage H, Tempst P, Berger I, Kornberg GL, et al. Architecture of the Mediator head module. Nature. 2011;475:240–243. doi: 10.1038/nature10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang JS, Kim SH, Hwang MS, Han SJ, Lee YC, Kim YJ. The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 2001;276:42003–42010. doi: 10.1074/jbc.M105961200. [DOI] [PubMed] [Google Scholar]
- 62.Takagi Y, Calero G, Komori H, Brown JA, Ehrensberger AH, Hudmon A, Asturias F, Kornberg RD. Head module control of mediator interactions. Mol. Cell. 2006;23:355–364. doi: 10.1016/j.molcel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 63.Baidoobonso SM, Guidi BW, Myers LC. Med19(Rox3) regulates Intermodule interactions in the Saccharomyces cerevisiae mediator complex. J. Biol. Chem. 2007;282:5551–5559. doi: 10.1074/jbc.M609484200. [DOI] [PubMed] [Google Scholar]
- 64.Cai G, Chaban YL, Imasaki T, Kovacs JA, Calero G, Penczek PA, Takagi Y, Asturias FJ. Interaction of the mediator head module with RNA polymerase II. Structure. 2012;20:899–910. doi: 10.1016/j.str.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim S, Gross DS. Mediator recruitment to heat shock genes requires dual hsf1 activation domains and Mediator tail subunits med15 and med16. J. Biol. Chem. 2013;288:12197–12213. doi: 10.1074/jbc.M112.449553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Choi JH, Lou W, Vancura A. A novel membrane-bound glutathione S-transferase functions in the stationary phase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:29915–29922. doi: 10.1074/jbc.273.45.29915. [DOI] [PubMed] [Google Scholar]
- 67.Biddick RK, Law GL, Chin KK, Young ET. The transcriptional coactivators SAGA, SWI/SNF, and Mediator make distinct contributions to activation of glucose-repressed genes. J. Biol. Chem. 2008;283:33101–33109. doi: 10.1074/jbc.M805258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol. Cell. Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein Mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 70.Valay JG, Simon M, Faye G. The KIN28 protein kinase is associated with a cyclin in Saccharomyces cerevisiae. J. Mol. Biol. 1993;234:307–310. doi: 10.1006/jmbi.1993.1587. [DOI] [PubMed] [Google Scholar]
- 71.Feaver WJ, Svejstrup J, Henry NL, Kornberg RD. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 72.Valay JG, Simon M, Dubois MF, Bensaude O, Facca C, Faye G. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J. Mol. Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- 73.Gibbons BJ, Brignole EJ, Azubel M, Murakami K, Voss NR, Bushnell DA, Asturias FJ, Kornberg RD. Subunit architecture of general transcription factor TFIIH. Proc. Natl Acad. Sci. USA. 2012;109:1949–1954. doi: 10.1073/pnas.1105266109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meyer PA, Fu J. Mutual remodeling and conformation grid: a Mediator code? Structure. 2012;20:755–757. doi: 10.1016/j.str.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leroy C, Cormier L, Kuras L. Independent recruitment of Mediator and SAGA by the activator Met4. Mol Cell Biol. 2006;26:3149–3163. doi: 10.1128/MCB.26.8.3149-3163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee JH, Cai G, Panigrahi AK, Dunham-Ems S, Nguyen TN, Radolf JD, Asturias FJ, Gunzl A. A TFIIH-associated Mediator head is a basal factor of small nuclear spliced leader RNA gene transcription in early-diverged trypanosomes. Mol. Cell. Biol. 2010;30:5502–5513. doi: 10.1128/MCB.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nonet ML, Young RA. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taatjes DJ, Naar AM, Andel F, III, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- 79.Taatjes DJ, Schneider-Poetsch T, Tjian R. Distinct conformational states of nuclear receptor-bound CRSP-Med complexes. Nat. Struct. Mol. Biol. 2004;11:664–671. doi: 10.1038/nsmb789. [DOI] [PubMed] [Google Scholar]
- 80.Bernecky C, Grob P, Ebmeier CC, Nogales E, Taatjes DJ. Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly. PLoS Biol. 2011;9:e1000603. doi: 10.1371/journal.pbio.1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bernecky C, Taatjes DJ. Activator-Mediator binding stabilizes RNA polymerase II orientation within the human Mediator-RNA polymerase II-TFIIF assembly. J. Mol. Biol. 2012;417:387–394. doi: 10.1016/j.jmb.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsai KL, Sato S, Tomomori-Sato C, Conaway RC, Conaway JW, Asturias FJ. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat. Struct. Mol. Biol. 2013;20:611–619. doi: 10.1038/nsmb.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bushnell DA, Kornberg RD. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc. Natl Acad. Sci. USA. 2003;100:6969–6973. doi: 10.1073/pnas.1130601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


