Abstract
Background
Microarray analysis has been used as the first-tier genetic testing to detect chromosomal imbalances and copy number variants (CNVs) for pediatric patients with intellectual and developmental disabilities (ID/DD). To further investigate the candidate genes and underlying dosage-sensitive mechanisms related to ID, cytogenomic mapping of critical regions and bioinformatic mining of candidate brain-expressed genes (BEGs) and their functional interactions were performed. Critical regions of chromosomal imbalances and pathogenic CNVs were mapped by subtracting known benign CNVs from the Databases of Genomic Variants (DGV) and extracting smallest overlap regions with cases from DatabasE of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER). BEGs from these critical regions were revealed by functional annotation using Database for Annotation, Visualization, and Integrated Discovery (DAVID) and by tissue expression pattern from Uniprot. Cross-region interrelations and functional networks of the BEGs were analyzed using Gene Relationships Across Implicated Loci (GRAIL) and Ingenuity Pathway Analysis (IPA).
Results
Of the 1,354 patients analyzed by oligonucleotide array comparative genomic hybridization (aCGH), pathogenic abnormalities were detected in 176 patients including genomic disorders in 66 patients (37.5%), subtelomeric rearrangements in 45 patients (25.6%), interstitial imbalances in 33 patients (18.8%), chromosomal structural rearrangements in 17 patients (9.7%) and aneuploidies in 15 patients (8.5%). Subtractive and extractive mapping defined 82 disjointed critical regions from the detected abnormalities. A total of 461 BEGs was generated from 73 disjointed critical regions. Enrichment of central nervous system specific genes in these regions was noted. The number of BEGs increased with the size of the regions. A list of 108 candidate BEGs with significant cross region interrelation was identified by GRAIL and five significant gene networks involving cell cycle, cell-to-cell signaling, cellular assembly, cell morphology, and gene expression regulations were denoted by IPA.
Conclusions
These results characterized ID related cross-region interrelations and multiple networks of candidate BEGs from the detected genomic imbalances. Further experimental study of these BEGs and their interactions will lead to a better understanding of dosage-sensitive mechanisms and modifying effects of human mental development.
Keywords: Intellectual disability, Critical regions, Brain expressed genes, Cross-region gene interrelation, Functional network
Background
Intellectual disability (ID), also known as mental retardation, is characterized by significant limitations in intellectual functioning and adaptive behavior and is frequently associated with developmental delay (DD) and multiple congenital anomalies (MCA) [1]. Conventional cytogenetic evaluation of ID/DD/MCA showed an abnormality detection rate of 3.7% by karyotyping for large numerical and structural chromosomal abnormalities and up to 6.8% when combined with fluorescence in situ hybridization (FISH) analysis for targeted cryptic and subtelemeric rearrangements [2]. However, this molecular cytogenetic analysis was limited by the average Giemsa banding resolution of 5–10 megabase (Mb) and the number of targeted probes available in FISH analysis. For the past five years, genomic technologies using oligonucleotide array comparative genomic hybridization (aCGH) or a single nucleotide polymorphism (SNP) chip have been validated and recommended as first-tier genetic testing in replacing the conventional cytogenetic analysis [3,4]. Integration of genomic analysis into clinical cytogenetic services has filled the gap between the megabase-range G-band and the kilo-base level gene and proven to be highly effective in defining the genomic localizations and gene contents from recurrent genomic disorders, interstitial imbalances and subtelomeric rearrangements. The clinical application of aCGH for large case series of children and newborns has been reported; the diagnostic yield from cytogenomic analysis on pediatric patients with ID/DD/MCA and autism is about 12-20% [5-13].
It has been hypothesized that the general ID/DD phenotype from chromosomal segmental imbalances and genomic copy number variants (CNVs) is caused by dosage-sensitive mechanisms of involved genes. A chromosomal map which associated human brain congenital malformations with specific chromosomal regions had been constructed, but this low resolution map is of limited value for identifying dosage-sensitive genes [14]. Current cytogenomic analysis has facilitated accurate subtractive mapping of critical regions or intervals for identifying candidate genes in an increasing number of patients. For example, the breakpoints and gene content mediated by regional low copy number repeats (LCRs) of 15q12 for Prader-Willi or Angelman syndrome and of 22q11.2 for DiGeorge syndrome have been defined [15,16]. However, the lack of commonly accepted protocols for CNV prioritization and data concerning the gene dosage effect in the human genome makes it difficult to identify the disease-causing genes. To date it is still a technical challenge to predict candidate dosage-sensitive genes from the detected chromosomal and genomic abnormalities.
In the post-Genome era, many bioinformatic tools have been developed for biological interpretation of large gene lists derived from high-throughput experiments. Gene annotation enrichment analysis is one of the widely used approaches to statistically determine the most over-represented gene ontology (GO) terms and reveal the underlying biological processes in a group of genes [17]. Another strategy is to look for functionally related or interacting genes across genetic loci [18]. More recently, Gene Relationships Across Implicated Loci (GRAIL) has been applied to dissect common biological pathways affecting various phenotypes from SNP variants across genomic loci [19-21]. Similarly, Ingenuity Pathway Analysis (IPA), a web-delivered application, provides an integrated knowledge base comprising over 200,000 full text articles about the human genome and enables discovery and exploration of molecular interaction networks. It has been used to assess the functional biological networks and the candidate genes from the recurrent 16p11.2 microdeletions and the 1p34 microdeletion associated with autism [22,23].
In this study we used integrated cytogenomic mapping and bioinformatic mining to identify potential candidate genes and common gene interactions or networks from 176 genomic imbalances detected by aCGH from 1354 individuals with ID/DD/MCA and autism. Gene ontology enrichment of brain-expressed genes (BEGs) was noted by the gene functional classification analysis tool, Database for Annotation, Visualization, and Integrated Discovery (DAVID). Further gene-interaction analysis using GRAIL identified 108 candidate genes with related biological functions and strong interrelation cross imbalance regions. Pathway analysis using IPA denoted five significant gene networks involving cell cycle, cell-to-cell signaling, cellular assembly, cell morphology, and gene expression regulations. These findings provide insight into the cross-region interrelations of the BEGs and possibly the underlying dosage-sensitive mechanisms and modifying effects for human mental development.
Results
CNV characterization and classification
Of the 1354 pediatric patients referred for oligonucleotide aCGH analysis, 373 genomic imbalances were detected; 205 of these imbalances were denoted as pathogenic abnormalities in 176 patients (an individual may have more than one imbalance) and 168 imbalances were classified as VOUS (data not shown). The abnormality detection rate was 13.0% (176/1354). Excluding cases with simple aneuploidies, the 190 genomic abnormalities showed a mean genomic size of 6.6 Mb and a mean gene content of 47 genes per imbalance. The diagnosis yield of 14.0% by the 180 K microarray was slightly higher than the 12.5% by 44 K microarray. Approximately 60% of the genomic imbalances were deletions and 40% were duplications. Abnormalities detected at the terminal 10–15 Mb in large autosomes numbered 1 to 12 and terminal 5–10 Mb in small autosomes numbered 13 to 22 are usually considered to be subtelomeric rearrangements. Abnormalities found between centromere and subtelomeric, excluding the recurrent genomic disorders, are considered to be interstitial imbalances.
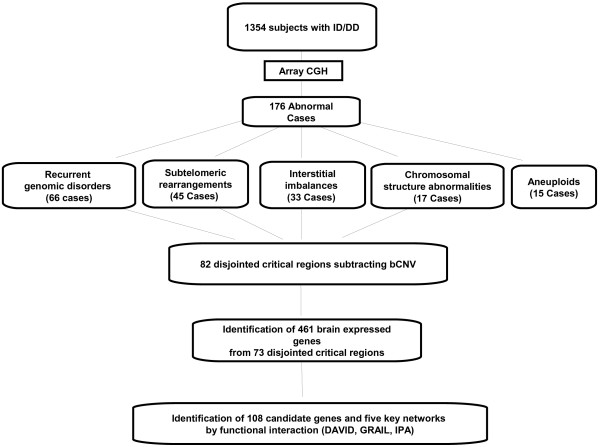
The workflow of cytogenomic mapping and bioinfomatic mining is outlined in Figure 1. Details of the aCGH results are summarized in Additional file 1: Table S1, Additional file 2: Table S2, Additional file 3: Table S3, Additional file 4: Table S4. The detected abnormalities were classified into five categories: recurrent genomic disorders mediated by regional low-copy repeats in 66 cases (37.5%, 66/176) (Additional file 1: Table S1), subtelomeric abnormalities resulting from terminal deletions or unbalanced rearrangements in 45 cases (25.6%) (Additional file 2: Table S2), sporadic interstitial deletions and duplications in 33 cases (18.8%) (Additional file 3: Table S3), chromosomal structure abnormalities in 17 cases (9.7%) (Additional file 4: Table S4) and simple aneuploidies in 15 cases (8.5%). A genome-wide view of the detected genomic disorders, subtelomeric rearrangements, interstitial imbalances and chromosomal structural abnormalities is shown in Figure 2. The most frequently detected genomic disorders were deletions of the 22q11.21 in 13 cases (OMIM188400) and the reciprocal duplication (OMIM 608363) in three cases. The subtelomeric rearrangements included simple terminal deletions and duplications identified in 31 cases and de novo or familial subtelomeric rearrangements in 14 cases. The size distribution of deletions and duplications from genomic disorders, subtelomeric and interstitial rearrangements is shown in Figure 3. The average size of deletions for subtelomeric and interstitial rearrangements was 4.9 Mb and 6.0 Mb while the average size of duplications was 5.2 Mb and 6.4 Mb, respectively. The 15 cases of simple aneuploidies included seven cases of trisomy 21, one trisomy 13 and seven cases of sex chromosome aneuploidies.
Figure 1.
Flowchart for abnormality classification and bioinformatic filtration.
Figure 2.
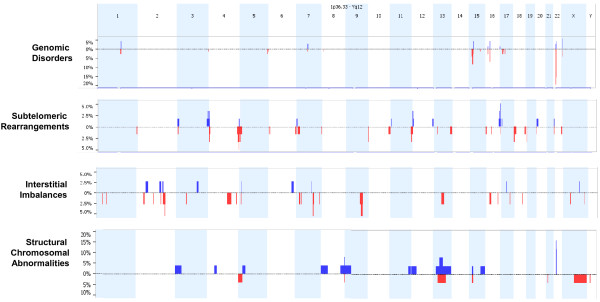
A genome-wide view of detected abnormalities. Detected recurrent genomic disorders, subtelomeric rearrangements, interstitial imbalances, and structural chromosomal abnormalities are shown in its category with genomic location and relative frequency (downward red bar for deletion and upward blue bar for duplication).
Figure 3.
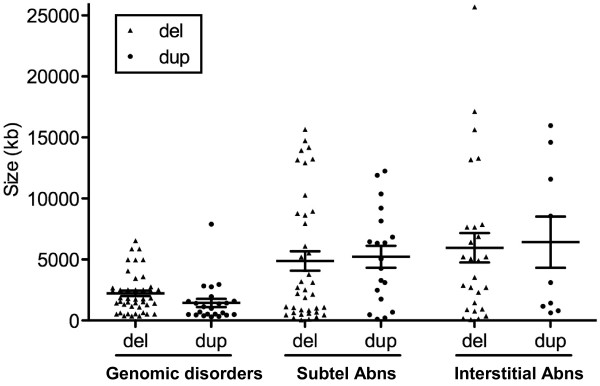
The size distribution of genomic disorders, subtelomeric rearrangements and interstitial imbalances. The average sizes for deletions and duplications are 2.2 Mb and 1.4 Mb for genomic disorders, 4.9 Mb and 5.2 Mb for subtelomeric abnormalities (abns), and 6.0 Mb and 6.2 Mb interstitial abnormalities, respectively.
Functional annotation enrichment
Excluding the 15 cases with simple aneuploidies, cytogenomic mapping was performed on the remaining 161 cases. After subtracting the bCNV, fine-mapping the critical regions and combining overlapping regions, 82 disjointed critical regions belonging to recurrent genomic disorders, subtelomeric and interstitial regions were defined; a total of 4080 genes were retrieved from the UCSC RefGene table (Additional file 5: Table S5). Functional annotation tool DAVID was used to analyze each gene list from recurrent genomic disorders, subtelomeric rearrangements, interstitial imbalances and structural chromosomal abnormalities with gene tissue expression annotations derived from uniprot (UP_TISSUE). Gene lists in the genomic disorder, subtelomeric and interstitial critical regions were noted to be enriched for brain specific genes (p < 0.05). To avoid biased tissue information from Uniprot and to understand the expression pattern comprehensively, the tissue expression enrichment was further investigated using GNF_U133A, a world-class tissue expression database. The results showed that genes in these four lists are all enriched in central nervous systems (CNS) (Additional file 6: Figure S1). Genes in recurrent genomic disorders and subtelomeric regions are preferentially expressed in medulla oblongata (p < 0.005), hypothalamus (p < 0.05) and pituitary (p < 0.1), while genes in interstitial and structural abnormal regions are enriched in cingulate cortex (p < 0.05), cerebellum (p < 0.1) and occipital lobe (p < 0.1). The enrichments were confirmed by 1000 runs of permutation. This observation indicated that there is brain functional gene enrichment in the defined critical regions.
Relationship between the numbers of the BEGs and the size of the abnormal loci
Based on findings that genes preferentially expressed in variable sites of the CNS are enriched in the critical regions, candidate BEGs were sorted out using the method described by Raychaudhuri, which examined and compared gene expression pattern in different tissues with the expression pattern in brain or spinal cord [18]. Genes with p < 0.01 were identified as preferentially expressed in the CNS system. A total of 461 BEGs from 73 disjointed critical regions in 133 cases were defined (Additional file 7: Table S6). In nine disjointed critical regions, there were no BEGs using this approach.
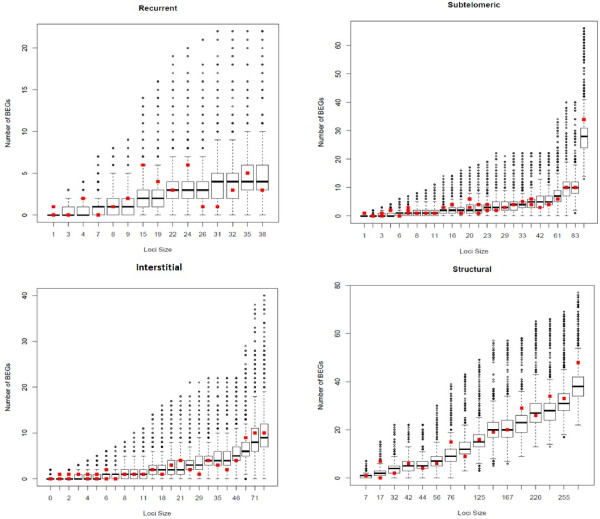
To evaluate the distribution of the number of BEGs in the critical regions and test whether the number of BEGs was over-represented in comparison to random loci with the same number of genes, permutation-based empirical distributions were conducted for the 16 regions in recurrent genomic disorders, 39 disjointed regions in subtelomeric rearrangements, 34 disjointed regions in interstitial imbalances and 17 disjointed regions in structural abnormalities. For each abnormal region, the number of BEGs in the random loci was determined and compared with that of the abnormal regions by boxplot. Although variations were noted in some recurrent genomic disorder regions, the overall distribution showed that the number of BEGs in detected abnormal regions is positively correlated to the size of the region (Figure 4). This observation indicated that the number of BEGs likely increases with the size of gene content of the imbalance regions.
Figure 4.
Boxplots showing the permutation-based empirical distributions of the number of BEGs observed in the critical regions of different gene content sizes (measured by gene numbers). For each of the 82 targeted abnormal regions, a set of 10,000 random genomic loci with gene content size defined by the same number of genes as the target regions was drawn from the genome. The X-axis represents the number of genes located in each regions, and the Y-axis stands for the numbers of BEGs observed. The actual numbers of BEGs detected in each abnormal region was shown as big solid red square.
Identification of candidate genes and functional interaction
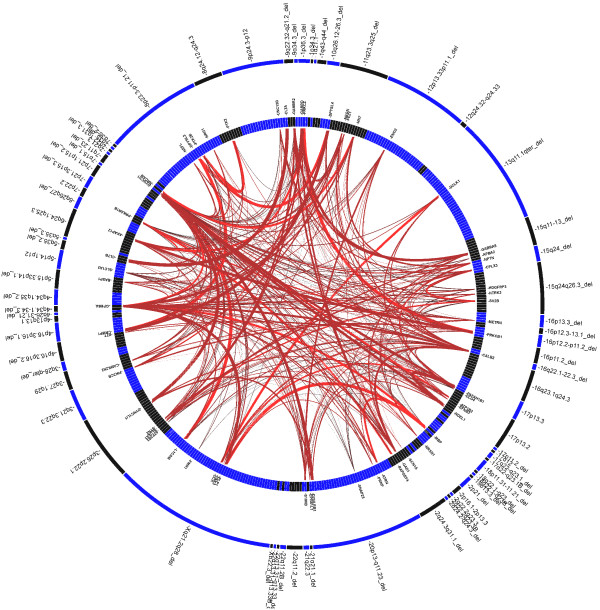
To test the hypothesis that there may be functional interrelations of the BEGs across critical regions and identify possible candidate genes, a statistical method GRAIL was applied to assess the degree of relatedness of the listed 461 BEGs [21]. A text-based similarity metric derived from PubMed literature was used to assess the relatedness between genes across the 73 regions. A subset of 108 genes were implicated inter-connection (ptext < 0.01) (Table 1), and the words that strongly linked the significant genes in each region were defined as keywords. The top keywords are ‘neuron’, ‘brain’, ‘synaptic’ ‘gaba’ and ‘kinase’, which indicate the common biological functions of these genes. In addition to PubMed literature, other two gene relatedness metrics – Gene Ontology (GO) annotations and gene expression patterns across multiple human tissues were also used. A total of 45 genes obtained pannotation < 0.01 with GO similarity and a subset of 317 genes achieved pexpression < 0.01 with expression similarity. The results from these three data sources showed that 27 genes were common to all three functional data sources and 74 genes were shared in two functional data sources (Additional file 6: Figure S2). Interestingly, among these candidate genes, CRMP1, PAFAH1B1 and PDE4D are spindle related genes, implying that the proper centrosome function is critical for brain development. The top five terms which show strong functional enrichments were neurological process, transmission of nerve pulse, synaptic transmission, cell junction and central nervous system neuron axonogenesis (p < 0.001). The functional connections among implicated genes in the 73 regions were shown in Figure 5 and Additional file 6: Figures S3 and S4.
Table 1.
List of candidate genes identified by GRAIL
| G-band | Candidate gene | G-band | Candidate gene | G-band | Candidate gene | G-band | Candidate gene |
|---|---|---|---|---|---|---|---|
| 1p36.3 |
PRKCZ |
5q35.2 |
CLTB |
11q23.3q25 |
HNT |
17p13.2 |
ATP1B2 |
| 1p36.3 |
GABRD |
5q35.3 |
MAPK9 |
11q23.3q25 |
NRGN |
17p13.2 |
EFNB3 |
| 1q43-q44 |
AKT3 |
6q24.1q25.3 |
AKAP12 |
11q23.3q25 |
JAM3 |
17p13.3 |
PAFAH1B1 |
| 2p21 |
NRXN1 |
6q26q27 |
PDE10A |
12p13.33p11.1 |
KRAS |
18q22.1-q23 |
CBLN2 |
| 2q22.2q23.3B |
KIF5C |
6q26q27 |
RPS6KA2 |
12p13.33p11.1 |
PTPRO |
18q22.1-q23 |
MBP |
| 2q24.3q31.1 |
STK39 |
7p22.2 |
PRKAR1B |
12p13.33p11.1 |
ENO2 |
20p13-q11.23 |
DYNLRB1 |
| 2q24.3q31.1 |
GAD1 |
7q21.3 |
DYNC1I1 |
13q11.1qter |
DCLK1 |
20p13-q11.23 |
PTPRA |
| 2q24.3q31.1 |
RAPGEF4 |
7q22.1q31.3 |
NRCAM |
13q11.1qter |
MYCBP2 |
20p13-q11.23 |
EPB41L1 |
| 2q24.3q31.1 |
SCN1A |
7q35 |
CNTNAP2 |
13q11.1qter |
ITM2B |
20p13-q11.23 |
SNTA1 |
| 3p26.2p22.1 |
DYNC1LI1 |
7q36.2 |
DPP6 |
13q11.1qter |
RAP2A |
20p13-q11.23 |
CHGB |
| 3p26.2p22.1 |
MOBP |
8p23.3-p11.21 |
NEFL |
13q11.1qter |
SACS |
20p13-q11.23 |
PAK7 |
| 3p26.2p22.1 |
ATP2B2 |
8p23.3-p11.21 |
NEFM |
13q11.1qter |
SLITRK5 |
20p13-q11.23 |
SNPH |
| 3p26.2p22.1 |
SYN2 |
8p23.3-p11.21 |
NRG1 |
13q11.1qter |
CLDN10 |
20p13-q11.23 |
ATRN |
| 3p26.2p22.1 |
SLC6A1 |
8p23.3-p11.21 |
STMN4 |
15q11-13 |
APBA2 |
20p13-q11.23 |
PRNP |
| 3q21.3q22.3 |
PIK3CB |
8p23.3-p11.21 |
DPYSL2 |
15q11-13 |
TJP1 |
20p13-q11.23 |
SNAP25 |
| 3q27.1q29 |
CAMK2N2 |
8p23.3-p11.21 |
PTK2B |
15q11-13 |
GABRA5 |
21q22.3 |
S100B |
| 3q27.1q29 |
SST |
8q24.12-q24.3 |
PTK2 |
15q24 |
CPLX3 |
22q11.2 |
CLDN5 |
| 3q28-qter |
APOD |
9p24.3-p12 |
SLC1A1 |
15q24 |
NPTN |
22q11.2 |
SEPT5 |
| 4p16.3p16.1 |
CRMP1 |
9p24.3-p12 |
DCTN3 |
15q24q26.3 |
SV2B |
22q13.31-q13.33 |
MLC1 |
| 4p16.3p16.1 |
WFS1 |
9p24.3-p12 |
CLTA |
15q24q26.3 |
NTRK3 |
Xq21.2q28 |
L1CAM |
| 4p16.3p16.2 |
HTT |
9p24.3-p12 |
PTPRD |
15q24q26.3 |
CHRNA3 |
Xq21.2q28 |
PAK3 |
| 4q34.1-34.3 |
GPM6A |
9p24.3-p12 |
UNC13B |
15q24q26.3 |
HDGFRP3 |
Xq21.2q28 |
DCX |
| 4q34.1-34.3 |
SCRG1 |
9q22.32-q21.2 |
GABBR2 |
16p12.2-p11.2 |
PRKCB1 |
Xq21.2q28 |
ATP6AP1 |
| 5p14.1p12 |
SLC1A3 |
10q26.12-26.3 |
DPYSL4 |
16p13.3 |
METRN |
Xq21.2q28 |
PCDH19 |
| 5p15.33p14.1 |
MYO10 |
11q23.3q25 |
SORL1 |
16q22.1-22.3 |
CALB2 |
Xq21.2q28 |
PLXNB3 |
| 5p15.33p14.1 |
BASP1 |
11q23.3q25 |
SCN3B |
17p13.2 |
DLG4 |
Xq21.2q28 |
FMR1 |
| 5q35.2 | SNCB | 11q23.3q25 | FEZ1 | 17p13.2 | NDEL1 | Xq21.2q28 | PLP1 |
Figure 5.
Functional connections between genes at the 73 critical regions inferred by GRAIL. The double circle plots depict the gene relationships across the abnormal regions by GRAIL based on data source PubMed Text. Outer circle represents abnormal regions, of which the G-band regions with '_del' are denoted deletions and the other regions are duplications. The inner circle stands for genes located in the corresponding regions in the outer circle, and for clear visualization only significant genes (p < 0.001) and their connections are shown. The lines represent pair-wise gene connections, with the thickness of the lines indicating the degree functional similarity.
Gene network and pathway analysis
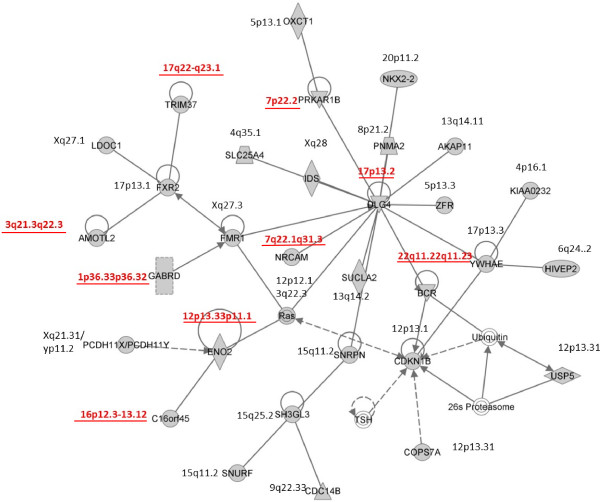
To understand the potential functional interactions of the 461 BEGs, Ingenuity Pathway Analysis (IPA) was performed to construct a network of genes involved in a specific biological process. The highest rated network of genes identified by IPA contains 31 significant genes which function in cell cycle, cell-to-cell signaling and interaction, nervous system development and function (Figure 6). In this network, eight genes including PRKAR1B, DLG4, NRCAM, GABRD, BCR, AMOTL2, ENO2, C16orf45 were also derived from GRAIL analysis. As shown in Figure 6, DLG4, the hub gene in network 1, interacts with 12 genes and has the highest degree (degree = 12, defined as the number of linked genes). IPA also denoted the other top four gene networks related to cellular assembly and organization, cellular function and maintenance and cell morphology. Three of them enriched for genes involved in nervous system development and function (Additional file 6: Figures S5-S8).
Figure 6.
The primary functional network of BEGs identified by the ingenuity pathway analysis. Nodes represent genes and a solid line means a direct interaction between the two genes and a dotted line stands for an indirect relationship. Shaded nodes represent genes within the abnormal regions (genes denoted with underlined G-band position are in the candidate gene list of Table 1)
Discussion
Recently, genomic analysis using array-based CGH or SNP chip was recommended as the first-tier genetic test for individuals with ID/DD/MCA and autism because of its high sensitivity and specificity for detecting submicroscopic deletions and duplications [3]. In this study, of the 1354 individuals with ID/DD/MCA/autism analyzed using Agilent’s 44 K and 180 K oligonucleotide arrays, 176 patients (13.0%) were detected with pCNVs, chromosomal structural abnormalities and aneuploidies. This diagnostic yield is in consistent with the reported range of 12%-20% by several studies [5-13,24-26]. Of all the abnormal cases, the largest proportion (37.5%) belongs to recurrent genomic disorders and the other two significant groups are subtelomeric rearrangements (25.6%) and sporadic interstitial abnormalities (18.8%). Recurrent genomic disorders are well recognized to be responsible for syndromic phenotypes. Recently, two case–control studies with large case series further define the phenotypes of recurrent genomic disorders and suggested further effort to develop a genome-wide map for dosage sensitive genes [9,24]. Subtelomeric and interstitial abnormalities showed larger variations in their size distribution and associated phenotypes. Therefore, it will be more difficult to identify the disease-causing genes.
Cytogenomic mapping of disjointed critical regions from the detected genomic imbalances allowed further bioinformatic mining for potential disease-causing genes. Of the 4080 genes sorted from the defined critical regions, about 11.3% (461/4080) of the genes showed brain specific expression by bioinformatic mining. Although the number of BEGs increases with the gene content of genomic regions, the enrichment of genes preferentially expressed in CNS tissues was noted by the gene annotation analysis. This observation suggested that the functional interactions rather than the number of the BEGs are more likely related to the ID phenotype. From the 461 BEGs, a subset of genes was identified with high relatedness through a statistical text mining approach. This enables us to recognize the biological connections across the disease-causing regions. In this subset of highly related genes, five genes have been established as causative genes for well-known mental retardation syndromes. These are GABRD for 1p36 deletion and duplication syndrome, NPTN for 15q24 BP0-BP1 deletion syndrome, PAFAH1B1 for 17p13.3 deletion and duplication syndrome, MECP2 for Rett syndrome, and FMR1 for fragile X syndrome. The identification of the known causative genes for ID/DD demonstrated the validity of this data mining approach.
ID/DD and autism are complex neurodevelopmental disorders. Recent evidence indicated that defects in synaptogenesis and synaptic activities and deregulation of neuronal development and function may contribute to pathogenesis of ID/DD [1]. Many of the candidate genes recovered by GRAIL are involved in neurological system process, synaptic transmission, cell junction and neuron axonogenesis. Part of the cross-loci interactions of gene ontology from detected deletions and duplications have been presented previously [27]. For example, the DLG4 gene, a member of synaptic molecules, is a major functional bridge in coupling the N-methyl-D-aspartate receptor to pathways that control bidirectional synaptic plasticity and learning. The phenotypes of DLG4 knockout mouse have been related to nervous system abnormalities [28]. The NRXN1 gene which interacts with its postsynaptic binding partner at the synapse has been associated with neurological diseases [29,30]. It is also noted that some of the BEGs did not show significant p-values (e.g, STS gene in Xp22.3 has a p-value of 0.83 when text is used). Actually, this exploratory bioinformatic mining was performed on a set of 4,080 genes, about one fifth of the estimated over 20,000 human genes. The inclusion of more abnormal cases and an increase in gene coverage will lead to more comprehensive mining of functional interactions and networks of candidate genes.
The pathway analyses by IPA also found that the BEGs are enriched in neurological disease and endocrine system disorders. The DLG4, FMR1, FXR2 and YWHAE genes with at least four neighbors (degree > = 4) in Network 1 are all related to neural disorders. The DLG4 gene is a homologue of a known disease-causing gene DLG1. The FMR1, FXR2 and YWHAE genes have been reported in ID related studies. Moreover, SNRPN, one of the deleted genes for Prader-Willi syndrome, is also directly connected to the DLG4 gene. This network of genes showed strong functional homogeneity and should be considered a significant network underlying the ID/DD mechanism. Network 2 can be approximately divided into four sub networks connected by the S100B gene which is related to neurodegeneration. One of the sub networks involved in cellular assembly and organization included spindle related genes such as CRMP1, indicating the important role of centrosome genes in neuron development. Another interesting finding is the detection of significant enrichment in the extracellular signal-regulated kinases (ERK) and mitogen-activated protein kinase (MAPK) signaling pathway by IPA. It has been demonstrated that ERK/MAPK is involved in human genomic disorders [31]. A previous bioinformatic analysis on a small case series showed gene prioritization within MAPK and neuroactive ligand-receptor interaction pathways [26]. A study on 1275 autism cases discovered an enrichment of CNVs disrupting functional gene sets involved in cellular proliferation, projection and motility and GTPase/Ras signaling [32]. Our study identified many significant genes and function pathways overlapped with those previous studies. Further experimental analysis to connect the functional genomics and molecular pathways is required for a better understanding of human brain and nervous system development [33].
Conclusions
In summary, we performed oligonucleotide aCGH to identify a spectrum of cytogenomic abnormalities including genomic disorders, subtelomeric rearrangements, interstitial imbalances, and chromosomal structural and numerical abnormalities. Further cytogenomic mapping and bioinformatic mining identified a set of candidate BEGs and reveal the underlying functional interrelations and networks likely contributing to the ID/DD phenotypes. The identification of cross-region interrelations of the BEGS could provide clues of related ID phenotypes and possible modifying effect for phenotype variations for better disease classification. The finding of five major networks suggested complex polygenic and multipathway models underlying haploinsufficiency and triple-sensitive regulation of mental development. However, this bioinformatic mining cannot differentiate the driving genetic factors from the modifying effects for the ID phenotypes. Further experimental analysis on animal or cellular models to characterize this complex genetic regulation is needed.
Methods
Subjects
The Yale Laboratory of Molecular Cytogenetics and Genomics is CLIA-approved and provides diagnostic services for pediatric patients with indications of ID, DD, MCA, learning disability, speech delay and autism. From 2006 to 2011, aCGH analyses were performed on 1354 pediatric patients using two array platforms: Agilent 44 K oligonucleotide array on 718 patients and Agilent 180 K oligonucleotide array on 636 patients. For cytogenomic mapping and bioinformatic mining of potential candidate genes from the abnormal findings, all patient information had been de-identified. There were no additional pre-study requirements on the patient's specimen and clinical indications and there were no post-study interaction and intervention with the patients. This project was categorized as a chart review retrospective study and deemed exemption from Institutional Review Boards (IRB) approval and waiver of consent based on the policy of Yale University IRB.
Oligonucleotide array comparative genomic hybridization (aCGH)
Genomic DNA was extracted from peripheral blood lymphocytes using the Gentra Puregene Kit (Qiagen, Valencia, CA). The DNA concentration was measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA) and high molecular weight DNA was verified by agarose gel electrophoresis. For each sample, 2–3 ug of genomic DNA was used following the manufacturer’s protocol for the Agilent Human Genome CGH microarray 44 K kit (containing 44,913 60-mer oligonucleotides) and 180 K kit (177,873 60-mer oligonucleotides) (Agilent Technologies, Inc., Santa Clara, CA). This aCGH procedure can achieve 99% sensitivity and 99% specificity using a sliding window of five to seven contiguous oligonucleotides, indicating an analytical resolution of 300–500 kilobase (Kb) for the 44 K platform and 100–150 Kb for the 180 K platform [4]. The base pair designations of 44 K and 180 K aCGH were based on the May 2004 Assembly (NCBI35/hg17) and the March 2006 Assembly (NCBI36/hg18) of the UCSC Human Genome browser (http://genome.ucsc.edu/), respectively.
The genomic imbalances detected by aCGH were classified as (1) chromosome abnormalities, (2) pathogenic CNV (pCNV), (3) variant of uncertain clinical significance (VOUS) and (4) benign CNV (bCNV) [34]. As previously described, large chromosome abnormalities (>5 Mb) were confirmed by high resolution karyotyping on metaphases prepared from cultured peripheral blood lymphocytes using the laboratory’s standardized protocols; small imbalances were confirmed by FISH using available commercial probes or targeted BAC probes [4]. Raw data of all detected chromosomal imbalances were loaded onto the Nexus5 Software (BioDiscovery, Los Angles, California, USA) to evaluate the genome-wide distribution and relative frequency of chromosomal and genomic abnormalities.
Fluorescence in situ hybridization (FISH)
To confirm the subtelomeric chromosome rearrangements and common genomic disorders detected by aCGH, FISH using selected ToTelVysion probes and locus-specific probes (Vysis/Abbott, Abbott Park, IL) as well as home brew targeted BAC probes was performed as previously described [4].
Cytogenomic mapping and bioinformatic analysis
A two-step cytogenomic mapping procedure was used to define critical regions from detected genomic abnormalities. First, segments overlapping with known bCNVs that have been documented at least three times in the Databases of Genomic Variants (DGV) within a pCNV were subtracted. Secondly, pCNVs were compared with other overlapping pCNVs associated with ID/DD from DECIPHER, and the smallest overlapped regions were extracted as critical regions. The gene content within each critical region was obtained based on gene coordinate (hg18) from the UCSC database browser [35]. Subsequent data mining focused on the BEGs from defined critical regions assuming that their functional interactions and networks likely contribute to human mental development.
Gene functional classification analysis
The tissue expression analysis was performed using the DAVID Gene Functional Classification Tool with the ‘GNF_U133A_QUARTILE’ option [36]. DAVID would report the tissues in which the gene expresses higher than 3rd quartile of its expression across all tissues. Moreover, the tissue expression patterns were further confirmed by 1,000 runs of permutation tests. For each run, a set of random genes with the same number of genes as that in the critical regions was generated and the number of genes expressed in a specific tissue was recorded.
Identification and distribution of brain-expressed genes (BEGs)
A list of BEGs was obtained following the procedures of Raychaudhuri et al. (2009a) [18]. Briefly, a widely used large human normal tissue expression dataset (Human U133A/GNF1H Gene Atlas) [37] was downloaded from the BioGPS website (http://www.biogps.org/) [38]. The downloaded expression data were gcRMA-normalized and averaged across replicates. Genes with expression values < 100 across all the tissues were excluded, since they are considered not expressed. One-tailed Mann–Whitney rank-sum p-values were then obtained to determine whether the genes express higher in the brain or spinal cord (21 tissues) than in the remaining tissue profiles. Genes with p < 0.01 were considered as BEGs.
For every defined critical region, a set of 10,000 random genomic loci was produced, each of which comprised of the same number of genes as in the tested critical region, and permutation of the random gene sets was conducted. For a specific region R, 10,000 consecutive regions of N genes (where N is the number of genes in R) were sampled from the set of the genes profiled on Human U133A/GNF1H platforms. The number of BEGs in the random gene set was recorded and then compared with that in true abnormal region R by boxplots.
Identification of candidate genes by gene interaction and networking
To examine the interrelations among the listed BEGs from different critical regions, GRAIL was used to assess the functional similarities by using PubMed abstracts, gene ontology terms or expression patterns in normal human tissues [18]. All critical regions containing BEGs were submitted as both seed and query regions and gene relationships among the abnormal regions were visualized by tools provided on the website (http://www.broadinstitute.org/mpg/grail/ ). The listed BEGs were also imported into the IPA software (Ingenuity Systems Inc; http://www.ingenuity.com/) to identify interactions between the focus genes and other gene objects.
Web resources
Database of Genomic Variants (DGV): http://projects.tcag.ca/variation/.
DatabasE of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER): http://decipher.sanger.ac.uk/.
BioGPS website (http://www.biogps.org/).
Gene Relationships Across Implicated Loci (http://www.broadinstitute.org/mpg/grail/.
Ingenuity Pathways Analysis (Ingenuity Systems Inc., RedwoodCity, California, USA; http://www.ingenuity.com/).
Abbreviations
aCGH: Array comparative genomic hybridization; BEGs: Brain-expressed genes; CNV: Copy number variant (pCNV, pathogenic; bCNV: Benign; VOUS: Variant of uncertain clinical significance); DD: Developmental disability; ID: Intellectual disability; LCRs: Low copy number repeats; MCA: Multiple congenital anomalies.
Competing interests
All authors declare that they have no competing interests.
Authors’ contributions
FX carried out the 180 K aCGH and organized cytogenomic findings, LL performed most bioinformatic analysis. FX and LL drafted the manuscript. VPS and PGG performed the IPA gene networking analysis. BX carried out the 44 K aCGH analysis, HZ supervised the bioinformatic and statistical analyses. HZ, PGG and PL participated in the design and coordinate of this study. All authors read and approved the final version of the manuscript.
Supplementary Material
Detected recurrent genomic disorders.
Subtelomeric chromosomal and genomic rearrangements.
Sporadic interstitial chromosomal and genomic imbalances.
Chromosome structural abnormalities.
Disjointed critical regions defined by cytogenomic mapping.
The distribution for the number of genes in a specific tissue. Arrow points to number of genes from abnormal regions. Figure S2. The number of candidate genes overlapping between the three data sources. Figure S3. Functional connections between genes at the 73 critical regions inferred by GRAIL based on Gene Ontology (significant genes with p < 0.01). Figure S4. Functional connections between genes at the 73 critical regions inferred by GRAIL based Human Expression Alta (significant genes with p < 0.0000000001). Figure S5. The secondary functional network of BEGs identified by the Ingenuity pathway analysis (genes denoted with red colored G-band position are in the candidate gene list of Table 1). Figure S6. The third functional network of BEGs identified by the Ingenuity pathway analysis (genes denoted with red colored G-band position are in the candidate gene list of Table 1). Figure S7. The fourth functional network of BEGs identified by the Ingenuity pathway analysis (genes denoted with red colored G-band position are in the candidate gene list of Table 1). Figure S8. The fifth functional network of BEGs identified by the Ingenuity pathway analysis (genes denoted with underlined G-band position are in the candidate gene list of Table 1).
List of brain expressed genes.
Contributor Information
Fang Xu, Email: fang.xu@yale.edu.
Lun Li, Email: irislilun@gmail.com.
Vincent P Schulz, Email: vincent.schulz@yale.edu.
Patrick G Gallagher, Email: patrick.gallagher@yale.edu.
Bixia Xiang, Email: bixiaxiang@gmail.com.
Hongyu Zhao, Email: hongyu.zhao@yale.edu.
Peining Li, Email: peining.li@yale.edu.
Acknowledgements
We would like to thank Dr. Maurice Mahoney for reviewing the ethical issues of this project, Kangmo Lu and Krista Sfiridis for their technical support, and Audrey Meusel for editing and proofreading this manuscript. Additionally, a fellowship award from the China Scholarship Council to Lun Li supported part of this work.
References
- Chelly J, Khelfaoui M, Francis F, Cherif B, Bienvenu T. Genetics and pathophysiology of mental retardation. Eur J Hum Genet. 2006;14:701–713. doi: 10.1038/sj.ejhg.5201595. [DOI] [PubMed] [Google Scholar]
- Shaffer LG. American College of Medical Genetics guideline on the cytogenetic evaluation of the individual with developmental delay or mental retardation. Genet Med. 2005;7:650–654. doi: 10.1097/01.gim.0000186545.83160.1e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ. et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang B, Li A, Valentin D, Nowak NJ, Zhao H, Li P. Analytical and clinical validity of whole-genome oligonucleotide array comparative genomic hybridization for pediatric patients with mental retardation and developmental delay. Am J Med Genet A. 2008;146A:1942–1954. doi: 10.1002/ajmg.a.32411. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Imoto I, Aizu Y, Okamoto N, Mizuno S, Kurosawa K, Honda S, Araki S, Mizutani S, Numabe H. et al. Clinical application of array-based comparative genomic hybridization by two-stage screening for 536 patients with mental retardation and multiple congenital anomalies. J Hum Genet. 2010;56:110–124. doi: 10.1038/jhg.2010.129. [DOI] [PubMed] [Google Scholar]
- Manolakos E, Vetro A, Kefalas K, Rapti SM, Louizou E, Garas A, Kitsos G, Vasileiadis L, Tsoplou P, Eleftheriades M. et al. The use of array-CGH in a cohort of Greek children with developmental delay. Mol Cytogenet. 2010;3:22. doi: 10.1186/1755-8166-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld JA, Ballif BC, Torchia BS, Sahoo T, Ravnan JB, Schultz R, Lamb A, Bejjani BA, Shaffer LG. Copy number variations associated with autism spectrum disorders contribute to a spectrum of neurodevelopmental disorders. Genet Med. 2010;12:694–702. doi: 10.1097/GIM.0b013e3181f0c5f3. [DOI] [PubMed] [Google Scholar]
- Xiang B, Zhu H, Shen Y, Miller DT, Lu K, Hu X, Andersson HC, Narumanchi TM, Wang Y, Martinez JE. et al. Genome-wide oligonucleotide array comparative genomic hybridization for etiological diagnosis of mental retardation: a multicenter experience of 1499 clinical cases. J Mol Diagn. 2010;12:204–212. doi: 10.2353/jmoldx.2010.090115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky EB, Kaul V, Paschall J, Church DM, Bunke B, Kunig D, Moreno-De-Luca D, Moreno-De-Luca A, Mulle JG, Warren ST. et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet Med. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iourov IY, Vorsanova SG, Kurinnaia OS, Zelenova MA, Silvanovich AP, Yurov YB. Molecular karyotyping by array CGH in a Russian cohort of children with intellectual disability, autism, epilepsy and congenital anomalies. Mol Cytogenet. 2012;5:46. doi: 10.1186/1755-8166-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JW, Bint S, Bergbaum A, Mann K, Hall RP, Ogilvie CM. Array CGH as a first line diagnostic test in place of karyotyping for postnatal referrals - results from four years' clinical application for over 8,700 patients. Mol Cytogenet. 2013;6:16. doi: 10.1186/1755-8166-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Jung EH, Ryu RS, Kang HW, Chung HD, Kang HY. The clinical application of array CGH for the detection of chromosomal defects in 20,126 unselected newborns. Mol Cytogenet. 2013;6:21. doi: 10.1186/1755-8166-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Xu F, Li P. Technology-driven and evidence-based genomic analysis for integrated pediatric and prenatal genetics evaluation. J Genet Genomics. 2013;40:1–14. doi: 10.1016/j.jgg.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Tyshchenko N, Lurie I, Schinzel A. Chromosomal map of human brain malformations. Hum Genet. 2008;124:73–80. doi: 10.1007/s00439-008-0528-2. [DOI] [PubMed] [Google Scholar]
- Bittel DC, Yu S, Newkirk H, Kibiryeva N, Holt A, Butler MG, Cooley LD. Refining the 22q11.2 deletion breakpoints in DiGeorge syndrome by aCGH. Cytogenet Genome Res. 2009;124:113–120. doi: 10.1159/000207515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo T, Shaw CA, Young AS, Whitehouse NL, Schroer RJ, Stevenson RE, Beaudet AL. Array-based comparative genomic hybridization analysis of recurrent chromosome 15q rearrangements. Am J Med Genet A. 2005;139A:106–113. doi: 10.1002/ajmg.a.31000. [DOI] [PubMed] [Google Scholar]
- Khatri P, Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, Sklar P, Scolnick EM, Xavier RJ, Altshuler D, Daly MJ. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Thomson BP, Remmers EF, Eyre S, Hinks A, Guiducci C, Catanese JJ, Xie G, Stahl EA, Chen R. et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41:1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH Jr, Dobyns WB, Christian SL. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- Kumar RA, Sudi J, Babatz TD, Brune CW, Oswald D, Yen M, Nowak NJ, Cook EH, Christian SL, Dobyns WB. A de novo 1p34.2 microdeletion identifies the synaptic vesicle gene RIMS3 as a novel candidate for autism. J Med Genet. 2009;47:81–90. doi: 10.1136/jmg.2008.065821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V. et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolen DA, Pfundt R, de Leeuw N, Hehir-Kwa JY, Nillesen WM, Neefs I, Scheltinga I, Sistermans E, Smeets D, Brunner HG. et al. Genomic microarrays in mental retardation: a practical workflow for diagnostic applications. Hum Mutat. 2009;30:283–292. doi: 10.1002/humu.20883. [DOI] [PubMed] [Google Scholar]
- Qiao Y, Harvard C, Tyson C, Liu X, Fawcett C, Pavlidis P, Holden JJ, Lewis ME, Rajcan-Separovic E. Outcome of array CGH analysis for 255 subjects with intellectual disability and search for candidate genes using bioinformatics. Hum Genet. 2010;128:179–194. doi: 10.1007/s00439-010-0837-0. [DOI] [PubMed] [Google Scholar]
- Xu F, Li P. In: Developmental disabilities - molecules involved, diagnosis, and clinical care. Salehi A, editor. : InTech; 2013. Cytogenomic abnormalities and dosage-sensitive mechanisms for intellectual and developmental disabilities. [Google Scholar]
- Feyder M, Karlsson RM, Mathur P, Lyman M, Bock R, Momenan R, Munasinghe J, Scattoni ML, Ihne J, Camp M. et al. Association of mouse Dlg4 (PSD-95) gene deletion and human DLG4 gene variation with phenotypes relevant to autism spectrum disorders and Williams' syndrome. Am J Psychiatry. 2010;167:1508–1517. doi: 10.1176/appi.ajp.2010.10040484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, Lally E, Weiss LA, Najm J, Kutsche K. et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujescu D, Ingason A, Cichon S, Pietilainen OP, Barnes MR, Toulopoulou T, Picchioni M, Vassos E, Ettinger U, Bramon E. et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Niihori T, Narumi Y, Kure S, Matsubara Y. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum Mutat. 2008;29:992–1006. doi: 10.1002/humu.20748. [DOI] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS. et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Konopka G. Neuroscience in the era of functional genomics and systems biology. Nature. 2009;461:908–915. doi: 10.1038/nature08537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680–685. doi: 10.1097/GIM.0b013e3182217a3a. [DOI] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A. et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Huang W, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G. et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW 3rd, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detected recurrent genomic disorders.
Subtelomeric chromosomal and genomic rearrangements.
Sporadic interstitial chromosomal and genomic imbalances.
Chromosome structural abnormalities.
Disjointed critical regions defined by cytogenomic mapping.
The distribution for the number of genes in a specific tissue. Arrow points to number of genes from abnormal regions. Figure S2. The number of candidate genes overlapping between the three data sources. Figure S3. Functional connections between genes at the 73 critical regions inferred by GRAIL based on Gene Ontology (significant genes with p < 0.01). Figure S4. Functional connections between genes at the 73 critical regions inferred by GRAIL based Human Expression Alta (significant genes with p < 0.0000000001). Figure S5. The secondary functional network of BEGs identified by the Ingenuity pathway analysis (genes denoted with red colored G-band position are in the candidate gene list of Table 1). Figure S6. The third functional network of BEGs identified by the Ingenuity pathway analysis (genes denoted with red colored G-band position are in the candidate gene list of Table 1). Figure S7. The fourth functional network of BEGs identified by the Ingenuity pathway analysis (genes denoted with red colored G-band position are in the candidate gene list of Table 1). Figure S8. The fifth functional network of BEGs identified by the Ingenuity pathway analysis (genes denoted with underlined G-band position are in the candidate gene list of Table 1).
List of brain expressed genes.