Abstract
Surveying the fungi of alkaline soils in Siberia, Trans-Baikal regions (Russia), the Aral lake (Kazakhstan), and Eastern Mongolia, we report an abundance of alkalitolerant species representing the Emericellopsis-clade within the Acremonium cluster of fungi (order Hypocreales). On an alkaline medium (pH ca. 10), 34 acremonium-like fungal strains were obtained. One of these was able to develop a sexual morph and was shown to be a new member of the genus Emericellopsis, described here as E. alkalina sp. nov. Previous studies showed two distinct ecological clades within Emericellopsis, one consisting of terrestrial isolates and one predominantly marine. Remarkably, all the isolates from our study sites show high phylogenetic similarity based on six loci (LSU and SSU rDNA, RPB2, TEF1-α, β-tub and ITS region), regardless of their provenance within a broad geographical distribution. They group within the known marine-origin species, a finding that provides a possible link to the evolution of the alkaliphilic trait in the Emericellopsis lineage. We tested the capacities of all newly isolated strains, and the few available reference ex-type cultures, to grow over wide pH ranges. The growth performance varied among the tested isolates, which showed differences in growth rate as well as in pH preference. Whereas every newly isolated strain from soda soils was extremely alkalitolerant and displayed the ability to grow over a wide range of ambient pH (range 4–11.2), reference marine-borne and terrestrial strains showed moderate and no alkalitolerance, respectively. The growth pattern of the alkalitolerant Emericellopsis isolates was unlike that of the recently described and taxonomically unrelated alkaliphilic Sodiomyces alkalinus, obtained from the same type of soils but which showed a narrower preference towards high pH.
Keywords: Acremonium, Emericellopsis, alkaline soils, molecular phylogeny, pH tolerance, soda soils
INTRODUCTION
Alkaline soils (or soda soils) and soda lakes represent a unique environmental niche. There are few studies available on the fungal biodiversity therein. The eye-catching characteristic of these soils is a high pH maintained mainly by the buffering capacities of soluble carbonates present. Soda accumulation is thought to be a common process associated with savannas, steppes and desert regions across the world (Jones et al. 1998). Some examples of such extreme occurrences include the Magadi Lake in Kenya and the Natron Lake in Tanzania where the pH values of water are as high as 11–12. Seventy fungi have been isolated from The Dead Sea in Israel, almost half Eurotiales, where the salt levels are 340–350 g salt/L (Buchalo et al. 2009). In Russia, alkaline soils are mostly restricted to areas adjacent to saline lake basins in south-western Siberia (Sorokin et al. 2008).
Naturally, high salts concentration and high environmental pH impose a substantial amount of stress to any living organism. Some have adapted and therefore evolved metabolic pathways in order to thrive in such harsh conditions, such as high osmotic pressures, low water potentials, and, clearly, elevated ambient pHs (>9). The vast majority of so-called alkaliphiles, with a growth optimum at pH above 9, include prokaryotes (Duckworth et al. 1996). However, some filamentous fungi have been shown to be able to grow optimally at pH values exceeding 9 (Nagai et al. 1995, 1998, Grum-Grzhimaylo et al. 2013). Alkaliphily in filamentous fungi is uncommon, while alkalitolerance, on the other hand, is far more widespread. Alkalitolerant fungi, i.e. fungi that can grow to some extent at an alkaline pH but with their optimum still being at neutral pH values, are not only of basic scientific interest for the molecular mechanisms of adaptation, but also in the search for potentially biotechnologically valuable enzymes. It has become more obvious that alkalitolerant fungi may be encountered in many neutral soils (Kladwang et al. 2003, Elíades et al. 2006). The relative abundance of alkalitolerant fungi has facilitated studies on both their biodiversity and their enzymatic properties. And yet, truly alkaliphilic filamentous fungi have been isolated infrequently. The few existing descriptive studies on alkalitolerant and alkaliphilic fungi show a bias towards fungi with simple conidial morphology, commonly asexual Acremonium or Verticillium species, and typically, without the development of the any sexual morph (Okada et al. 1993, Kladwang et al. 2003). Substantial difficulties in identifying Acremonium species imposed by their simple morphology have stimulated the use of molecular phylogeny in their identification. The array of fungi with acremonium-like conidiation has been shown to be highly polyphyletic, occupying several lineages throughout Ascomycota (Summerbell et al. 2011). However, most Acremonium species belong to Hypocreales (subphylum Hypocreomycetidae). One of the well-defined subclades within the hypocrealean acremonia is the Emericellopsis-clade (family Bionectriaceae), which includes isolates derived from various ecological niches. Notably, previous studies have shown a phylogenetic separation of marine-derived and terrestrial isolates within the Emericellopsis-clade (Zuccaro et al. 2004). The marine clade also contains fungi derived from soda soils. The current study confirms the evolutionary relationships between marine-borne and soda soil fungi of the genus Emericellopsis. Here, we analyse acremonium-like strains isolated from soda soils in western Siberia, the Trans-Baikal area (Russia), the Aral Sea (Kazakhstan) and the Gobi Desert (Mongolia) and elucidate their phylogenetic relationships, with an emphasis on the Emericellopsis-clade. A new Emericellopsis species, E. alkalina sp. nov., is described. We also analysed the newly isolated strains for growth at various pH values, in comparison with reference ex-type strains, and show that the alkalitolerant strains group within the known Emericellopsis isolates originated from the marine habitats. We discuss a possible origin of alkalitolerance in this particular lineage of mostly sea-borne fungi.
MATERIALS AND METHODS
Soil samples, strains and media
Soil samples were collected from several locations on the edge of the soda lakes (Table 1). We used alkaline agar (AA) with the antibiotic rifampicin (2 g/L) as a selective medium for alkalitolerant species isolation. For routine subculturing on AA of the newly isolated strains, the antibiotic was not used. The AA medium was prepared as described previously (Grum-Grzhimaylo et al. 2013). Several reference ex-type Emericellopsis strains were obtained from the KNAW-CBS Fungal Biodiversity Centre (CBS) as well as from the All-Russian Collection of Microorganisms (VKM). For the colony morphology characterization we used several types of media: WA, CZ, MYA, PDA, OA and AA (Mueller et al. 2004). The elucidation of the pH optimum was performed in duplicate using race tubes with the media ranging in pH as described previously (Grum-Grzhimaylo et al. 2013), with the following modification. Instead of using acetic buffer to generate pH 4 and 5.2, we used a citric acid buffer system. Race tubes and plates were incubated in the dark at 28 °C, and the growth rates were recorded once a week over 2 mo.
Table 1.
Strains used in the current study and characteristics of the sites of isolation. Newly isolated strains are given in bold.
| Strain | VKM number | CBS no. | Isolation area | Isolation place | pH of the soil | Total salts (g/kg) | Saltification type |
|---|---|---|---|---|---|---|---|
| Acremonium sclerotigenum A101 | - | - | Trans-Baikal, Russia | near Alla River | 8 | - | sulfate |
| Acremonium sclerotigenum A130 | - | - | Trans-Baikal, Russia | near Alla River | 8 | - | sulfate |
| Acremonium sp. A104 | - | - | Kulunda steppe, Altai, Russia | - | taken from Atriplex verrucifera MB. | soda | |
| Acremonium sp. A105 | - | - | Trans-Baikal, Russia | Orongoyskoe Lake | 7.8 | 26 | soda-sulfate |
| Acremonium sp. A106 | - | - | Trans-Baikal, Russia | Sulfatnoe Lake | 8.5 | 3.7 | sulfate-soda |
| Acremonium sp. A107 | - | - | Trans-Baikal, Russia | Chedder Lake | 9.1 | - | soda |
| Acremonium sp. A108 | - | - | Aral lake, Kazakhstan | Cape Aktumsyk | taken from Sueda salsa | chloride-sulfate | |
| Acremonium sp. A109 | - | - | Trans-Baikal, Russia | Kuchiger area | 9 | - | sulphate |
| Acremonium sp. A110 | - | - | Trans-Baikal, Russia | Sulfatnoe Lake | 10.3 | 139.4 | sulfate-soda |
| Acremonium sp. A111 | - | - | Aral lake, Kazakhstan | Cape Aktumsyk | 8 | - | chloride-sulfate |
| Acremonium sp. E102 | - | - | Kulunda steppe, Altai, Russia | Bezimyannoe Lake | 9.1 | 47 | chloride |
| Emericellopsis alkalina A103 | - | - | Kulunda steppe, Altai, Russia | Mirabilit Lake | 9.6 | 100 | soda-chloride-sulfate |
| Emericellopsis alkalina A112 | - | - | North-East Mongolia | Burd Lake | 10.1 | 33 | soda |
| Emericellopsis alkalina A113 | FW-1476 | - | Choibalsan area, North-East Mongolia | - | 11 | 57 | soda |
| Emericellopsis alkalina A114 | FW-1473 | - | Kulunda steppe, Altai, Russia | Solyonoe Lake | 10 | 187 | chloride |
| Emericellopsis alkalina A115 | FW-1474 | - | Kulunda steppe, Altai, Russia | - | 9.6 | 225 | chloride-sulfate |
| Emericellopsis alkalina A116 | - | - | Kulunda steppe, Altai, Russia | Mirabilit Lake | 9.6 | 100 | soda-chloride-sulfate |
| Emericellopsis alkalina A117 | FW-1471 | - | Kulunda steppe, Altai, Russia | Shukurtuz Lake | 9.9 | 53 | chloride-sulfate |
| Emericellopsis alkalina A118 | - | - | Kulunda steppe, Altai, Russia | Zheltir’ Lake | 9.6 | 137 | soda-chloride |
| Emericellopsis alkalina A119 | - | - | Kulunda steppe, Altai, Russia | Bezimyannoe Lake | 10.1 | 38 | chloride-sulfate |
| Emericellopsis alkalina A120 | - | - | Kulunda steppe, Altai, Russia | Bezimyannoe Lake | 9.9 | 310 | soda |
| Emericellopsis alkalina A121 | - | - | Kulunda steppe, Altai, Russia | Tanatar Lake | 10.2 | 73 | soda |
| Emericellopsis alkalina A122 | - | - | Kulunda steppe, Altai, Russia | - | 9.5 | 65 | chloride |
| Emericellopsis alkalina A123 | - | - | Kulunda steppe, Altai, Russia | - | taken from Salicornia europaea L. | soda | |
| Emericellopsis alkalina A124 | - | - | Kulunda steppe, Altai, Russia | south, Berdabay | 10.1 | 60 | soda |
| Emericellopsis alkalina A125 | - | - | Trans-Baikal, Russia | Nuhe-Nur Lake | 10.1 | 7.1 | soda |
| Emericellopsis alkalina A126 | - | - | Trans-Baikal, Russia | Nuhe-Nur Lake | 10.1 | 1.9 | soda |
| Emericellopsis alkalina A127 | - | - | Trans-Baikal, Russia | Nuhe-Nur Lake | 10.1 | 1.9 | soda |
| Emericellopsis alkalina A128 | - | - | Trans-Baikal, Russia | Sulfatnoe Lake | 10.3 | 139.4 | sulfate-soda |
| Emericellopsis alkalina E101 T | F-4108 | CBS 127350 | Kulunda steppe, Altai, Russia | Tanatar Lake | 10.1 | 73 | soda |
| Emericellopsis alkalina M14 | F-3905 | CBS 120043 | Kulunda steppe, Altai, Russia | Bezimyannoe Lake | 9.9 | 310 | soda |
| Emericellopsis alkalina M20 | FW-3040 | CBS 120044 | Kulunda steppe, Altai, Russia | Zheltir’ Lake | 9.6 | 137 | soda-chloride |
| Emericellopsis alkalina M71 | F-3907 | CBS 120049 | Trans-Baikal, Russia | Sulfatnoe Lake | 10.3 | 139 | sulfate-soda |
| Emericellopsis maritima T | F-1082 | CBS 491.71 | Black sea Sevastopol area, Crimea, Ukraine | sea water | - | - | - |
| Emericellopsis minima | F-1057 | CBS 871.68 | Germany | wheat field soil | - | - | - |
| Emericellopsis minima T | F-1484 | CBS 190.55 | Inhaca, Mozambique | mangrove soil | - | - | - |
| Emericellopsis pallida T | F-925 | CBS 490.71 | Black sea Sevastopol area, Crimea, Ukraine | sea water | - | - | - |
| Sarocladium sp. A131 | - | - | Aral lake, Kazakhstan | Cape Aktumsyk | 8.3 | - | chloride-sulfate |
Morphology
We used light microscopy (LM) and scanning electron microscopy (SEM) for morphological characterization of the strains, as described previously (Grum-Grzhimaylo et al. 2013).
DNA extraction, PCR, and sequencing
Total genomic DNA (gDNA) was extracted from mycelium using DNeasy Plant Mini kit (Qiagen, Chatsworth, CA). We amplified and sequenced six nuclear loci (large and small subunit rDNA, internal transcribed spacers 1 and 2, including 5.8S rDNA, RPB2, TEF1-α and β-tub) from gDNA using the standard primers set. Primer sets, thermo cycling programs and sequencing procedures were performed as described previously (Grum-Grzhimaylo et al. 2013). The amplification of beta-tubulin intron 3 (hereafter named as “β-tub”) was as in Zuccaro et al. (2004).
Phylogenetic analyses
We used five nuclear loci for phylogenetic analysis: large subunit rDNA (LSU), ITS region, RPB2, TEF1-α, and β-tub. The gene for small subunit rRNA (SSU), although sequenced, was not included in our phylogenetic reconstructions since it carried too little phylogenetic signal to contribute to clade differentiation. We constructed separate alignments for each of the analysed genes using the online MAFFT v. 7 service (Katoh & Standley 2013). Ambiguous regions were removed manually from the alignments with BioEdit v. 7.1.3.0 (Hall 1999). Two data sets for different phylogenetic analyses were constructed in order to achieve different degrees of resolution within the studied groups. Appropriate reference sequences were obtained from GenBank. The first analysis included a single LSU gene in order to build a large-scale taxonomy for hypocrealean acremonia. The second, a four-gene (ITS, β-tub, RPB2, and TEF1-α) concatenated super-matrix, was implemented to resolve the recent evolutionary relationships in the Emericellopsis-clade and our newly isolated alkalitolerant strains. The four-gene concatenated data set was constructed using Mesquite v. 2.75 (Maddison & Maddison 2011) and divided into four partitions corresponding to each individual gene. The best-fit model for nucleotide substitution for each partition was chosen according to the corrected Akaike Information Criterion (AICc) as implemented in jModelTest v. 2.1.1 (Guindon & Gascuel 2003, Darriba et al. 2012) (Table 2). GARLI v. 2.0 (Zwickl 2006) was used for Maximum Likelihood (ML) bootstrap analyses; for both phylogenetic analyses the number of searches was set to five for each of the 200 bootstrap replicates. A 50 % majority rule consensus trees were constructed using SumTrees v. 3.3.1 application within DendroPy v. 3.11.0 package (Sukumaran & Holder 2010) running under Python v. 2.6 platform. Bayesian analysis (BI) was performed using MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001). Two independent searches and four chains were set to run for 10 M generations for both phylogenetic analyses sampling every 100th generation. The convergence of the runs was checked in TRACER v. 1.5 (Rambaut & Drummond 2007). The first 30 % (50 % for four-gene analysis) of the resulting trees was eliminated from the further analysis. The rest were used to generate a 50 % majority rule consensus tree and calculate posterior probabilities (PP). The consensus tree was visualized and edited with TreeGraph v. 2.0.47-206 beta (Stöver & Müller 2010) and Adobe Illustrator CS6 (Adobe Systems, San Jose, CA). The node supports were considered to be strong if they received joint scores of ML>90 and PP>0.94. Newly generated sequences from the studied strains were deposited in GenBank with accessions listed in Table 3. Phylogenetic analyses were deposited in TreeBase (submission ID 14196).
Table 2.
Loci and substitution models used for the phylogenetic analyses.
| Phylogenetic analysis | Locus | Model for each partition | Characters | Informative characters | Uninformative variable characters | Invariable characters |
|---|---|---|---|---|---|---|
| 1 | LSU | TIM1+I+G (GTR+I+G)* | 962 | 162 | 62 | 738 |
| 2 | ITS | TIM1+G (GTR+I+G)* | 503 | 101 | 62 | 340 |
| β-tub | TrN+G (HKY+G)* | 333 | 80 | 29 | 224 | |
| RPB2 | TIM3+G (GTR+G)* | 1070 | 73 | 134 | 863 | |
| TEF1-α | TIM3+G (GTR+G)* | 904 | 54 | 58 | 792 |
* - for MrBayes.
Table 3.
List of taxa used for phylogenetic reconstructions. Strains used in the growth experiments and newly generated accessions are in bold.
| Taxon | Voucher | Appearance in phylogenetic analysis (1,2) | LSU | ITS | β-tub | RPB2 | TEF1-α | SSU |
|---|---|---|---|---|---|---|---|---|
| Acremonium acutatum T | CBS 682.71 | 1 | HQ231965 | |||||
| Acremonium alternatum T | CBS 407.66 | 1 | HQ231988 | |||||
| Acremonium biseptum T | CBS 750.69 | 1 | HQ231998 | |||||
| Acremonium brachypenium T | CBS 866.73 | 1 | HQ232004 | |||||
| Acremonium breve T | CBS 150.62 | 1 | HQ232005 | |||||
| Acremonium chrysogenum T | CBS 144.62 | 1 | HQ232017 | |||||
| Acremonium curvulum T | CBS 430.66 | 1 | HQ232026 | |||||
| Acremonium curvulum | CBS 229.75 | 1 | HQ232021 | |||||
| Acremonium exuviarum T | UAMH 9995 | 1,2 | HQ232036 | AY882946 | AY882947 | - | - | |
| Acremonium flavum T | CBS 596.70 | 1 | HQ232037 | |||||
| Acremonium fuci T | CBS 112868 | 2 | AY632653 | AY632690 | - | - | ||
| Acremonium fuci | CBS 113889 | 2 | AY632652 | - | - | - | ||
| Acremonium fuci | UAMH 6508 | 1 | HQ232038 | |||||
| Acremonium gamsii T | CBS 726.71 | 1 | HQ232040 | |||||
| Acremonium inflatum T | CBS 212.69 | 1 | HQ232050 | |||||
| Acremonium inflatum | CBS 439.70 | 1 | HQ232051 | |||||
| Acremonium persicinum T | CBS 310.59 | 1 | HQ232077 | |||||
| Acremonium persicinum | CBS 101694 | 1 | HQ232085 | |||||
| Acremonium persicinum | CBS 102349 | 1 | HQ232086 | |||||
| “Acremonium potronii” | CBS 189.70 | 1 | HQ232094 | |||||
| “Acremonium potronii” | CBS 379.70F | 1,2 | HQ232096 | AY632655 | AY632691 | - | - | |
| Acremonium radiatum T | CBS 142.62 | 1 | HQ232104 | |||||
| Acremonium roseolum T | CBS 289.62 | 1 | HQ232123 | |||||
| Acremonium rutilum T | CBS 396.66 | 1 | HQ232124 | |||||
| Acremonium salmoneum T | CBS 721.71 | 1 | HQ232125 | |||||
| Acremonium salmoneum | JS-NJ01 | 2 | HM747162 | - | - | - | ||
| Acremonium sclerotigenum T | CBS 124.42 | 1 | HQ232126 | |||||
| Acremonium sclerotigenum A101 | 1 | KC987215 | KC987139 | KC987101 | KC998999 | KC998961 | KC987177 | |
| Acremonium sclerotigenum A130 | 1 | KC987242 | KC987166 | KC987128 | KC999024 | KC998988 | KC987204 | |
| Acremonium sclerotigenum | CBS 786.69 | 1 | HQ232130 | |||||
| Acremonium sordidulum T | CBS 385.73 | 1 | HQ232136 | |||||
| Acremonium sp. A104 | 1,2 | KC987217 | KC987141 | KC987103 | KC999001 | KC998963 | KC987179 | |
| Acremonium sp. A105 | 1,2 | KC987218 | KC987142 | KC987104 | KC999002 | KC998964 | KC987180 | |
| Acremonium sp. A106 | 1,2 | KC987219 | KC987143 | KC987105 | KC999003 | KC998965 | KC987181 | |
| Acremonium sp. A107 | 1,2 | KC987220 | KC987144 | KC987106 | KC999004 | KC998966 | KC987182 | |
| Acremonium sp. A108 | 1,2 | KC987221 | KC987145 | KC987107 | KC999005 | KC998967 | KC987183 | |
| Acremonium sp. A109 | 1 | KC987222 | KC987146 | KC987108 | KC999006 | KC998968 | KC987184 | |
| Acremonium sp. A110 | 1,2 | KC987223 | KC987147 | KC987109 | KC999007 | KC998969 | KC987185 | |
| Acremonium sp. A111 | 1,2 | KC987224 | KC987148 | KC987110 | KC999008 | KC998970 | KC987186 | |
| Acremonium sp. E102 | 1,2 | KC987248 | KC987172 | KC987134 | KC999030 | KC998994 | KC987210 | |
| Acremonium tubakii T | CBS 790.69 | 1 | HQ232148 | |||||
| Acremonium tubakii | CBS 111360 | 2 | AY632654 | AY632689 | - | - | ||
| “Acremonium tubakii” | CBS 824.69 | 1 | HQ232149 | |||||
| Acremonium verruculosum T | CBS 989.69 | 1 | HQ232150 | |||||
| “Cephalosporium malorum” T | CBS 117.25 | 1 | HQ232015 | |||||
| “Cephalosporium purpurascens” T | CBS 149.62 | 1 | HQ232071 | |||||
| Didymostilbe echinofibrosa | AR 2824 | 1 | AY489706 | |||||
| Emericellopsis alkalina A103 | 1,2 | KC987216 | KC987140 | KC987102 | KC999000 | KC998962 | KC987178 | |
| Emericellopsis alkalina A112 | 1,2 | KC987225 | KC987149 | KC987111 | KC999009 | KC998971 | KC987187 | |
| Emericellopsis alkalina A113 | FW-1476 | 1,2 | KC987226 | KC987150 | KC987112 | KC999010 | KC998972 | KC987188 |
| Emericellopsis alkalina A114 | FW-1473 | 1,2 | KC987227 | KC987151 | KC987113 | KC999011 | KC998973 | KC987189 |
| Emericellopsis alkalina A115 | FW-1474 | 1,2 | KC987228 | KC987152 | KC987114 | KC999012 | KC998974 | KC987190 |
| Emericellopsis alkalina A116 | 1,2 | KC987229 | KC987153 | KC987115 | - | KC998975 | KC987191 | |
| Emericellopsis alkalina A117 | FW-1471 | 1,2 | KC987230 | KC987154 | KC987116 | KC999013 | KC998976 | KC987192 |
| Emericellopsis alkalina A118 | 1,2 | KC987231 | KC987155 | KC987117 | KC999014 | KC998977 | KC987193 | |
| Emericellopsis alkalina A119 | 1,2 | KC987232 | KC987156 | KC987118 | KC999015 | KC998978 | KC987194 | |
| Emericellopsis alkalina A120 | 1,2 | KC987233 | KC987157 | KC987119 | KC999016 | KC998979 | KC987195 | |
| Emericellopsis alkalina A121 | 1,2 | KC987234 | KC987158 | KC987120 | KC999017 | KC998980 | KC987196 | |
| Emericellopsis alkalina A122 | 1,2 | KC987235 | KC987159 | KC987121 | KC999018 | KC998981 | KC987197 | |
| Emericellopsis alkalina A123 | 1,2 | KC987236 | KC987160 | KC987122 | KC999019 | KC998982 | KC987198 | |
| Emericellopsis alkalina A124 | 1,2 | KC987237 | KC987161 | KC987123 | KC999020 | KC998983 | KC987199 | |
| Emericellopsis alkalina A125 | 1,2 | KC987238 | KC987162 | KC987124 | KC999021 | KC998984 | KC987200 | |
| Emericellopsis alkalina A126 | 1,2 | KC987239 | KC987163 | KC987125 | KC999022 | KC998985 | KC987201 | |
| Emericellopsis alkalina A127 | 1,2 | KC987240 | KC987164 | KC987126 | - | KC998986 | KC987202 | |
| Emericellopsis alkalina A128 | 1,2 | KC987241 | KC987165 | KC987127 | KC999023 | KC998987 | KC987203 | |
| Emericellopsis alkalina E101 T | CBS 127350 (=VKM F-4108) | 1,2 | KC987247 | KC987171 | KC987133 | KC999029 | KC998993 | KC987209 |
| Emericellopsis alkalina M14 | CBS 120043 (=VKM F-3905) | 1,2 | KC987244 | KC987168 | KC987130 | KC999026 | KC998990 | KC987206 |
| Emericellopsis alkalina M20 | CBS 120044 (=VKM F-3040) | 1,2 | KC987245 | KC987169 | KC987131 | KC999027 | KC998991 | KC987207 |
| Emericellopsis alkalina M71 | CBS 120049 (=VKM F-3907) | 1,2 | KC987246 | KC987170 | KC987132 | KC999028 | KC998992 | KC987208 |
| Emericellopsis donezkii T | CBS 489.71 | 2 | AY632658 | AY632674 | - | - | ||
| Emericellopsis glabra T | CBS 119.40 | 2 | AY632657 | AY632673 | - | - | ||
| Emericellopsis glabra | A.R. 3614 | 2 | HM484860 | HM484879 | - | HM484843 | ||
| Emericellopsis humicola T | CBS 180.56 | 2 | AY632659 | AY632675 | - | - | ||
| Emericellopsis maritima T | CBS 491.71 (=VKM F-1082) | 1,2 | KC987251 | KC987175 | KC987137 | KC999033 | KC998997 | KC987213 |
| Emericellopsis microspora T | CBS 380.62 | 2 | AY632663 | AY632679 | - | - | ||
| Emericellopsis minima T | CBS 190.55 (=VKM F-1484) | 1,2 | KC987249 | KC987173 | KC987135 | KC999031 | KC998995 | KC987211 |
| Emericellopsis minima | CBS 111361 | 2 | AY632661 | AY632677 | - | - | ||
| Emericellopsis minima | CBS 871.68 (=VKM F-1057) | 1,2 | KC987250 | KC987174 | KC987136 | KC999032 | KC998996 | KC987212 |
| Emericellopsis minima | CCFC226707 | 1 | AY283560 | |||||
| Emericellopsis mirabilis | CBS 177.53 | 2 | AY632656 | - | - | - | ||
| Emericellopsis pallida T | CBS 490.71 (=VKM F-925 | 1,2 | KC987252 | KC987176 | KC987138 | KC999034 | KC998998 | KC987214 |
| Emericellopsis pallida | CBS 624.73 | 2 | AY632667 | AY632683 | - | - | ||
| Emericellopsis robusta | CBS 489.73 | 2 | AY632664 | AY632680 | - | - | ||
| Emericellopsis salmosynnemata | CBS 382.62 | 2 | AY632666 | AY632682 | - | - | ||
| Emericellopsis stolkiae T | CBS 159.71 | 2 | AY632668 | AY632684 | - | - | ||
| Emericellopsis synnematicola T | CBS 176.60 | 2 | AY632665 | AY632681 | - | - | ||
| Emericellopsis terricola T | CBS 120.40 | 1,2 | U57082 | U57676 | - | - | - | |
| Emericellopsis terricola | CBS 229.59 | 1,2 | AY305034 | AY632662 | AY632678 | - | - | |
| Emericellopsis terricola | CCF3815 | 2 | FJ430737 | - | - | - | ||
| Emericellopsis terricola | NRRL 54109 | 2 | HQ698592 | - | - | - | ||
| Geosmithia lavendula | IFO 7729 | 1 | D88325 | |||||
| Geosmithia putterillii | IFO 31131 | 1 | AB047215 | |||||
| Gliomastix masseei T | CBS 794.69 | 1 | HQ232060 | |||||
| Gliomastix murorum | CBS 157.72 | 1 | HQ232067 | |||||
| Gliomastix polychroma T | CBS 181.27 | 1 | HQ232091 | |||||
| Gliomastix roseogrisea T | CBS 134.56 | 1 | HQ232121 | |||||
| Glomerella cingulata | FAU 553 | 1 | AF543786 | |||||
| Hapsidospora irregularis | ATCC 22087 | 1 | AF096192 | |||||
| Heleococcum aurantiacum | CBS 201.35 | 1 | JX158442 | |||||
| Heleococcum japonense | CBS 397.67 | 1 | JX158441 | |||||
| Hydropisphaera erubescens | ATCC 36093 | 1 | AY545726 | |||||
| Melanopsamma pomiformis | ATCC 18873 | 1 | AY489709 | |||||
| Mycoarachis inversa | ATCC 22107 | 1 | GQ505991 | |||||
| Mycopepon smithii | SMH 1609 | 1 | AF279400 | |||||
| Myrothecium verrucaria | BBA 70749 | 1 | AJ301999 | |||||
| Nigrosabulum globosum | ATCC 22102 | 1 | AF096195 | |||||
| Peethambara spirostriata | CBS 110115 | 1 | AY489724 | |||||
| Peethambara sundara | CBS 646.77 | 1 | AF193245 | |||||
| Roumegueriella rufula | CBS 346.85 | 1 | DQ518776 | |||||
| Sarocladium attenuatum T | CBS 399.73 | 1 | HQ232165 | |||||
| Sarocladium bacillisporum T | CBS 425.67 | 1 | HQ231992 | |||||
| Sarocladium bactrocephalum T | CBS 749.69 | 1 | HQ231994 | |||||
| Sarocladium glaucum T | CBS 796.69 | 1 | HQ232041 | |||||
| Sarocladium kiliense T | CBS 122.29 | 1 | HQ232052 | |||||
| Sarocladium ochraceum T | CBS 428.67 | 1 | HQ232070 | |||||
| Sarocladium oryzae | CBS 180.74 | 1 | HQ232166 | |||||
| Sarocladium sp. A131 | 1 | KC987243 | KC987167 | KC987129 | KC999025 | KC998989 | KC987205 | |
| Sarocladium strictum T | CBS 346.70 | 1 | HQ232141 | |||||
| Sarocladium zeae T | CBS 801.69 | 1 | HQ232152 | |||||
| Scopinella solani | CBS 770.84 | 1 | AY015632 | |||||
| Selinia pulchra | AR 2750 | 1 | AF193246 | |||||
| Selinia pulchra | AR 2812 | 2 | HM484859 | HM484884 | - | HM484841 | ||
| Stachybotrys chartarum | ATCC 9182 | 1 | AY489714 | |||||
| Stanjemonium grisellum | NRRL 26548 | 1 | AF049171 | |||||
| Stanjemonium grisellum T | CBS 655.79 | 2 | AY632671 | AY632687 | - | - | ||
| Stanjemonium ochroroseum T | CBS 656.79 | 2 | AY632672 | AY632688 | - | - | ||
| Stilbella fimetaria | D99026 | 2 | AY952467 | - | - | - | ||
| Stilbella fimetaria | DAOM 229279 | 1 | HQ232176 | |||||
| Stilbella fimetaria | MH178 | 2 | FJ430712 | - | - | - | ||
| Stilbella fimetaria | SES201 | 2 | FJ939394 | - | - | - | ||
| Verrucostoma freycinetiae T | MAFF 240100 | 2 | HM484866 | HM484885 | - | HM484853 |
RESULTS
Isolated strains
On the selective AA medium buffered at pH 10 and containing antibiotic, we isolated 34 strains of filamentous fungi from soda soils adjacent to the soda lake basins. Several of the isolated strains were deposited in CBS and VKM. All strains showed asexual acremonium-like sporulation and one displayed comprehensive sexual morphological features and was found to be a new species of the Emericellopsis lineage based on molecular, morphological and growth data (see below).
Molecular phylogenetic analyses
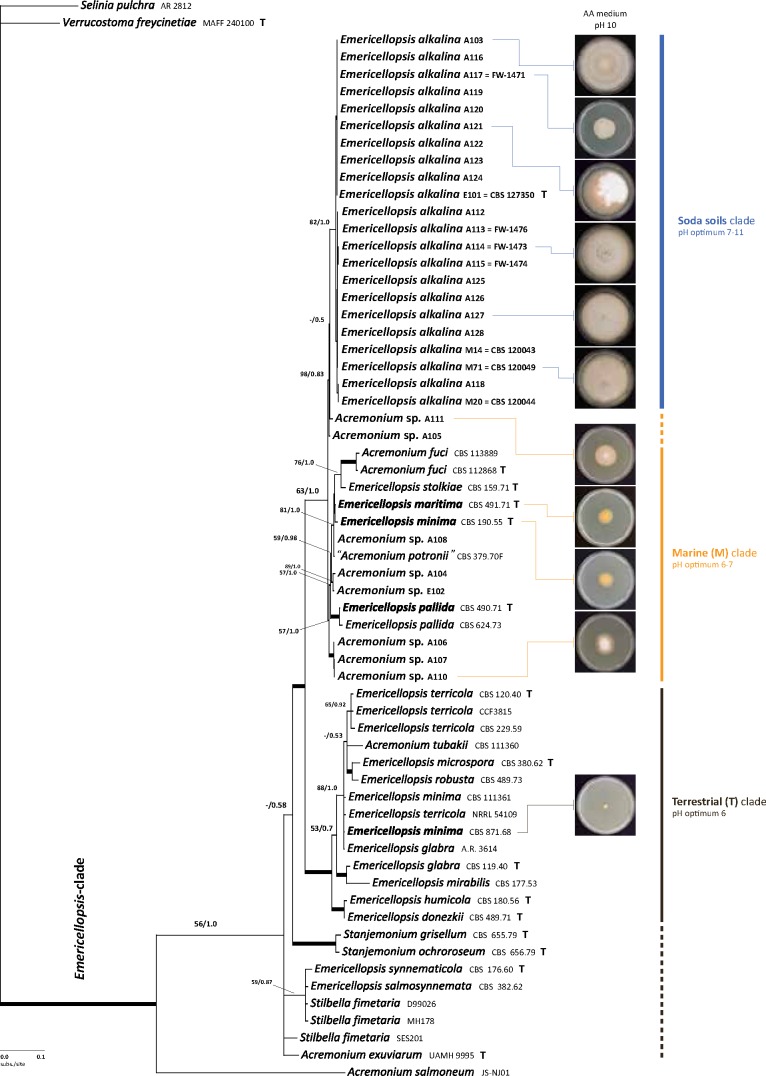
The alignment for the first phylogenetic analysis using the LSU gene contained 962 characters, with 162 (17 %) being phylogenetically informative (Table 2). The negative log likelihoods (-Ln) of the ML and BI consensus trees were 4696.03 and 5111.82, respectively. The phylogenetic reconstruction based on LSU sequences of our isolates from soda lakes along with the pertinent reference sequences from hypocrealean acremonia is consistent with the topology described by Summerbell et al. (2011), hence we follow the clade delineation outlined in that study. As seen in Fig. 1, the new isolates from the soda soils (in coloured boxes) almost exclusively fall into a strongly supported (97/1.0) Emericellopsis-clade (Bionectriaceae). This clade is known to include marine-borne fungi such as Acremonium fuci, A. tubakii, E. maritima, as well as terrestrial isolates like E. terricola, some Stilbella species, and the Stanjemonium species. The lizard-associated ex-type-strain of A. exuviarum (UAMH 9995), producing chains of conidia, has been shown before to have affinity to the Emericellopsis-clade (Sigler et al. 2004). Thirty of our new isolates in the Emericellopsis-clade stand together within a weakly supported clade (76/0.99) that also includes the ex-type strains of E. minima (CBS 190.55), E. maritima (CBS 491.71), and E. pallida (CBS 490.71), as well as “A. potronii” (isolate CBS 379.70F); the latter is a single isolate of an undescribed species that has so far only been isolated from a dolphin skin lesion, apparently not as an agent of infection (Zuccaro et al. 2004). The marine species from Fucus, a brown seaweed, A. fuci (UAMH 6508), also grouped with our isolates from soda soils. There is not enough phylogenetic signal from our LSU-based phylogenetic reconstruction to resolve the Emericellopsis-clade further. Four new isolates from soda soils appeared to be in the sister clades, namely, two in the sclerotigenum-clade, one in the Sarocladium-clade and one in the inflatum-clade. They are hence identified accordingly.
Fig. 1.
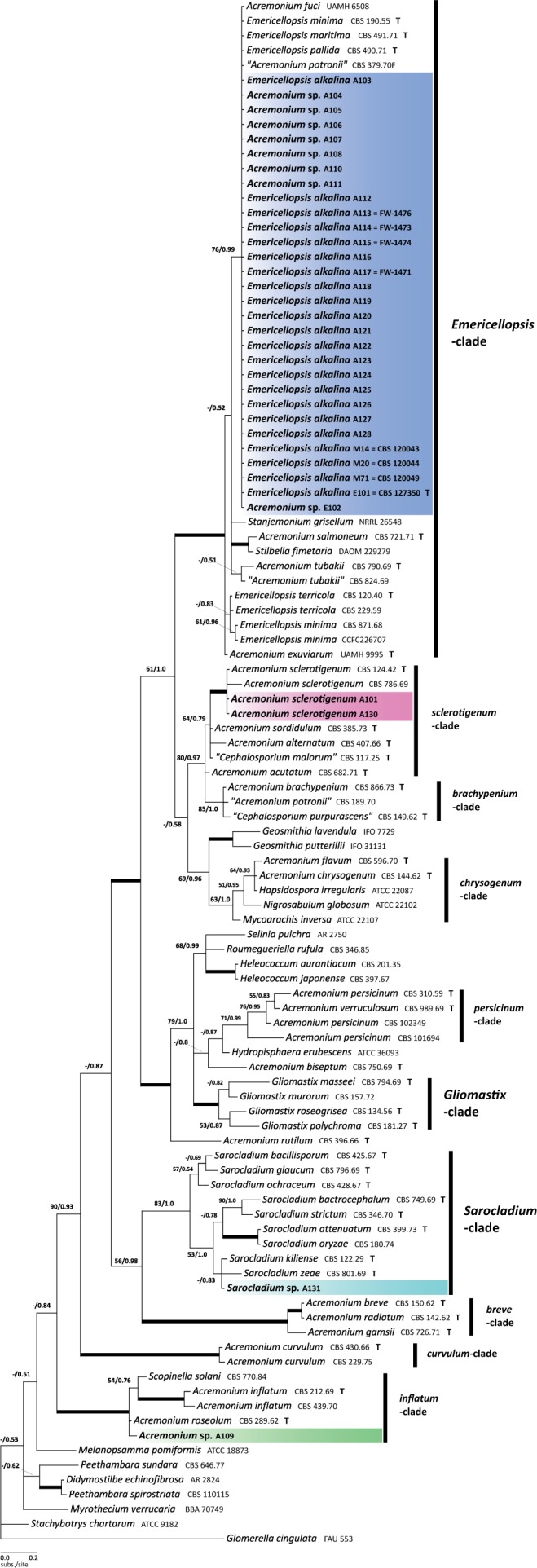
Phylogenetic reconstruction of Acremonium species in Bionectriaceae as inferred from the partial LSU gene sequences. New isolates from the soda soils are marked with colour boxes. Clade delineation is from Summerbell et al. (2011). Bayesian topology with the ML/PP support values over each node is displayed. Thickened branches indicate strong combined support (ML>90, PP>0.94). T – type/ex-type strains.
The second phylogenetic analysis included partial sequences of four genes (ITS, β-tub, RPB2, TEF1-α) known to have a higher mutation rate than LSU. We sampled a different set of taxa for this low-level taxonomic analysis. The sequences for the Emericellopsis-clade had a high degree of similarity, and were easily aligned and edited. The most variable locus in this set was the β-tub region containing introns, and this region thus contributed significantly to the reliability of the resulting tree. The alignment for this analysis had 2 810 characters of which 308 (11 %) were phylogenetically informative (Table 2). The MCMC runs in Bayesian analysis reached stationary status with a deviation of 0.008 after 5M generations. The negative log likelihoods (-Ln) of the ML and BI consensus trees were 8487.81 and 8645.85, respectively.
The tree that was generated for the Emericellopsis-clade is displayed in Fig. 2. Here, unlike in the first analysis, the Emericellopsis-clade is deeply resolved, displaying several major clades consistent with the previous study by Zuccaro et al. (2004). The basal group consists of a highly supported asexual Stanjemonium clade, asexual Stilbella fimentaria haplotypes, and the soil-derived ex-type isolates of E. synnematicola, CBS 176.60, and E. salmosynnemata, CBS 382.62. The ex-type isolate of Acremonium exuviarum, mentioned earlier, seems to be more distally basal to the rest of the core tree members. Our phylogenetic analysis confirms the presence of the two ecological groups in the Emericellopsis lineage, both of which were supported by the molecular studies. The clades designated as marine (M) and terrestrial (T), outlined previously by Zuccaro et al. (2004), also appear in our phylogenetic analysis. The T clade (98/1.0) almost exclusively contains terrestrial species of Emericellopsis, such as E. robusta, E. terricola, and E. microspora. There are a few exceptions, namely, E. donezkii CBS 489.71, E. minima CBS 111361, and A. tubakii CBS 111360, which were found in aquatic environments. The very weakly supported M clade (57/1.0) predominantly contains isolates from marine and soda lake habitats, with the exception of E. pallida CBS 624.73 and the ex-type isolate of E. minima, CBS 190.55. Interestingly, eight of our new isolates from the soda soils fall into the M clade while the majority (22 strains) form a well-supported sister clade (82/1.0). We name that clade the “soda soils” clade. It comprises 22 of our isolates that collectively represent a new species named E. alkalina sp. nov. here. Of those 22 strains, one formed ascomata, while the others only displayed asexual structures. These structures were identical to those seen in CBS 127350, the sexual strain from which we derived the type of E. alkalina.
Fig. 2.
Four-gene phylogeny of the new alkalitolerant isolates within the Emericellopsis-clade based on partial sequences for ITS (including 5.8S rDNA), β-tub, RPB2 and TEF1-α genes. All strains studied are in bold. Bayesian topology is displayed with the ML/PP support values over each node. Thickened branches indicate strong combined support (ML>90, PP>0.94). T – type/ex-type strains. Representative strains from each delineated clade are shown on AA medium plates (11-d-old).
SSU sequences showed almost no variation among our newly isolated strains in the Emericellopsis-clade. We found only two variable sites among 1 637 base pairs.
Growth patterns
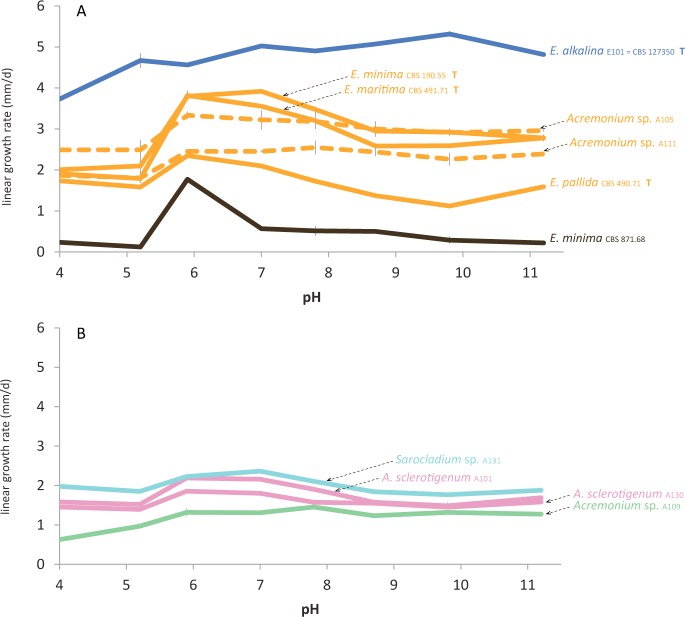
In order to link our phylogenetic data to ecological preferences, we conducted a growth experiment testing the growth ability of all studied strains at different ambient pH values. As seen in Fig. 3A, the pH preferences vary among the members of the different clades within the Emericellopsis lineage. A reference member of the T clade, E. minima (CBS 871.68), displayed a very narrow growth optimum at pH 6 with no ability to cope with both lower and higher pH values. Three reference members of the M clade, the ex-type strains of E. maritima (CBS 491.71), E. minima (CBS 190.55), and E. pallida (CBS 490.71), had an optimum growth at pH 6–7, but were able to tolerate higher pH values. Identical growth patterns were seen in our strains Acremonium sp. A104, A105, A106, A107, A108, A110, A111, and E102 (data not shown) which also fall into the M clade. Two strains (A105 and A111) seem to be paraphyletic to the M clade, but based on their growth patterns they belong to the M clade (dashed line). Members of the M clade grew faster than E. minima (CBS 871.68) from the T clade. All new isolates of E. alkalina (except A117, which had very low growth rate and no pH preference) showed a higher growth rate than that seen in the members of the M and T clades. They had a broad pH optimum in the 7–11 range, and displayed a wide tolerance across the pH scale.
Fig. 3.
Growth patterns of the representative strains at pH 4 through 11.2 based on MYA medium. A. strains from the T, M and soda soils clades within the Emericellopsis lineage including intermediate Acremonium sp. isolates A105 and A111; B. isolated alkalitolerant strains from the sister clade of the Emericellopsis lineage.
Isolates Acremonium sp. A109, A. sclerotigenum A101, A130 and Sarocladium sp. A131, which fall into a sister-clade to the Emericellopsis-clade, had an overall slow growth rate with a slight preference for neutral pH combined with the ability to tolerate higher pH values. This pattern somewhat resembled that seen in the M clade (Fig. 3B).
TAXONOMY
Emericellopsis alkalina Bilanenko & Georgieva, sp. nov.
MycoBank MB804572
Fig. 4.
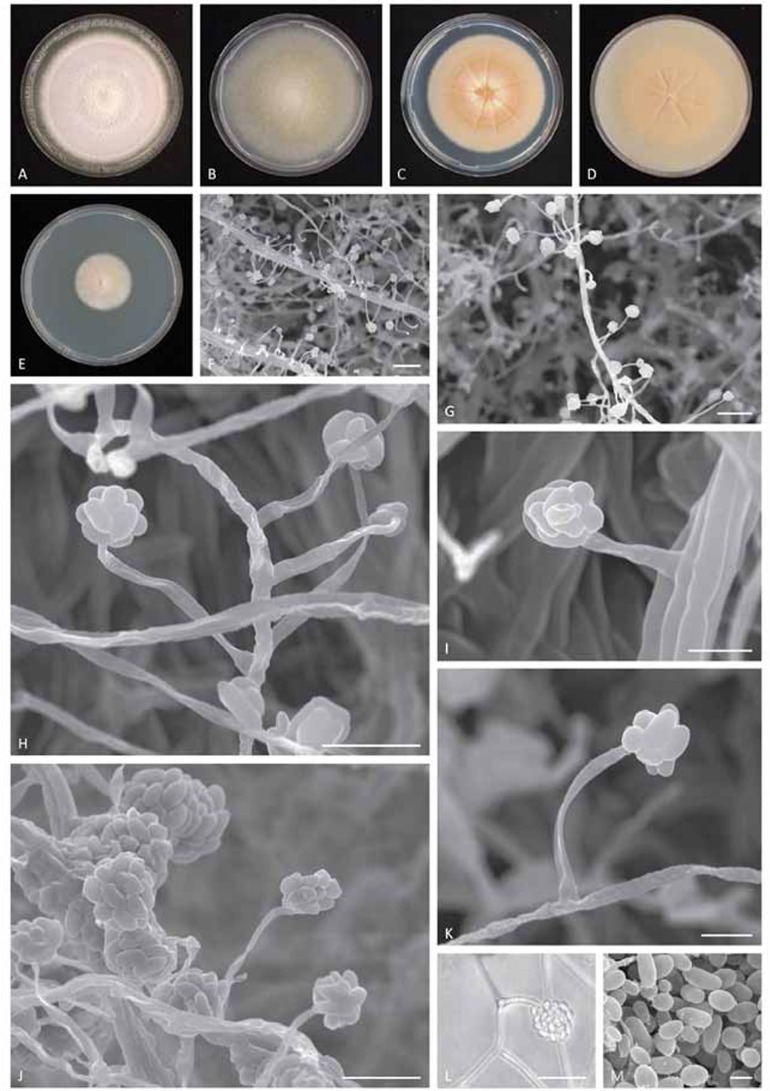
Emericellopsis alkalina (CBS 127350). A–E. 11-d-old (28 °C, dark regime, 9 cm Petri dish) colony on alkaline agar (AA), Czapek agar (CZ), potato dextrose agar (PDA), oatmeal agar (OA), malt yeast extract agar (MYA). F–G. Hyphal bundles with acremonium-like conidiation (SEM). H. Conidiogeous cells emerging from single hypha (SEM). I. Conidial head on a single conidiogenous cell emerging from the hyphal bundle (SEM). J. Matured conidial heads (SEM). K. Single conidiogenous cell with young conidial head (SEM). L. Conidial head (LM). M. Conidia (SEM). Bars F–G = 20 μm; H, J and L = 10 μm; I and K = 5 μm; and M = 2 μm.
Fig. 5.
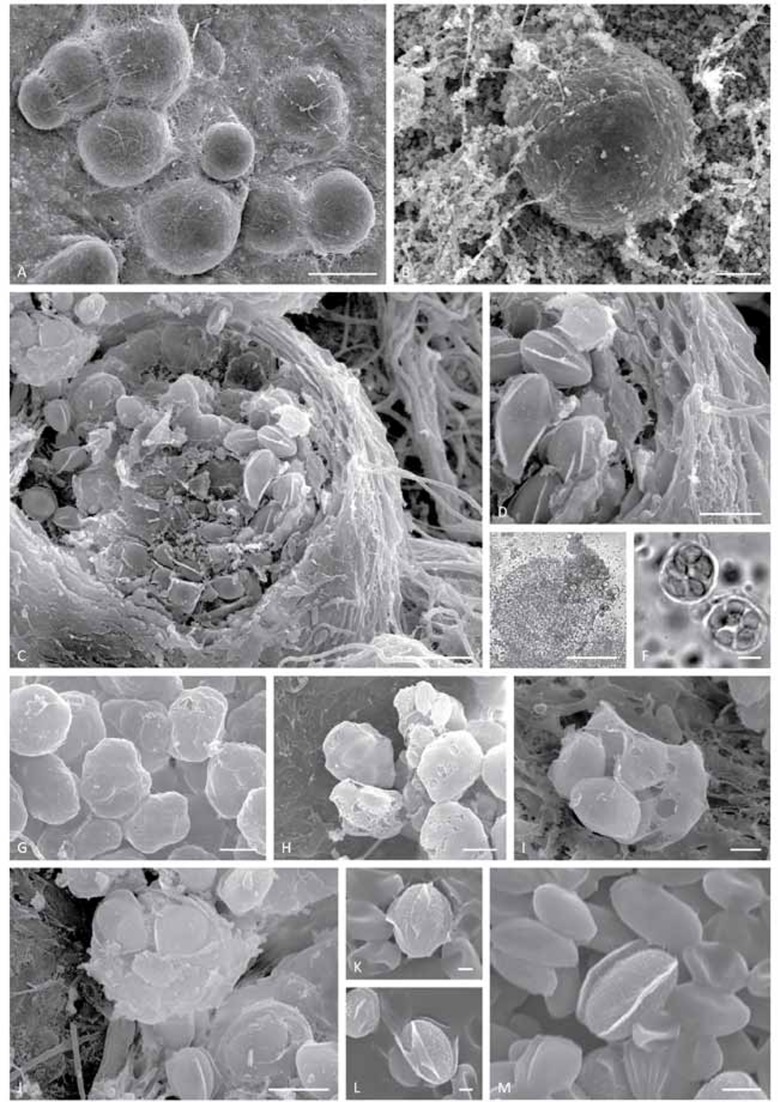
Emericellopsis alkalina (CBS 127350). A. Cleistothecia (SEM). B. Cleistothecium surrounded by the asexual sporulation (SEM). C. Open cleistothecium (SEM). D. Magnified view on the multilayered peridium (SEM). E. Open cleistothecium (LM). F. Young asci (LM). G. Young asci (SEM). H–J. Lysing asci (SEM). K–M. Ascospores with alar appendages (SEM). Bars: A and E = 100 μm; B = 20 μm; C = 10 μm; D, F–H, and J = 5 μm; I and M = 2 μm; and K–L = 1 μm.
Etymology: Epithet taken from the ability to grow at high ambient pH.
Diagnosis: Asci saccate, 12–15 μm long, unitunicate. Ascospores ellipsoid, pale brown, with uneven surfaces, 4.5–5.5 × 2.5–3.0 μm, surrounded by 3, but frequently 5 longitudinal, subhyaline, smooth-edged alar appendages, width up to 1.0 μm. Asexual morph acremonium-like.
Type: Russia: Altai, Kulunda steppe, soda soil (total salts 73 g kg−1, pH 10.1) on the edge of the basin of Tanatar Lake, August 2002, D. Sorokin (CBS H-21412 – holotype; culture ex-type E101 = CBS 127350 = VKM F-4108).
Description: Ascomata dark brown, superficial on the substratum, globose, 50–120(–180) μm diam, non-ostiolate, wall 6–10 μm thick. Peridium multi-layered, pseudoparenchymatous, composed of 3–5 layers of compressed cells. Asci saccate, 12–15 μm long, with thin deliquescent wall, soon dissolving, unitunicate, scattered irregularly in the ascocarp. Ascospores ellipsoid, pale brown, with uneven surfaces, 4.5–5.5 × 2.5–3.0 μm, surrounded by 3, but frequently 5 longitudinal, subhyaline, smooth-edged alar appendages, width up to 1.0 μm. Asexual morph acremonium-like. Conidiation abundant, mostly plectonematogenous, partially nematogenous. Conidiophores mostly simple orthotropic. Conidiogenous cells 20–35 μm long, tapering from 1.5–1.8 μm at the base to 0.7–0.8 μm at the apex, sometimes lateral branches form. Conidia narrowly ellipsoid, smooth-surfaced, 3.5–6.0 × 1.8–2.2 μm, about the same length as ascospores but narrower, hyaline, adhering in slimy heads. Chlamydospores absent.
Culture characteristics: Colonies on alkaline agar (AA, pH 10.0–10.2) fast-growing, reaching 70–80 mm diam in 10 d at 25°C. On MEA (pH 6.5) growing slower, reaching 32–38 mm diam in 10 d. Colonies orange-salmon-pink, later darkening in centre due to the formation of ascomata with tufted aerial mycelium sometimes forming concentric zones upon exposure to light. Reverse colourless. Exudate absent. Decumbent vegetative hyphae thin-walled, hyaline, 0.5–2.0 μm wide. Mycelium consisting of hyaline, smooth-walled, septate hyphae, 1–3 μm wide, often fasciculate.
Additional specimens examined: A103, A112, A113 (= VKM FW-1476), A114 (= VKM FW-1473), A115 (= VKM FW-1474), A116, A117 (= VKM FW-1471), A118, A119, A120, A121, A122, A123, A124, A125, A126, A127, A128, M14 (= VKM F-3905 = CBS 120043), M20 (= VKM FW-3040 = CBS 120044), M71 (= VKM F-3907 = CBS 120049).
Notes: The current study shows a well-supported clade (82/1.0) as inferred from four phylogenetic loci (ITS, β-tub, RPB2, TEF1-α) containing 22 isolates including the type E101. Although only the type E101 strain formed a sexual morph, we assign the remaining 21 isolates to E. alkalina as well, based on sequence similarity and the identity of asexual morphology. All 22 isolates of E. alkalina showed essentially the same growth patterns with a wide pH tolerance culminating in an optimum at pH 7–11. Isolate A117 is the only exception, showing a highly reduced growth rate in general, and no obvious pH optimum.
Morphological differences from sister species: The ascomata of the type of Emericellopsis alkalina (CBS 127350), have a multilayered peridium, composed mostly of five layers of flattened cells. The peridium of E. pallida ex-type isolate CBS 490.71 is thinner, 1–2 layered. The ascospore morphology of the type of E. alkalina (CBS 127350) looks similar to that of E. pallida and E. minima. However, E. alkalina ascospores have an uneven surface with (3–)5 alar appendages, while E. pallida, as represented by ex-type CBS 490.71, has smooth ascospores often with three alar appendages. The ex-type of E. minima (CBS 190.55), unfortunately did not produce ascomata during our investigation. A non-type isolate of E. minima, CBS 871.68, has wider (2 μm) alar appendages with flexuose rims, while E. alkalina (CBS 127350) has narrow (1 μm) appendages with smooth rims.
DISCUSSION
Here we provide phylogenetic evidence that our newly isolated alkalitolerant fungi from geographically diverse soda soils, are derived from marine-borne species within the genus Emericellopsis. Based on pH growth preference, the highly alkalitolerant strains form a “soda soils” clade distinct from the moderately alkalitolerant “marine” clade and the neutrophilic “terrestrial” clade. The genus Emericellopsis, previously considered to belong to Eurotiales, was erected in 1940, based on the isolation of E. terricola and its variant E. terricola var. glabra (eventually renamed E. glabra; Backus & Orpurt 1961). Van Beyma (1939–40) described E. terricola based on an isolate from soil collected near the town of Baarn in The Netherlands. The generic name came from the close morphological resemblance of the ascospore ornamentation to that of Emericella nidulans, which was originally thought to be taxonomically related. Subsequent studies described additional soil-borne Emericellopsis species from various parts of the world (Stolk 1955, Gilman 1957, Mathur & Thirumalachar 1960, 1962, Backus & Orpurt 1961). At the beginning of the 1960s, the genus contained five species and one variety. Ascospore size and shape constituted the major criteria used to distinguish species (Durrell 1959).
The beginning of the 1970s marked a new period in the study of Emericellopsis with the establishment of marine mycology. New Emericellopsis species were discovered in the sediments of soda lakes and along the seacoasts. Emericellopsis stolkiae, for instance, was isolated from the soil on the edge of the soda lake in south-western Wyoming, USA (Davidson & Christensen 1971). That species had larger ascospores than previously known Emericellopsis species, and also had distinct alar appendages. Tubaki (1973) suggested the conidial genus Cephalosporium was characteristic of aquatic sediments, and he linked Emericellopsis as the corresponding sexual state.
Emericellopsis was revised by the Russian mycologist Belyakova (1974) who analysed the morphological features of the then known Emericellopsis species and compiled an identification key for 12 species. She also described three new aquatic species: Emericellopsis donezkii isolated from the basin of the North Donetz River (Ukraine), and E. maritima and E. pallida from the intertidal zone of the Black Sea in the Crimean peninsula (Ukraine) (Belyakova 1970, 1974).
At the moment, Emericellopsis comprises homothallic saprobic cleistothecial species with acremonium-like conidiation; one species, E. synnematicola, also forms stilbella-like synnemata. However, different authors accept different numbers of species in the genus. Currently, 16 species with four varieties are listed in the MycoBank database (Crous et al. 2004). All authors have so far supported the opinion that the main distinguishing features among species are the morphology of the ascospores and their alar appendages. Molecular studies conducted in the late 1990s placed Emericellopsis in Hypocreales (Glenn et al. 1996). Analysis of SSU and LSU revealed it as a member of the family Hypocreaceae (Ogawa et al. 1997), as it was then defined, although it was subsequently assigned to Bionectriaceae (Rossman et al. 1999, 2001). The genus appears to be monophyletic, with strong support values obtained in the analysis of the ITS and beta-tubulin sequences (Zuccaro et al. 2004). The Emericellopsis lineage s. lat. also harbours the asexual genera Stilbella and Stanjemonium, along with the marine species Acremonium tubakii and A. fuci (Summerbell et al. 2011).
The accumulated knowledge on the genus Emericellopsis suggests a wide ecological amplitude and worldwide distribution. This includes typical species of soils undergoing periodic flooding (e.g. rice paddies), as well as species found in bogs, the sediments of freshwater and seawater basins, and even the soils around subterranean wasp nests where humidity and alkalinity are elevated (Batra et al. 1973, Tubaki 1973, Domsch et al. 2007). Some species have a broad ecological distribution, such as E. terricola, which has been isolated from alkaline soils at the Mono Lake in California as well as from both acidic and saline soils in the Czech National Park (Steiman et al. 2004, Hujslová et al. 2010). A survey of ascomycetous fungi in limestone soils in Argentina formed by mollusc shells yielded E. minima, with its ability to grow from pH 5 to 11 (Elíades et al. 2006). The pattern of marine and other salt-associated isolations has suggested that marine habitats might harbour a large number of the Emericellopsis species. The ability to survive in high salinity and pH does not always coincide with the ability to develop the full life-cycle in those conditions, making the salts-adapted species difficult to discriminate from “transit” species and hampering efforts to estimate their ecological contribution (Kohlmeyer & Volkmann-Kohlmeyer 2003). A study by Zuccaro et al. (2004) revealed the presence of distinct marine and terrestrial clades within Emericellopsis, as noted above. The M clade contained isolates from saline habitats, including the recently described A. fuci from the thalli of the seaweed Fucus serratus and F. distichus. Members of the marine clade within Emericellopsis showed an ability to utilize sugars present in seaborne brown algae (e.g. fucoidan, fucose). The presence of marine water appeared to be necessary for conidial germination in A. fuci.
Involvement of additional loci in our phylogenetic analysis confirms the presence of the M and T clades (Fig. 2). Our new alkalitolerant isolates are exclusively linked to the M clade, with our 22 E. alkalina isolates displaying an extreme alkalitolerant phenotype. Both growth patterns and molecular data suggest that the E. alkalina group originated from the marine isolates of the M clade, linking evolutionary development in the marine habitat with that of the soda soils. Clearly, these environments share high salinity and elevated ambient pH values. As far as we know, however, such an ecological overlap has not been demonstrated for other marine fungal lineages. To address this issue, we need systematic biodiversity research on the fungi from soda lakes.
That the intron of the β-tub gene contributed extensively to the phylogenetic signal in our study suggests a relatively recent divergence of E. alkalina from the M clade. Our Acremonium sp. strains A105 and A111 seem to be intermediate isolates situated in a statistically ambiguous position between the alkaline and marine lineages. The growth pattern of these isolates contributed significantly to our decision to include them within the M clade.
Emericellopsis alkalina grew well at pHs from 4 to 11.2, with a slight preference towards 7–11. However, a few isolates of this species, namely A113, A118, A122, A126, A127, and M20, displayed a significant dip in growth rate at neutral pH values (data not shown). This feature could be seen as a physiological trade-off that has evolved in some strains of E. alkalina that thrive along with alkalitolerant strains from the M clade. Interestingly, A128 from the soda soils clade, and A110, were isolated from the same soil sample at Sulfatnoe Lake. And yet, this trend does not extend to all E. alkalina strains that were jointly isolated with M clade strains. It is unclear what makes the majority of E. alkalina strains grow more vigorously than the M clade members essentially at every pH value we tested. That E. alkalina performs well along a large section of the pH scale makes it difficult to specify the ecology of this species in conventional terms. It is technically not correct to label it an ‘alkaliphile’, since it is capable of growth at low pH as well as at high pH. Nor is the term ‘alkalitolerant’ entirely true, since the optimal growth pH is above neutral. The term ‘pH-tolerant’ with the preference towards alkaline conditions might be suitable. As opposed to the soda soils clade, members of the M clade can be appropriately called ‘alkalitolerant’, while E. minima (CBS 871.68) from clade T can safely be termed a ‘neutrophile’.
A link between marine and soda soil inhabitants has previously been observed in bacteria. In metabolic studies of fungi, specifically Fusarium oxysporum, it has been shown that the expression of the gene ena1 encoding P-type Na+-ATPase, which is believed to be an important player in the halotolerance adaptation cascade response, is up-regulated as the ambient pH goes up (Caracuel et al. 2003). Therefore, halophilic or halotolerant species may hold a clue towards elucidating the mechanisms of the ability to thrive at high pH. The molecular aspects of the ability to cope with high ambient pH have not been studied in filamentous fungi. Future work aimed at revealing these molecular properties could be carried out by contrasting the genomics of neutrophiles and alkaliphiles. Such a project might provide answers to the intriguing questions inherent in the alkaliphily phenomenon.
Acknowledgments
We are grateful to Dmitry Sorokin for providing soil samples, Bertha Koopmanschap and Marijke Slakhorst for technical assistance, and Denis Landin for processing the images. The work was funded by the Laboratory of Genetics at Wageningen University (The Netherlands) and an RFBR grant No. 11-04-01576 (Russia).
REFERENCES
- Backus MP, Orpurt PA. (1961) A new Emericellopsis from Wisconsin, with notes on other species. Mycologia 53(1): 64–83 [Google Scholar]
- Batra LR, Batra SWT, Bohart GE. (1973) The mycoflora of domesticated and wild bees (Apoidea). Mycopathologia et Mycologia applicata 49: 13–44 [Google Scholar]
- Belyakova LA. (1970) Novij vid roda Emericellopsis (Euroticaeae). Mikologiya i Fitopatologiya 4: 530–531 [In Russian.] [Google Scholar]
- Belyakova LA. (1974) Rod Emericellopsis van Beyma (Eurotiaceae). Mikologiya i Fitopatologiya 8: 385–395 [In Russian.] [Google Scholar]
- Buchalo AS, Wasser SP, Nevo E. (2009) Fungi in saline water bodies with special attention to the hypersaline Dead Sea mycobiota. In: Fungi from Different Environments (Misra JK, Deshmukh SK, eds): 56–80 Enfield, NH: Science Publishers [Google Scholar]
- Cain RF. (1956) Studies of soil fungi. Saturnomyces a new genus of the Aspergillaceae. Canadian Journal of Botany 34: 135–141 [Google Scholar]
- Caracuel Z, Casanova C, Roncero MIG, Di Pietro A, Ramos J. (2003) pH response transcription factor PacC controls salt stress tolerance and expression of the P-type Na+-ATPase ena1 in Fusarium oxysporum. Eukaryot Cell 2: 1246–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. (2004) MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22 [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson DE, Christensen M. (1971) Emericellopsis stolkiae sp.nov. from saline soils in Wyoming. Transactions of the British Mycolological Society 57: 385–391 [Google Scholar]
- Domsch KH, Gams W, Anderson T-H. (2007) Compendium of Soil Fungi. 2nd edn Eching: IHW-Verlag [Google Scholar]
- Duckworth AW, Grant WD, Jones BE, Steenbergen RV. (1996) Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiology Ecology 19: 181–191 [Google Scholar]
- Durrell LW. (1959) Some studies of Emericellopsis. Mycologia 51(1): 31–43 [Google Scholar]
- Elíades LA, Cabello MN, Voget CE. (2006) Contribution to the study of alkalophilic and alkalitolerant Ascomycota from Argentina. Darwiniana 44: 64–73 [Google Scholar]
- Gilman JC. (1957) A Manual of Soil Fungi. 2nd edn Ames, IA: Iowa State College Press [Google Scholar]
- Glenn AE, Bacon CW, Price R, Hanlin RT. (1996) Molecular phylogeny of Acremonium and its taxonomic implications. Mycologia 88: 369–383 [Google Scholar]
- Grum-Grzhimaylo AA, Debets AJM, van Diepeningen AD, Georgieva ML, Bilanenko EN. (2013) Sodiomyces alkalinus, a new holomorphic alkaliphilic ascomycete within the Plectosphaerellaceae. Persoonia 31: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. (2003) A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Systematic Biology 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98 [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MrBayes: bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Hujslová M, Kubátová A, Chudíčková M, Kolařík M. (2010) Diversity of fungal communities in saline and acidic soils in the Soos National Natural Reserve, Czech Republic. Mycological Progress 9: 1–15 [Google Scholar]
- Jones BE, Grant WD, Duckworth AW, Owenson GG. (1998) Microbial diversity of soda lakes. Extremophiles 2: 191–200 [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladwang W, Bhumirattana A, Hywel-Jones N. (2003) Alkaline-tolerant fungi from Thailand. Fungal Diversity 13: 69–83 [Google Scholar]
- Kohlmeyer J, Volkmann-Kohlmeyer B. (2003) Fungi from coral reefs: a commentary. Mycological Research 107: 386–387 [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. (2011) Mesquite: a modular system for evolutionary analysis. Version 2.75. http://mesquiteproject.org
- Mathur PN, Thirumalachar MJ. (1960) A new Emericellopsis species with Stilbella-type of conidia. Mycologia 52: 694–697 [Google Scholar]
- Mathur PN, Thirumalachar MJ. (1962) Studies on some Indian soil fungi-II. On some new or interesting Ascomycetes. Sydowia 16: 1–6 [Google Scholar]
- Mueller GM, Bills GF, Foster MS. (eds) (2004) Biodiversity of Fungi: inventory and monitoring methods. Amsterdam: Elsevier Academic Press [Google Scholar]
- Nagai K, Sakai T, Rantiatmodjo R, Suzuki K, Gams W, Okada G. (1995) Studies on the distribution of alkalophilic and alkali-tolerant Soil Fungi I. Mycoscience 36: 247–256 [Google Scholar]
- Nagai K, Suzuki K, Okada G. (1998) Studies on the distribution of alkalophilic and alkali-tolerant soil fungi II: fungal flora in two limestone caves in Japan. Mycoscience 39: 293–298 [Google Scholar]
- Ogawa H, Yoshimura A, Sugiyama J. (1997) Polyphyletic origins of species of the anamorphic genus Geosmithia and the relationships of the cleistothecial genera: evidence from 18S, 5S and 28S rDNA sequence analyses. Mycologia 89: 756–771 [Google Scholar]
- Okada G, Niimura Y, Sakata T, Uchimura T, Ohara N, Suzuki H, Kozaki M. (1993) Acremonium alcalophilum, a new alkalophilic cellulolytic hyphomycete. Transactions of the Mycological Society of Japan 34: 171–185 [Google Scholar]
- Rambaut A, Drummond AJ. (2007) Tracer v. 1.5. Computer program and documentation distributed by the authors: http://beast.bio.ed.ac.uk/Tracer
- Rossman AY, McKemy JM, Pardo-Schultheiss RA, Schroers HJ. (2001) Molecular studies of the Bionectriaceae using large subunit rDNA sequences. Mycologia 93: 100–110 [Google Scholar]
- Rossman AY, Samuels GJ, Rogerson CT, Lowen R. (1999) Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Studies in Mycology 42: 1–248 [Google Scholar]
- Sigler L, Zuccaro A, Summerbell RC, Mitchell J, Paré JA. (2004) Acremonium exuviarum sp. nov., a lizard-associated fungus with affinity to Emericellopsis. Studies in Mycology 50: 409–413 [Google Scholar]
- Sorokin ID, Kravchenko IK, Tourova TP, Kolganova TV, Boulygina ES, Sorokin DY. (2008) Bacillus alkalidiazotrophicus sp. nov., a diazotrophic, low salt-tolerant alkaliphile isolated from Mongolian soda soil. International Journal of Systematic and Evolutionary Microbiology 58: 2459–2464 [DOI] [PubMed] [Google Scholar]
- Steiman R, Ford L, Ducros V, Lafond JL, Guiraud P. (2004) First survey of fungi in hypersaline soil and water of Mono Lake area (California). Antonie van Leeuwenhoek 85: 69–83 [DOI] [PubMed] [Google Scholar]
- Stolk AC. (1955) Emericellopsis minima sp. nov. and Westerdykella ornata gen. nov. sp. nov. Transactions of the British Mycological Society 38: 419–424 [Google Scholar]
- Stöver BC, Müller KF. (2010) TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran J, Holder MT. (2010) DendroPy: A Python library for phylogenetic computing. Bioinformatics 26: 1569–1571 [DOI] [PubMed] [Google Scholar]
- Summerbell RC, Gueidan C, Schroers HJ, De Hoog GS, Starink M, Rosete YA, Guarro J, Scott JA. (2011) Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Studies in Mycology 68: 139–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubaki K. (1973) Aquatic sediment as a habitat of Emericellopsis, with a description of an undescribed species of Cephalosporium. Mycologia 65: 938–941 [Google Scholar]
- van Beyma thoe Kingma FH. (1939-40) Beschreibung einiger neuer pilzarten aus dem Centraalbureau voor Schimmelcultures, Baarn (Nederland). Antonie van Leeuwenhoek 6: 263–290 [PubMed] [Google Scholar]
- Zuccaro A, Summerbell RC, Gams W, Schroers HJ, Mitchell JI. (2004) A new Acremonium species associated with Fucus spp., and its affinity with a phylogenetically distinct marine Emericellopsis clade. Studies in Mycology 50: 283–297 [Google Scholar]
- Zwickl DJ. (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD dissertation, University of Texas at Austin [Google Scholar]