Abstract
Background
Prion diseases are characterized by the accumulation of the pathogenic PrPSc protein, mainly in the brain and the lymphoreticular system. Although prions multiply/accumulate in the lymph nodes without any detectable pathology, transcriptional changes in this tissue may reflect biological processes that contribute to the molecular pathogenesis of prion diseases. Little is known about the molecular processes that occur in the lymphoreticular system in early and late stages of prion disease. We performed a microarray-based study to identify genes that are differentially expressed at different disease stages in the mesenteric lymph node of sheep naturally infected with scrapie. Oligo DNA microarrays were used to identify gene-expression profiles in the early/middle (preclinical) and late (clinical) stages of the disease.
Results
In the clinical stage of the disease, we detected 105 genes that were differentially expressed (≥2-fold change in expression). Of these, 43 were upregulated and 62 downregulated as compared with age-matched negative controls. Fewer genes (50) were differentially expressed in the preclinical stage of the disease. Gene Ontology enrichment analysis revealed that the differentially expressed genes were largely associated with the following terms: glycoprotein, extracellular region, disulfide bond, cell cycle and extracellular matrix. Moreover, some of the annotated genes could be grouped into 3 specific signaling pathways: focal adhesion, PPAR signaling and ECM-receptor interaction. We discuss the relationship between the observed gene expression profiles and PrPSc deposition and the potential involvement in the pathogenesis of scrapie of 7 specific differentially expressed genes whose expression levels were confirmed by real time-PCR.
Conclusions
The present findings identify new genes that may be involved in the pathogenesis of natural scrapie infection in the lymphoreticular system, and confirm previous reports describing scrapie-induced alterations in the expression of genes involved in protein misfolding, angiogenesis and the oxidative stress response. Further studies will be necessary to determine the role of these genes in prion replication, dissemination and in the response of the organism to this disease.
Keywords: Natural scrapie, Mesenteric lymph node, Microarray, Gene expression, Real time PCR, Prion
Background
Transmissible spongiform encephalopathies (TSEs) or prion diseases are a group of fatal neurodegenerative disorders of animals and humans. TSEs generally involve long incubation periods, which in humans can span several decades [1]. These diseases are characterized by the accumulation, mainly in nervous and lymphoreticular tissues, of an abnormal isoform (PrPSc) of a normal host-coded cell surface glycoprotein (PrPc). Pathogenesis typically involves a triad of histological events; vacuolation, neuronal loss and astrocytosis. PrPSc accumulation is currently considered the only reliable biochemical marker for this group of diseases [2].
Scrapie is a TSE that naturally affects sheep and goats. Ovine scrapie is a useful model in which to study the pathogenic mechanisms of the variant Creutzfeldt Jakob disease (vCJD), which affects humans [3]. Like vCJD, classical scrapie is associated with widespread tissue infectivity, mainly in the central nervous system (CNS), peripheral nervous system (PNS), and lymphoreticular system (LRS) [4].
Mesenteric lymph nodes are one of the first tissues of the LRS in which PrPSc accumulates during the preclinical disease stage in naturally infected sheep [5,6], although this process strongly depends on the animal’s PRNP genotype. Subsequently, other LRS tissues are rapidly and near simultaneously exposed to infectivity, presumably by circulating prions. In the clinical stages of the disease and in all susceptible PRNP genotypes except VRQ/ARR, infectivity and PrPSc accumulation are generally observed throughout all lymphoid tissues, except for the thymus [7]. Follicular dendritic cells (FDCs) may be necessary for prion propagation within the LRS [8], and macrophages and tingible body macrophages (TBMs) of the LRS have been identified as reservoirs of the infectious agents of TSEs [4,9].
The molecular mechanisms that underlie the uptake of the infectious prion agent and the progression of the disease remain largely unknown. Given the complexity and multifactorial nature of the spread and accumulation of the infectious agent, we and others have used gene expression analysis platforms to identify signaling pathways that are altered during PrPSc accumulation and subsequent neurodegeneration. This approach facilitates the identification of potential biomarkers and drug targets in natural ovine scrapie [10,11] and in experimental murine scrapie models [12-14].
Few gene expression-profiling studies have investigated changes in the lymphoreticular system in sheep with natural scrapie; most have been performed using murine models [15,16] or ileal Peyer’s patches from orally inoculated lambs [17]. In the present study, we used an ovine microarray technology to identify transcriptional changes in the mesenteric lymph node in both clinical and preclinical disease stages in sheep that were naturally infected with scrapie. This is the first transcriptome-wide expression profiling study of the lymphoreticular system in sheep with natural scrapie.
Methods
Ethics statement
This study was approved by the Ethics Committee for Animal Experiments of the University of Zaragoza (Permit Number: PI02/08) and was carried out in strict accordance with the recommendations for the care and use of experimental animals and in agreement with national law (R.D. 1201/2005).
Animals, necropsy, tissue collection and RNA isolation
All details of the animals used and the necropsy and RNA isolation procedures have been previously reported [10,11]. Briefly, we used a 17 female ARQ/ARQ Rasa Aragonesa sheep (aged 1-6 years). Eleven animals were obtained from scrapie-infected flocks from several locations in Aragon (Spain); 7 of these exhibited clinical signs of scrapie while 4 were preclinical. Six animals were selected from flocks located in scrapie-free areas and were used as breed, genotype and age-matched controls.
Sheep in the clinical disease stage were identified by assessing clinical signs associated with the disease [18]. Third eyelid [18] and rectal mucosa biopsies [19] were used to confirm this diagnosis and to identify animals in the preclinical disease stage. Postmortem examinations revealed no additional pathological findings. Mesenteric lymph node samples were divided into 2 halves; one was snap-frozen in liquid nitrogen and stored at −80°C until RNA extraction, and the other was formalin-fixed and paraffin-embedded for further histopathological analysis. Total RNA isolation, purification and quality control were performed as previously described [10,11].
Immunohistochemical detection of PrPSc
Immunohistochemical (IHC) analyses were performed using serial sections. Prion protein was detected, after formic acid treatment and proteinase K digestion, using the monoclonal primary antibody L42 (1:500; R-Biopharm), as previously described [20]. Negative controls were performed omitting the primary antibody from control and scrapie sections.
The preparations were microscopically examined and global PrPSc deposition was scored by quantifying the proportion of lymphoid follicles with PrPSc deposits as follows: 1, <20%; 2, 20-50%; 3, >50% [19]. Significant differences between clinical, preclinical and control groups were identified using the Kruskal-Wallis test.
Microarray hybridization
The custom CVI 4x44K microarrays used in this study contained custom eArray-designed 60-mer probes of previously sequenced, normalized and subtracted cDNA libraries of ovine Peyer’s Patch, obex and tonsil, supplemented with the publicly available Ovis aries transcripts from the NCBI/EBI databases and the Agilent O. aries transcript catalog. All arrays were printed using Sureprint technology (Agilent Technologies).
Preparation of the labeled cDNA probes and subsequent Genechip hybridizations were performed in accordance with the Agilent Technologies One-Color Microarray-Based Gene Expression Analysis guidelines, as previously described [10,11]. Hybridizations were performed in duplicate, resulting in 14 microarrays for the clinical samples, 8 microarrays for the preclinical samples and 12 for the negative control samples. Microarrays were scanned using the GenePix 4200AL Scanner (Axon Instruments) and GenePix Pro 6.0 software and the hybridization data were extracted using Agilent Feature Extraction software v9.5.3.1 (Agilent Technologies) before processing with GeneSpring GX 10.0.2 (Agilent Technologies). Chip values were normalized using the 75th percentile value and expression levels were calculated. Global mesenteric lymph node gene expression profiles for the clinical and preclinical disease stages were compared with the negative controls using a linear model that accounts for both technical (random animal effects) and biological replicates (disease effects). A multiple testing correction developed by Benjamini and Hochberg was also applied. Only genes that exhibited at least a 2-fold change (FC) in expression between healthy and infected animals and had a P-value ≤ 0.05 were selected. These genes were clustered according to their Euclidean distance coefficient using PermutMatrix software [21]. A Megablast search of the GenBank database nr/nt was performed to identify genes that were similar to the differentially expressed probes. The molecular functions of the genes were classified according to Gene Ontology (GO) using an on-line functional annotation tool (DAVID Bioinformatics Resources 6.7; NIAID/NIH, USA) [22,23].
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was performed to confirm the expression of 7 genes. Genes were selected based on previous studies describing their association with prion and other neurodegenerative diseases or their potential role in protein misfolding repair, the promotion of angiogenesis, or the systemic response of the organism to infection [24-30]. The PCR primer sequences used for the quantification of the genes encoding butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (BBOX1), ceruloplasmin (ferroxidase) (CP), prefoldin subunit 2 (PFDN2), proteasome subunit, alpha type, 7 (PSMA7), serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 (SERPINE1), ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) (UCHL1) and vascular endothelial growth factor A (VEGFA) are shown in Table 1. The RNA samples used for qRT-PCR were the same as those used for microarray experiments. The qRT-PCR assays were designed using Primer Express 2.0 software (Applied Biosystems) to select appropriate primer sequences from known ovine or bovine sequences. Whenever possible, the exon-exon border was included to prevent amplification of genomic DNA in the PCR reaction. Complementary DNA (cDNA) was synthesized from 1 μg RNA using random hexamer primers with the Superscript First Standard Synthesis System for RT-PCR (Invitrogen). To confirm the elimination of any remaining DNA, reverse transcription was performed with and without the enzyme.
Table 1.
Genes analyzed by quantitative real-time PCR
| Gene | Primer sequence | Size (bp) | Accession number |
|---|---|---|---|
|
BBOX1 |
F: TGCAAACAATGTGGCTTACACA |
85 |
NM_001101881.2** |
| |
R: AAGCTGAACCCCAGGTGGAT |
|
|
|
CP |
F: GCAGCCAGATACTGCAGGGAC |
97 |
NM_001009733.1* |
| |
R: CCCGCACTGGCTCACAGTATAT |
|
|
|
PFDN2 |
F: GCAGGTAATTGCTGGCTTCAA |
84 |
NM_001080221.2** |
| |
R: TTCAACTCCATCTCCAGCTCAG |
|
|
|
PSMA7 |
F: TAATGTCTGCATGGCATTCGC |
81 |
NM_001034233.2** |
| |
R: TGGCTCTGGCATTCCACC |
|
|
|
SERPINE1 |
F: TGTACGTGTCGCAGGCGC |
83 |
NM_174137.2** |
| |
R: TACAAGGGCTGTGGAGGAGGAC |
|
|
|
UCHL1 |
F: AACTTGATGGACGGATGCCTT |
84 |
NM_001046172.2** |
| |
R: TGCAGACCTTGGCAGCGT |
|
|
|
VEGFA |
F: GGGCTGCTGTAATGACGAAAGT |
81 |
NM_001025110.1* |
| R: GGTTTGATCCGCATAATCTGCA | |||
*Ovine cDNA.
**Bovine cDNA.
Primers (F, forward; R, reverse) used for gene amplification, amplicon size and GenBank accession numbers for the bovine cDNA sequences used for primer design.
Quantitative RT-PCR was performed using SYBR® Green (PE Applied Biosystems) assays as previously described [10,11]. To improve the normalization accuracy, a normalization factor (NF) was used to determine the expression level of each gene in each sample, as previously described [31]. The NF was calculated using the geometric mean expression of 3 housekeeping genes (GAPDH, G6PDH and RPL32). The primers and PCR conditions used for the amplification of these housekeeping genes have been described previously [31,32].
The quantitative results obtained from the qRT-PCR assays were expressed as FC with respect to controls. Significant differences between groups were determined using a Student’s t-test (P < 0.05).
The relationship between neuropathology and gene expression
We used 2 different models to quantify the relationships between the PrPSc deposition and gene expression. We first used a simple regression model,
where yi is the gene expression data for the ith individual for each gene, μ is the general mean, b is slope of the regression analysis, pi is the PrPSc deposition for the ith individual and ei is the residual. In addition, we used another model,
which includes a systematic effect associated with the 3 categories (Preclinical, Clinical and Healthy). Here yij represents the gene expression for the jth individual at the ith category, Ti the effects of the ith treatment, pij the PrPSc deposition and eij the residual. The statistical significance of the slope associated with PrPSc deposition was determined for both models. Analyses were performed using the R project for Statistical Computing.
Results
PrPSc deposition in lymphoid tissue
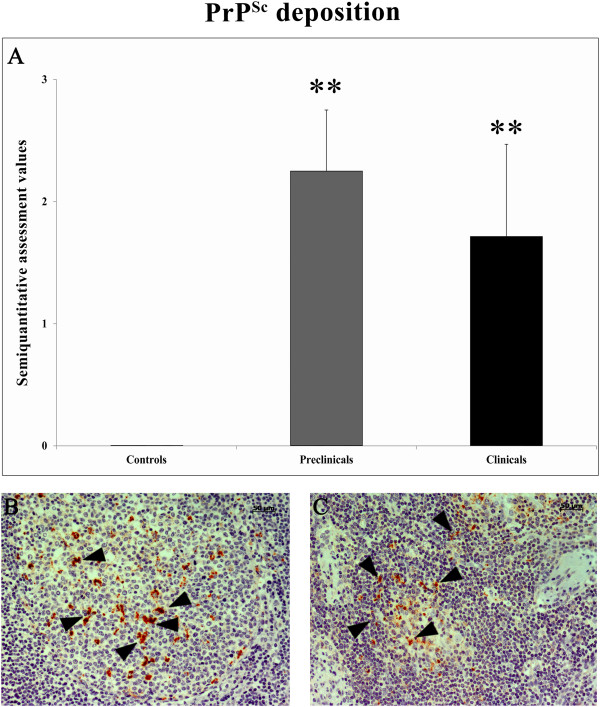
The accumulation of PrPSc in the LRS was consistent with the features of classical scrapie. PrPSc was detected within the primary and secondary lymphoid follicles of the lymph nodes. Some cells in the follicular mantle and paracortical area were also PrPSc-positive. Interfollicular and sinusal cells were rarely labeled. As seen in other LRS tissues, distinct granular immunolabeling was detected in the cytoplasm of tingible body macrophages in both the dark and light zones of the lymphoid follicles. A granular deposition was observed in the vicinity of round cells, which were morphologically similar to immature B lymphocytes (Figure 1). None of the control animals displayed PrPSc immunostaining and no statistically significant differences were observed between the 2 scrapie-infected groups (preclinical and clinical).
Figure 1.
Semi-quantitative assessment of PrPSc deposition in mesenteric lymph nodes samples from control, preclinical and clinical scrapie sheep. Deposition was evaluated on a scale of 0 (negative) to 5 (maximum staining intensity). Significant differences were detected between control and scrapie groups using a Kruskal-Wallis test (**P < 0.01) (A). PrPSc deposition in the mesenteric lymph node from preclinical (B) and clinical (C) scrapie sheep.
Identification of differentially expressed genes in the mesenteric lymph node of sheep with natural scrapie
A total of 234 probe sets displayed significant differences in expression of 2-fold or more between the control and scrapie (preclinical and clinical) groups. While the genes of Ovis aries are relatively poorly annotated, we used BLAST searches against publicly available databases to identify a set of 177 known genes from the complete set of 234 differentially expressed probes. Some of these 234 probes encoded the same gene. The microarray data were deposited in ArrayExpress and assigned the accession number E-MTAB-1346.
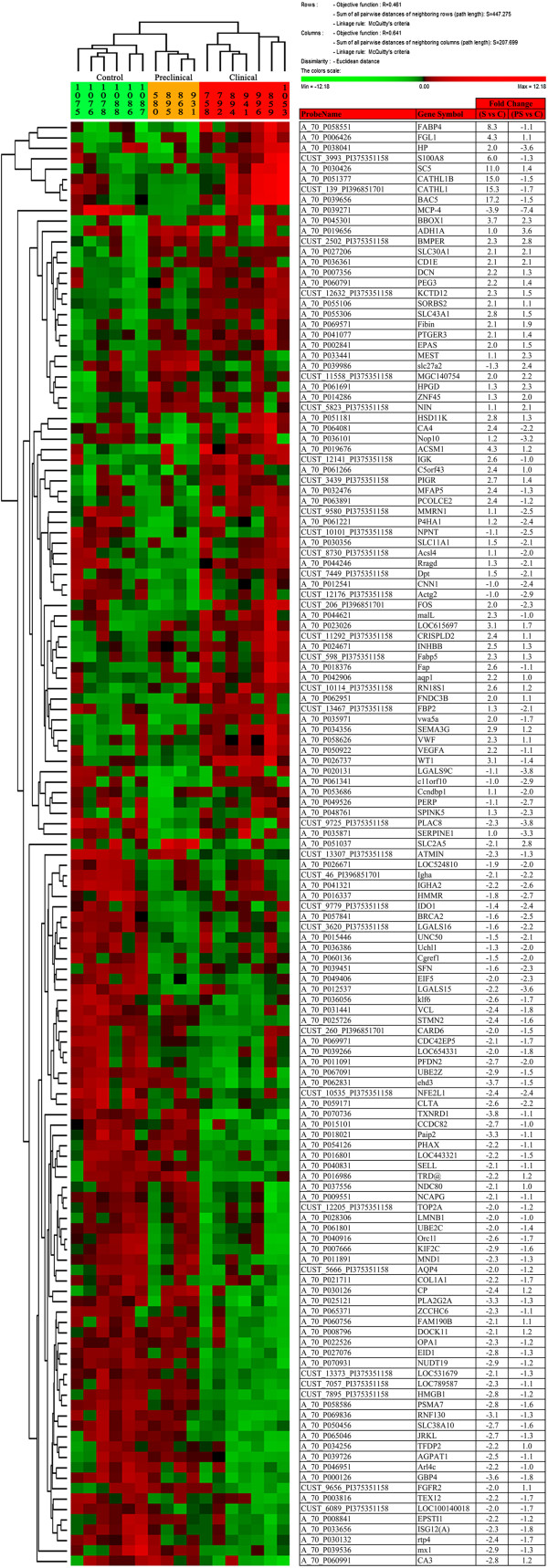
Based on the GO analysis of human homologs, 139 genes had known functions (Table 2), of which 105 were differentially regulated in clinical scrapie-infected sheep (43 upregulated and 62 downregulated) and 50 in preclinical scrapie-infected sheep (12 upregulated and 38 downregulated). The functional groups with the highest p values in the enrichment analysis are shown in Table 2; the most relevant terms were glycoprotein, extracellular region, disulfide bond and cell cycle. In addition, some of the annotated genes were grouped in 3 signaling pathways: focal adhesion, PPAR signaling pathway and ECM-receptor interaction (Table 3). After hierarchical clustering using the expression levels of all annotated genes, we were able to discriminate the 3 experimental groups: clinical, preclinical and healthy (Figure 2).
Table 2.
GO terms for differentially expressed genes
| Term | P value | ProbeName | FC (C vs NC) | FC (PC vs NC) | Gene symbol | Gene name | Citation |
|---|---|---|---|---|---|---|---|
| Glycoprotein |
5.16E-04 |
A_70_P042906 |
2.22 |
1.00 |
AQP1 |
Aquaporin 1 (Colton blood group) |
|
| CUST_5666_PI375351158 |
−2.02 |
−1.16 |
AQP4 |
Aquaporin 4 |
[10] |
||
| A_70_P041077 |
2.09 |
1.39 |
PTGER3 |
Prostaglandin E receptor 3 (subtype EP3) |
|
||
| A_70_P030356 |
1.48 |
−2.06 |
SLC11A1 |
Solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 |
|
||
| A_70_P051037 |
−2.09 |
2.85 |
SLC2A5 |
Solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
[10] |
||
| A_70_P036386 |
−1.29 |
−2.01 |
UCHL1* |
Ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
|
||
| Extracellular region |
5.37E-04 |
CUST_2502_PI375351158 |
2.26 |
2.76 |
BMPER |
BMP binding endothelial regulator |
|
| A_70_P006426 |
4.26 |
1.06 |
FGL1 |
Fibrinogen-like 1 |
|
||
| A_70_P069571 |
2.05 |
1.93 |
FIBIN |
Fin bud initiation factor homolog (zebrafish) |
|
||
| A_70_P038041 |
1.95 |
−3.63 |
HP |
Haptoglobin |
[10] |
||
| CUST_3439_PI375351158 |
2.70 |
1.38 |
PIGR |
Polymeric immunoglobulin receptor |
|
||
| A_70_P035871 |
1.03 |
−3.29 |
SERPINE1* |
Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
|
||
| A_70_P039451 |
−1.57 |
−2.28 |
SFN |
Stratifin |
|
||
| A_70_P050922 |
2.19 |
−1.06 |
VEGFA* |
Vascular endothelial growth factor A |
|
||
| Disulfide bond |
1.10E-03 |
CUST_598_PI375351158 |
2.28 |
1.33 |
FABP5 |
Fatty acid binding protein 5 (psoriasis-associated) |
|
| A_70_P070736 |
−3.83 |
−1.06 |
TXNRD1 |
Thioredoxin reductase 1 |
|
||
| Cell cycle |
1.17E-03 |
A_70_P058586 |
−2.85 |
−1.55 |
PSMA7* |
Proteasome (prosome, macropain) subunit, alpha type, 7 |
|
| A_70_P057841 |
−1.61 |
−2.51 |
BRCA2 |
Breast cancer 2, early onset |
|
||
| A_70_P053686 |
1.11 |
−2.01 |
CCNDBP1 |
Cyclin D-type binding-protein 1 |
|
||
| A_70_P060136 |
−1.48 |
−2.04 |
CGREF1 |
Cell growth regulator with EF-hand domain 1 |
|
||
| A_70_P027076 |
−2.79 |
−1.28 |
EID1 |
EP300 interacting inhibitor of differentiation 1 |
|
||
| A_70_P007666 |
−2.87 |
−1.61 |
KIF2C |
Kinesin family member 2C |
|
||
| A_70_P011891 |
−2.34 |
−1.29 |
MND1 |
Meiotic nuclear divisions 1 homolog (S. cerevisiae) |
|
||
| A_70_P009551 |
−2.13 |
−1.07 |
NCAPG |
Non-SMC condensin I complex, subunit G |
|
||
| A_70_P037556 |
−2.12 |
1.01 |
NDC80 |
NDC80 homolog, kinetochore complex component (S. cerevisiae) |
|
||
| A_70_P034256 |
−2.20 |
1.05 |
TFDP2 |
Transcription factor Dp-2 (E2F dimerization partner 2) |
|
||
| A_70_P061801 |
−2.01 |
−1.39 |
UBE2C |
Ubiquitin-conjugating enzyme E2C |
|
||
| Condensed chromosome |
1.71E-03 |
CUST_7895_PI375351158 |
−2.79 |
−1.15 |
HMGB1 |
High-mobility group box 1 |
|
| A_70_P003816 |
−2.20 |
−1.67 |
TEX12 |
Testis expressed 12 |
|
||
| Alternative splicing |
2.04E-03 |
A_70_P059171 |
−2.57 |
−2.17 |
CLTA |
Clathrin, light chain (Lca) |
|
| A_70_P033441 |
1.13 |
2.27 |
MEST |
Mesoderm specific transcript homolog (mouse) |
|
||
| A_70_P039536 |
−2.86 |
−1.34 |
MX1 |
Myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
|
||
| A_70_P060791 |
2.25 |
1.38 |
PEG3 |
Paternally expressed 3 |
|
||
| A_70_P015446 |
−1.52 |
−2.13 |
UNC50 |
Unc-50 homolog (C. elegans) |
|
||
| Chromosome |
2.20E-03 |
A_70_P065046 |
−2.68 |
−1.32 |
JRKL |
Jerky homolog-like (mouse) |
|
| A_70_P040916 |
−2.62 |
−1.67 |
ORC1L |
Origin recognition complex, subunit 1-like (yeast) |
|
||
| CUST_12205_PI375351158 |
−2.04 |
−1.20 |
TOP2A |
Topoisomerase (DNA) II alpha 170 kDa |
[33] |
||
| Extracellular matrix |
3.15E-03 |
A_70_P021711 |
−2.19 |
−1.69 |
COL1A1 |
Collagen, type I, alpha 1 |
[10,16] |
| CUST_11292_PI375351158 |
2.40 |
1.12 |
CRISPLD2 |
Cysteine-rich secretory protein LCCL domain containing 2 |
|
||
| A_70_P007356 |
2.20 |
1.26 |
DCN |
Decorin |
[10,16] |
||
| CUST_7449_PI375351158 |
1.46 |
−2.05 |
DPT |
Dermatopontin |
|
||
| A_70_P032476 |
2.45 |
−1.35 |
MFAP5 |
Microfibrillar associated protein 5 |
[10] |
||
| A_70_P058626 |
2.27 |
1.13 |
VWF |
Von Willebrand factor |
|
||
| Signal peptide |
3.25E-03 |
A_70_P064081 |
2.44 |
−2.18 |
CA4 |
Carbonic anhydrase IV |
|
| A_70_P061221 |
1.20 |
−2.43 |
P4HA1 |
Prolyl 4-hydroxylase, alpha polypeptide I |
[10,11] |
||
| A_70_P040831 |
−2.09 |
−1.07 |
SELL |
Selectin L |
|
||
| Tumor suppressor |
1.47E-02 |
A_70_P035971 |
2.05 |
−1.75 |
VWA5A |
Von Willebrand factor A domain containing 5A |
|
| A_70_P026737 |
3.11 |
−1.41 |
WT1 |
Wilms tumor 1 |
|
||
| A_70_P061691 |
1.26 |
2.26 |
HPGD |
Hydroxyprostaglandin dehydrogenase 15-(NAD) |
|
||
| Cytoplasm |
1.82E-02 |
A_70_P016337 |
−1.78 |
−2.72 |
HMMR |
Hyaluronan-mediated motility receptor (RHAMM) |
|
| CUST_12176_PI375351158 |
−1.04 |
−2.92 |
ACTG2 |
Actin, gamma 2, smooth muscle, enteric |
|
||
| A_70_P019656 |
1.01 |
3.59 |
ADH1A |
Alcohol dehydrogenase 1A (class I), alpha polypeptide |
|
||
| A_70_P045301 |
3.70 |
2.33 |
BBOX1* |
Butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
|
||
| A_70_P069971 |
−2.09 |
−1.73 |
CDC42EP5 |
CDC42 effector protein (Rho GTPase binding) 5 |
|
||
| CUST_5823_PI375351158 |
1.12 |
2.07 |
NIN |
Ninein (GSK3B interacting protein) |
|
||
| A_70_P018021 |
−3.25 |
−1.15 |
PAIP2 |
Poly(A) binding protein interacting protein 2 |
|
||
| A_70_P054126 |
−2.16 |
−1.11 |
PHAX |
Phosphorylated adaptor for RNA export |
|
||
| A_70_P044246 |
1.26 |
−2.14 |
RRAGD |
Ras-related GTP binding D |
|
||
| A_70_P055106 |
2.05 |
1.07 |
SORBS2 |
Sorbin and SH3 domain containing 2 |
|
||
| A_70_P025726 |
−2.39 |
−1.60 |
STMN2 |
Stathmin-like 2 |
|
||
| A_70_P060991 |
−2.80 |
1.21 |
CA3 |
Carbonic anhydrase III, muscle specific |
|
||
| Fatty acid metabolism |
2.26E-02 |
CUST_8730_PI375351158 |
1.06 |
−2.05 |
ACSL4 |
Acyl-CoA synthetase long-chain family member 4 |
|
| A_70_P039986 |
−1.27 |
2.39 |
SLC27A2 |
Solute carrier family 27 (fatty acid transporter), member 2 |
|
||
| Phosphatidic acid metabolic process |
2.86E-02 |
A_70_P039726 |
−2.50 |
−1.10 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
|
| A_70_P025121 |
−3.34 |
−1.30 |
PLA2G2A |
Phospholipase A2, group IIA (platelets, synovial fluid) |
|
||
| Protein dimerization activity |
2.91E-02 |
CUST_206_PI396851701 |
2.04 |
−2.28 |
FOS |
v-fos FBJ murine osteosarcoma viral oncogene homolog |
[10,11,33] |
| CUST_10535_PI375351158 |
−2.41 |
−2.41 |
NFE2L1 |
Nuclear factor (erythroid-derived 2)-like 1 |
|
||
| Zinc finger, C2H2-type |
3.09E-02 |
CUST_13307_PI375351158 |
−2.34 |
−1.26 |
ATMIN |
ATM interactor |
|
| A_70_P036056 |
−2.65 |
−1.71 |
KLF6 |
Kruppel-like factor 6 |
|
||
| A_70_P065371 |
−2.27 |
−1.13 |
ZCCHC6 |
Zinc finger, CCHC domain containing 6 |
|
||
| A_70_P014286 |
1.33 |
2.01 |
ZNF45 |
Zinc finger protein 45 |
|
||
| Cytoplasmic membrane-bounded vesicle |
3.21E-02 |
A_70_P062831 |
−3.71 |
−1.48 |
EHD3 |
EH-domain containing 3 |
|
| A_70_P044621 |
2.27 |
−1.04 |
MALL |
Mal, T-cell differentiation protein-like |
|
||
| Metal ion transport |
3.39E-02 |
A_70_P030126 |
−2.36 |
1.24 |
CP* |
Ceruloplasmin (ferroxidase) |
[10,16] |
| CUST_12632_PI375351158 |
2.28 |
1.50 |
KCTD12 |
Potassium channel tetramerisation domain containing 12 |
|
||
| A_70_P027206 |
2.15 |
2.10 |
SLC30A1 |
Solute carrier family 30 (zinc transporter), member 1 |
[10,11] |
||
| A_70_P050456 |
−2.65 |
−1.58 |
SLC38A10 |
Solute carrier family 38, member 10 |
|
||
| Antimicrobial |
5.05E-02 |
CUST_139_PI396851701 |
15.26 |
−1.74 |
CATHL1 |
Cathelicidin 1 |
|
| CUST_3993_PI375351158 |
5.96 |
−1.25 |
S100A8 |
S100 calcium binding protein A8 |
[10] |
||
| Apoptosis |
5.45E-02 |
CUST_260_PI396851701 |
−2.04 |
−1.55 |
CARD6 |
Caspase recruitment domain family, member 6 |
|
| A_70_P022526 |
−2.29 |
−1.16 |
OPA1 |
Optic atrophy 1 (autosomal dominant) |
|
||
| A_70_P049526 |
−1.12 |
−2.70 |
PERP |
PERP, TP53 apoptosis effector |
|
||
| A_70_P069836 |
−3.14 |
−1.31 |
RNF130 |
Ring finger protein 130 |
|
||
| A_70_P067091 |
−2.90 |
−1.55 |
UBE2Z |
Ubiquitin-conjugating enzyme E2Z |
|
||
| Immunoglobulin-like fold |
5.55E-02 |
A_70_P036361 |
2.06 |
2.09 |
CD1E |
CD1e molecule |
|
| CUST_9656_PI375351158 |
−2.03 |
1.11 |
FGFR2 |
Fibroblast growth factor receptor 2 |
|
||
| CUST_12141_PI375351158 |
2.64 |
−1.00 |
IGK |
Ig kappa chain |
|
||
| A_70_P016986 |
−2.23 |
1.16 |
TRD@ |
T cell receptor delta locus |
[11] |
||
| Secreted |
5.70E-02 |
A_70_P048761 |
1.34 |
−2.27 |
SPINK5 |
Serine peptidase inhibitor, Kazal type 5 |
|
| CUST_9580_PI375351158 |
1.13 |
−2.49 |
MMRN1 |
Multimerin 1 |
|
||
| CUST_10101_PI375351158 |
−1.07 |
−2.53 |
NPNT |
Nephronectin |
|
||
| A_70_P034356 |
2.88 |
1.19 |
SEMA3G |
Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
|
||
| A_70_P063891 |
2.38 |
−1.22 |
PCOLCE2 |
Procollagen C-endopeptidase enhancer 2 |
|
||
| GTP binding |
7.49E-02 |
A_70_P019676 |
4.33 |
1.16 |
ACSM1 |
Acyl-CoA synthetase medium-chain family member 1 |
[10] |
| A_70_P046951 |
−2.24 |
−1.02 |
ARL4C |
ADP-ribosylation factor-like 4C |
|
||
| A_70_P008796 |
−2.06 |
1.15 |
DOCK11 |
Dedicator of cytokinesis 11 |
|
||
| A_70_P000126 |
−3.63 |
−1.82 |
GBP4 |
Guanylate binding protein 4 |
|
||
| Disulfide bond |
7.58E-02 |
A_70_P018376 |
2.56 |
−1.08 |
FAP |
Fibroblast activation protein, alpha |
|
| A_70_P041321 |
−2.15 |
−2.57 |
IGHA2 |
Immunoglobulin heavy constant alpha 2 (A2m marker) |
|
||
| Structural molecule activity |
8.04E-02 |
A_70_P028306 |
−2.03 |
−1.05 |
LMNB1 |
Lamin B1 |
|
| CUST_13373_PI375351158 |
−2.15 |
−1.29 |
LOC531679 |
Ribosomal protein 17-like; similar to 60S ribosomal Protein L17 (L23); similar to ribosomal protein L17; similar to Rpl17 protein; similar to mCG3798 |
|
||
| CUST_7057_PI375351158 |
−2.33 |
−1.07 |
LOC789587 |
Similar to Ribosomal_L22 domain containing protein RGD1359290 |
|
||
| A_70_P031441 |
−2.43 |
−1.79 |
VCL |
Vinculin |
[11] |
||
| Positive regulation of inflammatory response |
9.48E-02 |
A_70_P058551 |
8.25 |
−1.12 |
FABP4 |
Fatty acid binding protein 4, adipocyte |
|
| CUST_9779_PI375351158 |
−1.40 |
−2.38 |
IDO1 |
Indoleamine 2,3-dioxygenase 1 |
|
||
| Other | CUST_13467_PI375351158 |
1.29 |
−2.14 |
FBP2 |
Fructose-1,6-bisphosphatase 2 |
|
|
| A_70_P055306 |
2.77 |
1.50 |
SLC43A1 |
Solute carrier family 43, member 1 |
|
||
| A_70_P049406 |
−1.99 |
−2.32 |
EIF5 |
Eukaryotic translation initiation factor 5 |
[11] |
||
| A_70_P070931 |
−2.87 |
−1.18 |
NUDT19 |
Nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
|
||
| A_70_P036101 |
1.22 |
−3.18 |
NOP10 |
NOP10 ribonucleoprotein homolog (yeast) |
[11] |
||
| A_70_P011091 |
−2.74 |
−2.01 |
PFDN2* |
Prefoldin subunit 2 |
|
||
| A_70_P039656 |
17.20 |
−1.52 |
BAC5 |
5 kDa bactinecin precursor |
|
||
| A_70_P061341 |
−1.03 |
−2.94 |
C11ORF10 |
Chromosome 11 open reading frame 10 |
|
||
| A_70_P061266 |
2.36 |
1.03 |
C5ORF43 |
Chromosome 5 open reading frame 43 |
|
||
| A_70_P051377 |
14.99 |
−1.48 |
CATHL1B |
Procyclic dodecapeptide |
|
||
| A_70_P015101 |
−2.70 |
−1.03 |
CCDC82 |
Coiled-coil domain containing 82 |
|
||
| A_70_P012541 |
−1.02 |
−2.36 |
CNN1 |
Calponin 1, basic, smooth muscle |
|
||
| A_70_P002841 |
2.03 |
1.48 |
EPAS |
Endothelial PAS domain protein 1 |
|
||
| A_70_P008841 |
−2.18 |
−1.23 |
EPSTI1 |
Epithelial stromal interaction 1 (breast) |
|
||
| A_70_P060756 |
−2.12 |
1.08 |
FAM190B |
KIAA1128 |
|
||
| A_70_P062951 |
2.05 |
1.12 |
FNDC3B |
Fibronectin type III domain containing 3B |
[33] |
||
| A_70_P051181 |
2.76 |
1.28 |
HSD11K |
NAD-dependent 11-beta-hydroxysteroid dehydrogenase |
|
||
| CUST_46_PI396851701 |
−2.06 |
−2.15 |
IGHA |
Immunoglobulin heavy constant alpha |
|
||
| A_70_P024671 |
2.46 |
1.31 |
INHBB |
Inhibin, beta B |
|
||
| A_70_P033656 |
−2.34 |
−1.77 |
ISG12(A) |
ISG12(a) protein-like |
|
||
| A_70_P012537 |
−2.19 |
−3.58 |
LGALS15 |
Galectin 15 |
|
||
| CUST_3620_PI375351158 |
−1.58 |
−2.22 |
LGALS16 |
Beta-galactoside-binding lectin |
|
||
| A_70_P020131 |
−1.10 |
−3.84 |
LGALS9C |
Lectin, galactoside-binding, soluble, 9C |
|
||
| CUST_6089_PI375351158 |
−2.02 |
−1.71 |
LOC100140018 |
Similar to cytochrome P450, family 2, subfamily J |
|
||
| A_70_P016801 |
−2.20 |
−1.48 |
LOC443321 |
Lysozyme 2a precursor |
|
||
| A_70_P026671 |
−1.91 |
−2.05 |
LOC524810 |
IgM |
|
||
| A_70_P023026 |
3.09 |
1.74 |
LOC615697 |
Similar to cytochrome P450 |
|
||
| A_70_P039266 |
−2.03 |
−1.83 |
LOC654331 |
Pancreatitis-associated protein I |
[10] |
||
| A_70_P039271 |
−3.94 |
−7.40 |
MCP-4 |
Mast cell proteinase-4 |
|
||
| CUST_11558_PI375351158 |
2.00 |
2.23 |
MGC140754 |
Hypothetical LOC508613 |
|
||
| CUST_9725_PI375351158 |
−2.26 |
−3.77 |
PLAC8 |
Placenta-specific 8 |
|
||
| CUST_10114_PI375351158 |
2.56 |
1.15 |
RN18S1 |
18S ribosomal RNA |
|
||
| A_70_P030132 |
−2.40 |
−1.71 |
RTP4 |
Receptor (chemosensory) transporter protein 4 |
[34] |
||
| A_70_P030426 | 10.95 | 1.39 | SC5 | Cathelin-related prepropeptide |
*Genes selected for validation by quantitative RT-PCR.
BLAST results for clones exhibiting significant alterations in gene expression (P ≤ 0.05 and ≥ 2-fold change) in preclinical and clinical stages of natural scrapie. The GO was determined using DAVID Bioinformatics Resources 6.7 (NIAID/NIH, USA), an on-line functional annotation tool. Only genes with a known GO are shown. (C, Clinical; FC, Fold change; NC, Negative control; PC, Preclinical). Previous studies describing altered expression of the differentially expressed genes reported here in the brain and lymphoid tissues of experimentally-infected sheep are cited.
Table 3.
Signaling pathways associated with differentially expressed genes
| Term | P value | ProbeName | FC (C vs NC) | FC (PC vs NC) | Gene symbol | Gene name |
|---|---|---|---|---|---|---|
| PPAR signaling pathway |
1.88E-02 |
CUST_598_PI375351158 |
2.28 |
1.33 |
FABP5 |
Fatty acid binding protein 5 (psoriasis-associated) |
| CUST_8730_PI375351158 |
1.06 |
−2.05 |
ACSL4 |
Acyl-CoA synthetase long-chain family member 4 |
||
| A_70_P058551 |
8.25 |
−1.12 |
FABP4 |
Fatty acid binding protein 4, adipocyte |
||
| A_70_P039986 |
−1.27 |
2.39 |
SLC27A2 |
Solute carrier family 27 (fatty acid transporter), member 2 |
||
| ECM-receptor interaction |
2.53E-02 |
A_70_P021711 |
−2.19 |
−1.69 |
COL1A1 |
Collagen, type I, alpha 1 |
| A_70_P058626 |
2.27 |
1.13 |
VWF |
von Willebrand factor |
||
| A_70_P016337 |
−1.78 |
−2.72 |
HMMR |
Hyaluronan-mediated motility receptor (RHAMM) |
||
| Focal adhesion | 6.73E-02 | A_70_P021711 |
−2.19 |
−1.69 |
COL1A1 |
Collagen, type I, alpha 1 |
| A_70_P058626 |
2.27 |
1.13 |
VWF |
von Willebrand factor |
||
| A_70_P050922 |
2.19 |
−1.06 |
VEGFA |
Vascular endothelial growth factor A |
||
| A_70_P031441 | −2.43 | −1.79 | VCL | Vinculin |
Signaling pathways with which differentially expressed genes (P ≤ 0.05 and ≥ 2-fold change) are associated were identified by enrichment analysis using the on-line functional annotation (DAVID Bioinformatics Resources 6.7; NIAID/NIH, USA). (C, Clinical; FC, Fold change; NC, Negative control; PC, Preclinical).
Figure 2.
Condition trees generated by cluster analysis. A hierarchical cluster analysis (Euclidean distance clustering algorithm) was performed using PermutMatrix [21], which identified 139 genes whose expression differed significantly (≥ 2 fold change with respect to controls) in at least at one of the 2 disease stages analyzed (preclinical and clinical). Colors represent the level of expression. Sample information is listed across the top. The names of the known genes are indicated. Note the distinct patterns of gene expression in control, preclinical and clinical animals.
The relatively low number of animals analyzed and the fact that they were obtained from different geographical locations may limit the statistical power of our study. However, it is difficult to perform this kind of study with a larger number of animals due to the paucity of natural scrapie cases. Moreover, finding negative age-matched controls with the same PRNP genotype within the same scrapie-affected flocks is not possible since the disease attack-rate in these animals is close to 100%.
Validation of gene expression profiling by quantitative RT-PCR
To confirm the microarray results, the expression of 7 genes (BBOX1, CP, PFDN2, PSMA7, SERPINE1, UCHL1 and VEGFA) was analyzed by qRT-PCR. The expression of 4 of the selected targets (BBOX1, PFDN2, PSMA7 and UCHL1) was altered in the same direction in both the clinical and preclinical stages of scrapie disease. Expression levels of the other 3 genes (CP, SERPINE1 and VEGFA) were altered in opposite directions in the clinical and preclinical stages, respectively. In all cases, qRT-PCR confirmed the changes observed in the microarray hybridizations (Figure 3), revealing statistically significant differences between control and scrapie groups.
Figure 3.
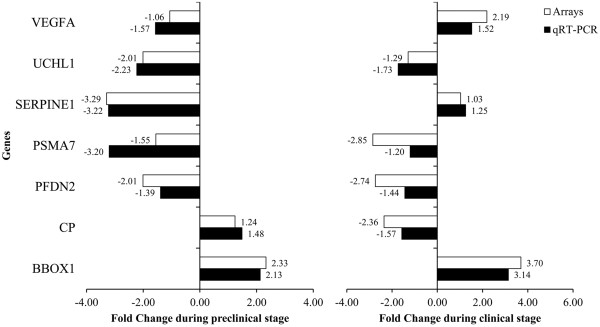
Confirmation of microarray results by quantitative real-time PCR. Differential expression of selected genes analyzed by microarray and quantitative RT-PCR in mesenteric lymph node samples from preclinical and clinical scrapie sheep: gamma-butyrobetaine hydroxylase (BBOX1), ceruloplasmin (CP), prefoldin subunit 2 (PFDN2), proteasome subunit alpha type-7 (PSMA7), plasminogen activator inhibitor-1 (SERPINE1), ubiquitin carboxy-terminal hydrolase L1 (UCHL1), and vascular endothelial growth factor (VEGFA).
Identification of prion deposition-related genes
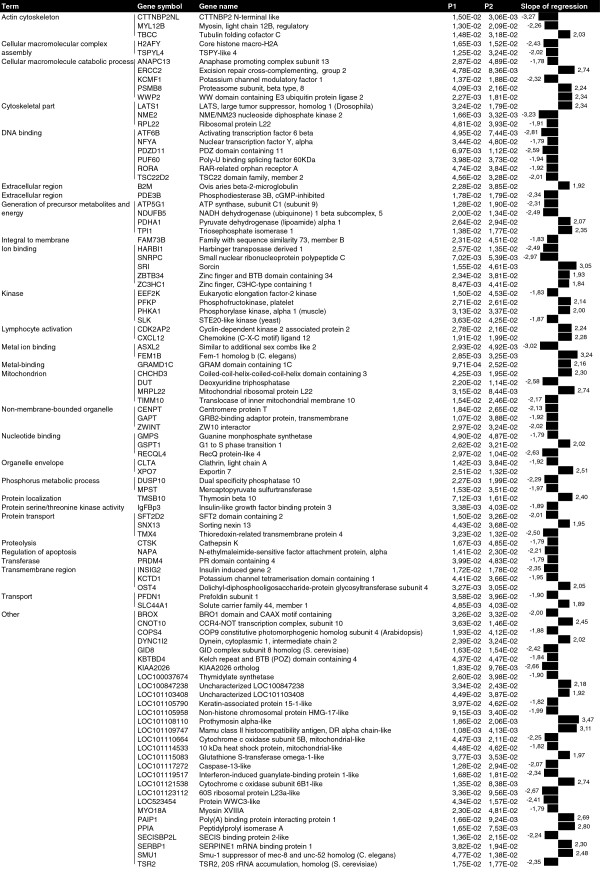
Two regression models were used to identify genes associated with prion deposition. One is a regression model that relates gene expression with prion deposition scores regardless of group (Preclinical, Clinical or Healthy), while the other includes a systematic effect associated with the 3 different animal categories and analyzes the remaining variation after subtracting the variability associated with each of the 3 groups. Natural conditions normally display high variability within groups. Using these regression analyses we identified genes whose expression was associated with prion deposition, even if the changes in expression were not significant. We identified 96 genes with known functions whose expression was significantly associated with PrPSc deposition in both regression models (Figure 4). The slope value represented was obtained using the second regression model. The gene ontology analysis revealed that genes associated with prion deposition were involved in DNA binding, the generation of precursor metabolites and energy, ion binding, catabolic process and kinase functions. Only statistically significant slopes (p < 0.05) for both association models are presented in Figure 4.
Figure 4.
Relationship between gene expression profiles and PrPSc deposition. Figure shows genes whose expression was statistically associated with PrPSc deposition. P1 is the probability calculated using the simple regression model yi = μ + bpi + ei, and P2 is the probability calculated using the second regression model yij = μ + Ti + bpij + eij (which includes a systematic effect associated with the 3 categories; Preclinical, Clinical and Healthy). The slope of regression describing the relationship between histopathological lesions and gene expression was calculated using the second model. Analyses were performed using R software (R project for Statistical Computing).
Discussion
Mesenteric lymph nodes are one of the first organs in which PrPSc accumulates [5]. To the best of our knowledge, ours is the first study to use gene expression microarray technology to analyze mesenteric lymph nodes from naturally-infected sheep during the preclinical and clinical stages of scrapie.
Our microarray hybridization analysis identified 139 annotated genes of known functions that exhibited changes in expression of 2-fold or more with respect to controls during at least one of the 2 disease stages analyzed. Of these 139 genes, 105 were differentially regulated in clinical scrapie-infected sheep and 50 in preclinical scrapie-infected sheep. These findings indicate that in the early disease stages, fewer genes are activated or the changes in expression are not sufficiently large to be detected by microarray. PrPSc accumulation in MLN peaked during the preclinical phase of scrapie (Figure 1), indicating that dysregulated genes that were not common to both disease stages could not be specifically linked to the prion accumulation process.
Prion infection of neural cell lines has revealed a limited transcriptional response and clear differences in transcriptional profiles between different cell lines and between cell cultures and whole brains from infected mice [35-37]. Similarly, our results differ from the profiles described in the brains of sheep with natural scrapie [10,11], indicating the absence of universal changes associated with prion infection.
The differentially expressed genes identified in the present study encode proteins located in almost all subcellular locations: membranes (glycoproteins), the extracellular matrix, the cytoplasm and chromosomes. These proteins participate in general cell processes such as the cell cycle, apoptosis, alternative splicing and chromosome condensation. Genes associated with glycoprotein metabolism, the extracellular matrix and the cell cycle have been previously detected in leukocyte-depleted splenic cells from mice infected with scrapie [16]. We found that gene encoding decorin (DCN), a protein located in the extracellular matrix, was overexpressed in scrapie animals, in line with previous reports of DCN upregulation at both the gene and protein level in cells isolated from scrapie-infected mouse spleen [16]. Our results confirm the differential regulation of DCN in the MLN of sheep with natural scrapie. Some of differentially transcribed genes that we identified encode proteins that may be involved in the pathogenesis of protein misfolding diseases. Examples include proteins involved in disulfide bonding, GTP binding and protein dimerization. Differential expression of some of these genes has been reported in other tissues in scrapie-infected mice [12,14,38] and in natural scrapie [10,11].
The GO enrichment analysis of our microarray data identified 3 signaling pathways that may be involved in the pathogenesis of scrapie; extracellular matrix (ECM)-receptor interaction, focal adhesion and the peroxisome proliferator-activated receptor (PPAR) signaling pathway. We and others have previously reported altered expression of COL1A2, COL3A1 and COL12A1 and other ECM related genes in the CNS of sheep infected with scrapie and other prion diseases [10,11,39,40] and in leukocyte-depleted splenic cells from scrapie-infected mice [16]. Moreover, prion protein regulates β1 integrin signaling activity in prion-infected neurons [41] and the expression of genes of the PPAR signaling pathway is altered in Alzheimer’s disease (AD) [42,43]. Our findings indicate alterations in the extracellular matrix, focal adhesion and in fatty acid metabolism via the PPAR signaling pathway in non-neural tissues susceptible to prion infection.
In addition to analyzing differential gene expression, we investigated the effect of prion deposition on LRS gene expression in order to further our understanding of the prion-specific pathogenic pathways in scrapie. Two regression models (see Methods section) were used to identify possible associations between gene expression data and the PrPSc deposition score, as determined by immunohistochemistry. These statistical association approaches can identify genes that may be implicated in prion-specific processes, although the differences in their expression levels between control and infected animals were not significant. Except for CLTA, none of the genes identified in the association analysis exhibited significant differences in expression in the array data. The gene ontology analysis revealed that genes associated with prion deposition are involved in DNA binding, the generation of precursor metabolites and energy, ion binding, catabolic processes and kinase functions. Among the genes that were closely associated with prion protein deposition were cathepsin K (CTSK) and clathrin, light chain A (CLTA). Several members of the cathepsin family (including cathepsins C, D, H, S and Z) are differentially expressed in prion-affected brain tissues [12,14,38,44,45]. We observed a negative association between CTSK expression and prion deposition, suggesting a possible proteolytic effect of cathepsin K on prion protein. Furthermore, CLTA expression was negatively associated with prion deposition. The expression of this gene is also altered in many neurodegenerative diseases such as Alzheimer’s and Huntington’s diseases [46,47], and clathrin-mediated pathways are intimately involved in amyloid formation in neurodegenerative diseases [48]. These observations suggest that CLTA may also be involved in prion amyloid formation.
Seven of the differentially expressed genes detected were selected for further analysis by qRT-PCR. These genes were chosen based on their involvement in protein misfolding pathways (PFDN2, UCHL1 and PSMA7), the regulation of angiogenesis (VEGFA and SERPINE1), iron metabolism and neurotoxicity (CP) and antioxidant mechanisms (BBOX1). Comparable fold changes in the expression of all 7 genes, in both preclinical and clinical disease stages, were observed in the microarray and qRT-PCR analyses, confirming the validity of both the array data and the alignment analysis.
Here, we will briefly discuss how these genes may be involved in PrPSc formation, dissemination and toxicity and in the response of the organism to prion infection. Genes encoding proteins involved in molecular pathways that regulate protein misfolding (PSMA7, UCHL1 and PFDN2) were downregulated in scrapie-infected animals. Furthermore, our association study revealed a positive association between proteasome subunit, beta type, 8 (PSMB8) and PrPSc deposition. The proteins encoded by PSMA7, UCHL1 and PSMB8 form part of the ubiquitin proteasome system (UPS), a complex that regulates the degradation of incomplete, damaged or misfolded proteins [49,50]. The UCHL1 protein has been implicated in protein misfolding diseases. UCHL1 is abundantly expressed in nervous system tissues [51], and gene variants and changes in the activity of UCHL1 have been associated with neurodegenerative disorders such as Alzheimer’s, Parkinson’s and Huntington’s diseases [27,52-55]. In prion diseases, the functional capacity of the UPS is impaired by the direct interaction of β-sheet-rich PrP with the 20S core particle of the proteasome complex, inhibiting substrate entry [56]. UPS impairment may enable the conversion of cytosolic PrPC to an abnormal, “PrPSc-like” form [57]. Increased levels of ubiquitin conjugates are found in scrapie-infected mouse brains, and are correlated with decreased proteasome function [58]. The downregulation of PSMA7 and UCHL1 genes and the positive association of PrPSc deposition with PSMB8 expression in both preclinical and clinical stages of natural scrapie in the LRS of sheep is in line with the impaired proteasome function described in prion diseases of the nervous system [28]. Moreover, prefoldin (PFD) is likely to bind to substrate proteins that exist in an unfolded state and to transfer these proteins to the cytosolic chaperonin-containing TCP-1 (CCT) for functional folding [59]. The role of this protein in protein misfolding diseases has been investigated and prefoldin subunit 2 (PFDN2) has been implicated in AD [60]. Our findings reveal downregulation of PFDN2 expression in both preclinical and clinical stages of natural scrapie in the LRS of sheep. The observed downregulation of PSMA7 and UCHL1 may facilitate prion conversion, implicating these genes in the pathogenic mechanism of the scrapie.
The expression of SERPINE 1 or plasminogen activator inhibitor-1 (PAI-1) was downregulated in preclinical scrapie animals and upregulated in the clinical phase of the disease, suggesting a dual role for this gene in scrapie. SERPINE1 is an inhibitor of 2 types of plasminogen activators; tissue-type plasminogen (tPA) and urokinase-type plasminogen (uPA) activators. Plasminogen was recently show to stimulate PrP misfolding by accelerating the conversion of PrPC to PrPSc[26]. The downregulation of SERPINE1 in preclinical scrapie may inhibit PrPSc formation in the early stages of the disease. SERPINE1 is also involved in angiogenesis, as evidenced by the defective angiogenesis seen in tumor-bearing SERPINE1-deficient mice [61]. Accordingly, decreased expression of SERPINE1 may represent a protective response to PrPSc propagation during the preclinical stage of natural scrapie. However, we observed a slight increase in SERPINE1 expression in clinical scrapie-infected sheep as compared with controls (FC = 1.25). The expression of vascular endothelial growth factor (VEGF, also known as VEGFA), which is also involved in angiogenesis, was altered in scrapie. This gene mediates a critical rate-limiting step in physiological angiogenesis and plays a key role in tumor progression and angiogenesis and its expression by primary tumor promotes lymphangiogenesis and facilitates metastasis to sentinel lymph nodes [62]. As observed for SERPINE1, we detected a slight decrease in VEGFA expression in lymphatic tissues in the preclinical stage of natural scrapie, followed by a moderate increase during the clinical stage of the disease. Lymphatic vessels are one of the proposed routes of prion dissemination [63]. Accordingly, the differential regulation of SERPINE1 and VEGFA observed in scrapie may reflect a role in prion propagation. However, further studies will be necessary to clarify the role of these proteins in prion dissemination.
In addition to genes involved in prion misfolding and propagation, we analyzed the expression of the ceruloplasmin (CP) gene, which is involved in neurotoxicity. CP is a ferroxidase that contains 95% of the copper present in blood plasma, and is thus a key mediator of copper transport and metabolism [64]. Copper plays an important role in prion integrity; in vitro, copper enhances renaturation and stabilization of PrPSc, restoring protease resistance and infectivity [64]. Increased CP expression has been reported in leukocyte-depleted splenic cells from clinical scrapie-infected mice [16]. However, we found that CP expression was significantly downregulated in the lymph nodes of scrapie-infected animals in the clinical stage of the disease. In patients with aceruloplasminemia, the predominant clinical symptoms are neurologic and include dystonia, dysarthria, and subcortical dementia due to progressive degeneration of the basal ganglia, as well as selective neuronal loss and increased iron content in the microglia, neurons and basal ganglia [29]. Ceruloplasmin also plays an essential role in normal brain iron metabolism and protects against neuronal loss [65]. Our findings suggest that CP deficiency may contribute to the neuronal degeneration observed in scrapie, although further analyses of brain expression and blood levels of CP will be required to determine its role in scrapie.
We detected upregulation of gamma-butyrobetaine hydroxylase (BBOX1), a gene involved in the response of the organism to oxidative stress. BBOX1 is the enzyme responsible for the biosynthesis of L-carnitine, a molecule involved in fatty acid metabolism with important anti-free radical and antioxidant activities [66]. The acetylated form of L-carnitine (acetyl-L-carnitine) has been proposed as a potential treatment for neurodegenerative diseases, including AD [30]. Our results demonstrate upregulation of BBOX1 in both clinical and preclinical stages of scrapie in the LRS, perhaps reflecting an increase in the levels of antioxidant molecules (L-carnitine and acetyl-L-carnitine) in response to the damage induced by the prion.
The known functions of the genes that were up- and downregulated in scrapie suggest potential roles in the pathogenesis of prion diseases. However, we cannot rule out the possibility that these changes in expression are a secondary response of the organism to the disease. The findings of this transcriptomic analysis thus need to be complemented by further studies analyzing the cellular localization, protein levels and function of the genes of interest in order to establish their specific roles in prion diseases.
Conclusions
Our genome-wide expression analysis of the LRS of sheep with natural scrapie, at both preclinical and clinical stages of the disease, has identified new genes that may be involved in the pathogenesis of scrapie. We confirmed the differential expression of 7 genes involved in prion or other neurodegenerative diseases. The downregulation of genes involved in repairing misfolded proteins (PFDN2, PSMA7 and UCHL1) may contribute to prion formation. Furthermore, the altered expression of genes that promote angiogenesis in the lymph nodes (SERPINE1 and VEGFA) may facilitate prion dissemination, while the observed decrease in CP expression may contribute to prion toxicity. Our results confirm the altered expression of BBOX1, a gene implicated in the systemic response of the organism to prion-induced oxidative stress, in scrapie infected animals. Although further studies are necessary to determine the specific roles of these genes in the pathogenesis of prion diseases, our findings represent an important first step towards identifying candidate genes that may serve as useful biomarkers for the development of early diagnostic tools and therapeutic strategies for scrapie.
Availability of supporting data
Microarray data are available in the ArrayExpress database: accession number E-MTAB-1346 (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-1346/).
Abbreviations
ARQ: Alanine arginine glutamine; BBOX1: Gamma-butyrobetaine hydroxylase; BSE: Bovine spongiform encephalopathy; CNS: Central nervous system; CP: Ceruloplasmin; DAB: 3,3′-diaminobenzidine; FDCs: Follicular dendritic cells; G6PDH: Glucose-6-phosphate dehydrogenase; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; GO: Gene ontology; IHC: Immunohistochemistry; LRS: Lymphoreticular system; MLN: Mesenteric lymph node; PFDN2: Prefoldin subunit 2; PRNP: Prion protein; PrPC: Cellular prion protein; PrPSc: Scrapie prion protein; PSMA7: Proteasome subunit alpha type-7; RIN: RNA integrity number; RPL32: Ribosomal protein l32; SERPINE1: Plasminogen activator inhibitor-1; TBMs: Tingible body macrophages; TSEs: Transmissible spongiform encephalopathies; UCHL1: Carboxy-terminal hydrolase L1; VEGFA: Vascular endothelial growth factor; VRQ: Valine arginine glutamine.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HF performed the experiments and drafted the manuscript. IMB helped design the study, participated in the molecular genetic studies and the sequence alignment study, and drafted the manuscript. FH participated in the microarray analysis. LV performed the association analysis. CH participated in the pathological characterization of the animals. DM performed the qRT-PCR experiments. MM helped design the study and drafted the manuscript. AB helped design the microarray and sequence alignment studies. JJB helped design and coordinate the study and to draft the manuscript. RB conceived the study, participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final version of the manuscript.
Contributor Information
Hicham Filali, Email: hicham@unizar.es.
Inmaculada Martín-Burriel, Email: minma@unizar.es.
Frank Harders, Email: frank.harders@wur.nl.
Luis Varona, Email: lvarona@unizar.es.
Carlos Hedman, Email: hedman@unizar.es.
Diego R Mediano, Email: mediano_87@hotmail.com.
Marta Monzón, Email: mmonzon@unizar.es.
Alex Bossers, Email: alex.bossers@wur.nl.
Juan J Badiola, Email: badiola@unizar.es.
Rosa Bolea, Email: rbolea@unizar.es.
Acknowledgements
The authors wish to acknowledge the excellent technical support of Belén Marín, Silvia Ruiz, Nuria Segovia, Silvia Castejón, and Sonia Gómez. This work was performed as part of the AGL2008-0256 project, financed by MICINN-FEDER. Hicham Filali was supported by doctoral grants from the Ministerio de Asuntos Exteriores y de Cooperación (MAEC/AECID). The arrays and sequenced libraries were made available from projects at CVI supported by grants from the Dutch Ministry of Agriculture, Nature Management, and Fisheries (LNV).
References
- Aguzzi A, Heikenwalder M. Pathogenesis of prion diseases: current status and future outlook. Nat Rev Microbiol. 2006;15:765–775. doi: 10.1038/nrmicro1492. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;15:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. Scrapie and experimental BSE in sheep. B Med Bull. 2003;15:171–183. doi: 10.1093/bmb/66.1.171. [DOI] [PubMed] [Google Scholar]
- Jeffrey M, Gonzalez L. Classical sheep transmissible spongiform encephalopathies: pathogenesis, pathological phenotypes and clinical disease. Neuropathol App Neurobiol. 2007;15:373–394. doi: 10.1111/j.1365-2990.2007.00868.x. [DOI] [PubMed] [Google Scholar]
- Andreoletti O, Berthon P, Marc D, Sarradin P, Grosclaude J, Van Keulen L, Schelcher F, Elsen J, Lantier F. Early accumulation of PrPsc in gut-associated lymphoid and nervous tissues of susceptible sheep from a Romanov flock with natural scrapie. J Gen Virol. 2000;15:3115–3126. doi: 10.1099/0022-1317-81-12-3115. [DOI] [PubMed] [Google Scholar]
- Van Keulen LJ, Vromans ME, Van Zijderveld FG. Early and late pathogenesis of natural scrapie infection in sheep. APMIS. 2002;15:23–32. doi: 10.1034/j.1600-0463.2002.100104.x. [DOI] [PubMed] [Google Scholar]
- Van Keulen LJ, Bossers A, Van Zijderveld F. TSE pathogenesis in cattle and sheep. Vet Res. 2008;15:24. doi: 10.1051/vetres:2007061. [DOI] [PubMed] [Google Scholar]
- Brown KL, Stewart K, Ritchie DL, Mabbott NA, Williams A, Fraser H, Morrison WI, Bruce ME. Scrapie replication in lymphoid tissues depends on prion protein-expressing follicular dendritic cells. Nat Med. 1999;15:1308–1312. doi: 10.1038/15264. [DOI] [PubMed] [Google Scholar]
- McGovern G, Jeffrey M. Scrapie-specific pathology of sheep lymphoid tissues. PloS One. 2007;15:e1304. doi: 10.1371/journal.pone.0001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filali H, Martin-Burriel I, Harders F, Varona L, Lyahyai J, Zaragoza P, Pumarola M, Badiola JJ, Bossers A, Bolea R. Gene expression profiling and association with prion-related lesions in the medulla oblongata of symptomatic natural scrapie animals. PloS One. 2011;15:e19909. doi: 10.1371/journal.pone.0019909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filali H, Martin-Burriel I, Harders F, Varona L, Serrano C, Acin C, Badiola JJ, Bossers A, Bolea R. Medulla oblongata transcriptome changes during presymptomatic natural scrapie and their association with prion-related lesions. BMC Genomics. 2012;15:399. doi: 10.1186/1471-2164-13-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner PJ, Abbassi H, Chesebro B, Race RE, Reilly C, Haase AT. Gene expression alterations in brains of mice infected with three strains of scrapie. BMC Genomics. 2006;15:114. doi: 10.1186/1471-2164-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AR, Webb J, Rebus S, Williams A, Fazakerley JK. Identification of up-regulated genes by array analysis in scrapie-infected mouse brains. Neuropathol App Neurobiol. 2004;15:555–567. doi: 10.1111/j.1365-2990.2004.00565.x. [DOI] [PubMed] [Google Scholar]
- Xiang W, Windl O, Wunsch G, Dugas M, Kohlmann A, Dierkes N, Westner IM, Kretzschmar HA. Identification of differentially expressed genes in scrapie-infected mouse brains by using global gene expression technology. J Virol. 2004;15:11051–11060. doi: 10.1128/JVI.78.20.11051-11060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillerme-Bosselut F, Forestier L, Jayat-Vignoles C, Vilotte JL, Popa I, Portoukalian J, Le Dur A, Laude H, Julien R, Gallet PF. Glycosylation-related gene expression profiling in the brain and spleen of scrapie-affected mouse. Glycobiology. 2009;15:879–889. doi: 10.1093/glycob/cwp062. [DOI] [PubMed] [Google Scholar]
- Huzarewich RL, Medina S, Robertson C, Parchaliuk D, Booth SA. Transcriptional modulation in a leukocyte-depleted splenic cell population during prion disease. J Toxicol Environ Health Part A. 2011;15:1504–1520. doi: 10.1080/15287394.2011.618979. [DOI] [PubMed] [Google Scholar]
- Skretting G, Espenes A, Ulvund MJ, Olsaker I. cDNA representational difference analysis of ileal Peyer’s patches in lambs after oral inoculation with scrapie. Biochem Biophys Res Commun. 2004;15:272–279. doi: 10.1016/j.bbrc.2004.02.040. [DOI] [PubMed] [Google Scholar]
- Vargas F, Lujan L, Bolea R, Monleon E, Martin-Burriel I, Fernandez A, De Blas I, Badiola JJ. Detection and clinical evolution of scrapie in sheep by 3rd eyelid biopsy. J Vet Intern Med. 2006;15:187–193. doi: 10.1111/j.1939-1676.2006.tb02840.x. [DOI] [PubMed] [Google Scholar]
- Monleon E, Garza MC, Sarasa R, Alvarez-Rodriguez J, Bolea R, Monzon M, Vargas MA, Badiola JJ, Acin C. An assessment of the efficiency of PrPsc detection in rectal mucosa and third-eyelid biopsies from animals infected with scrapie. Vet Microbiol. 2011;15:237–243. doi: 10.1016/j.vetmic.2010.06.028. [DOI] [PubMed] [Google Scholar]
- Monleon E, Monzon M, Hortells P, Vargas A, Acin C, Badiola JJ. Detection of PrPsc on lymphoid tissues from naturally affected scrapie animals: comparison of three visualization systems. J Histochem Cytochem. 2004;15:145–151. doi: 10.1177/002215540405200201. [DOI] [PubMed] [Google Scholar]
- Caraux G, Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics. 2005;15:1280–1281. doi: 10.1093/bioinformatics/bti141. [DOI] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;15:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;15:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Del Bo R, Ghezzi S, Scarpini E, Bresolin N, Comi GP. VEGF genetic variability is associated with increased risk of developing Alzheimer’s disease. J Neurol Sci. 2009;15:66–68. doi: 10.1016/j.jns.2009.02.318. [DOI] [PubMed] [Google Scholar]
- Fang Y, Zhang L, Zeng Z, Lian Y, Jia Y, Zhu H, Xu Y. Promoter polymorphisms of SERPINE1 are associated with the antidepressant response to depression in Alzheimer’s disease. Neurosci Lett. 2012;15:217–220. doi: 10.1016/j.neulet.2012.03.090. [DOI] [PubMed] [Google Scholar]
- Mays CE, Ryou C. Plasminogen stimulates propagation of protease-resistant prion protein in vitro. FASEB J. 2010;15:5102–5112. doi: 10.1096/fj.10-163600. [DOI] [PubMed] [Google Scholar]
- Saigoh K, Wang YL, Suh JG, Yamanishi T, Sakai Y, Kiyosawa H, Harada T, Ichihara N, Wakana S, Kikuchi T, Wada K. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat Genet. 1999;15:47–51. doi: 10.1038/12647. [DOI] [PubMed] [Google Scholar]
- Deriziotis P, Tabrizi SJ. Prions and the proteasome. Biochim Biophys Acta. 2008;15:713–722. doi: 10.1016/j.bbadis.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Morita H, Ikeda S, Yamamoto K, Morita S, Yoshida K, Nomoto S, Kato M, Yanagisawa N. Hereditary ceruloplasmin deficiency with hemosiderosis: a clinicopathological study of a Japanese family. Ann Neurol. 1995;15:646–656. doi: 10.1002/ana.410370515. [DOI] [PubMed] [Google Scholar]
- Aliev G, Li Y, Palacios HH, Obrenovich ME. Oxidative stress induced mitochondrial DNA deletion as a hallmark for the drug development in the context of the cerebrovascular diseases. Recent Pat Cardiovasc Drug Discov. 2011;15:222–241. doi: 10.2174/157489011797376942. [DOI] [PubMed] [Google Scholar]
- Lyahyai J, Serrano C, Ranera B, Badiola JJ, Zaragoza P, Martin-Burriel I. Effect of scrapie on the stability of housekeeping genes. Anim Biotechnol. 2010;15:1–13. doi: 10.1080/10495390903323851. [DOI] [PubMed] [Google Scholar]
- Garcia-Crespo D, Juste RA, Hurtado A. Selection of ovine housekeeping genes for normalisation by real-time RT-PCR; analysis of PrP gene expression and genetic susceptibility to scrapie. BMC Vet Res. 2005;15:3. doi: 10.1186/1746-6148-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossner A, Roupaka S, Foster J, Hunter N, Hopkins J. Transcriptional profiling of peripheral lymphoid tissue reveals genes and networks linked to SSBP/1 scrapie pathology in sheep. Vet Microbiol. 2011;15:218–228. doi: 10.1016/j.vetmic.2011.05.036. [DOI] [PubMed] [Google Scholar]
- Kim HO, Snyder GP, Blazey TM, Race RE, Chesebro B, Skinner PJ. Prion disease induced alterations in gene expression in spleen and brain prior to clinical symptoms. Adv Appl Bioinformatics Chem: AABC. 2008;15:29–50. doi: 10.2147/aabc.s3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius C, Hutter G, Wagner U, Seeger H, Kana V, Kranich J, Klohn PC, Weissmann C, Miele G, Aguzzi A. Transcriptional stability of cultured cells upon prion infection (vol 375, pg 1222, 2008) J Mol Biol. 2009;15:207–207. doi: 10.1016/j.jmb.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Stanton JB, Knowles DP, Call DR, Mathison BA, Baszler TV. Limited transcriptional response of ovine microglia to prion accumulation. Biochem Biophys Res Commun. 2009;15:345–350. doi: 10.1016/j.bbrc.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano C, Campana V, Griffiths B, Kelly G, Schiavo G, Zurzolo C. Gene expression profile of quinacrine-cured prion-infected mouse neuronal cells. J Neurochem. 2008;15:239–250. doi: 10.1111/j.1471-4159.2007.05140.x. [DOI] [PubMed] [Google Scholar]
- Booth S, Bowman C, Baumgartner R, Sorensen G, Robertson C, Coulthart M, Phillipson C, Somorjai RL. Identification of central nervous system genes involved in the host response to the scrapie agent during preclinical and clinical infection. J Gen Virol. 2004;15:3459–3471. doi: 10.1099/vir.0.80110-0. [DOI] [PubMed] [Google Scholar]
- Vidal E, Acin C, Foradada L, Monzon M, Marquez M, Monleon E, Pumarola M, Badiola JJ, Bolea R. Immunohistochemical characterisation of classical scrapie neuropathology in sheep. J Comp Pathol. 2009;15:135–146. doi: 10.1016/j.jcpa.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Costa C, Tortosa R, Domenech A, Vidal E, Pumarola M, Bassols A. Mapping of aggrecan, hyaluronic acid, heparan sulphate proteoglycans and aquaporin 4 in the central nervous system of the mouse. J Chem Neuroanat. 2007;15:111–123. doi: 10.1016/j.jchemneu.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Loubet D, Dakowski C, Pietri M, Pradines E, Bernard S, Callebert J, Ardila-Osorio H, Mouillet-Richard S, Launay JM, Kellermann O, Schneider B. Neuritogenesis: the prion protein controls beta1 integrin signaling activity. FASEB J. 2012;15:678–690. doi: 10.1096/fj.11-185579. [DOI] [PubMed] [Google Scholar]
- Kolsch H, Lehmann DJ, Ibrahim-Verbaas CA, Combarros O, Van Duijn CM, Hammond N, Belbin O, Cortina-Borja M, Lehmann MG, Aulchenko YS. et al. Interaction of insulin and PPAR-alpha genes in Alzheimer’s disease: the Epistasis Project. J Neural Transm. 2012;15:473–479. doi: 10.1007/s00702-011-0732-4. [DOI] [PubMed] [Google Scholar]
- Mandrekar-Colucci S, Karlo JC, Landreth GE. Mechanisms underlying the rapid peroxisome proliferator-activated receptor-gamma-mediated amyloid clearance and reversal of cognitive deficits in a murine model of Alzheimer’s disease. J Neurosci. 2012;15:10117–10128. doi: 10.1523/JNEUROSCI.5268-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandoy-Dron F, Guillo F, Benboudjema L, Deslys JP, Lasmezas C, Dormont D, Tovey MG, Dron M. Gene expression in scrapie: cloning of a new scrapie-responsive gene and the identification of increased levels of seven other mRNA transcripts. J Biol Chem. 1998;15:7691–7697. doi: 10.1074/jbc.273.13.7691. [DOI] [PubMed] [Google Scholar]
- Baker CA, Manuelidis L. Unique inflammatory RNA profiles of microglia in Creutzfeldt-Jakob disease. Proc Natl Acad Sci USA. 2003;15:675–679. doi: 10.1073/pnas.0237313100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Takeda M, Yoshimi K, Hattori H, Hariguchi S, Kitajima S, Hashimoto S, Nishimura T. Involvement of clathrin light chains in the pathology of Alzheimer’s disease. Acta Neuropathol. 1994;15:23–31. doi: 10.1007/BF00386251. [DOI] [PubMed] [Google Scholar]
- Legendre-Guillemin V, Metzler M, Lemaire JF, Philie J, Gan L, Hayden MR, McPherson PS. Huntingtin interacting protein 1 (HIP1) regulates clathrin assembly through direct binding to the regulatory region of the clathrin light chain. J Biol Chem. 2005;15:6101–6108. doi: 10.1074/jbc.M408430200. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Amouyel P. Genetics of Alzheimer’s disease: new evidences for an old hypothesis? Curr Opin Genet Dev. 2011;15:295–301. doi: 10.1016/j.gde.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Cook C, Petrucelli L. A critical evaluation of the ubiquitin-proteasome system in Parkinson’s disease. Biochim Biophys Acta. 2009;15:664–675. doi: 10.1016/j.bbadis.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;15:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Wilkinson KD, Lee KM, Deshpande S, Duerksen-Hughes P, Boss JM, Pohl J. The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science. 1989;15:670–673. doi: 10.1126/science.2530630. [DOI] [PubMed] [Google Scholar]
- Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem. 2004;15:13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- Naze P, Vuillaume I, Destee A, Pasquier F, Sablonniere B. Mutation analysis and association studies of the ubiquitin carboxy-terminal hydrolase L1 gene in Huntington’s disease. Neurosci Lett. 2002;15:1–4. doi: 10.1016/S0304-3940(02)00231-8. [DOI] [PubMed] [Google Scholar]
- Moore DJ, Dawson VL, Dawson TM. Role for the ubiquitin-proteasome system in Parkinson’s disease and other neurodegenerative brain amyloidoses. Neuromol Med. 2003;15:95–108. doi: 10.1385/NMM:4:1-2:95. [DOI] [PubMed] [Google Scholar]
- Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson’s disease. Science. 2003;15:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- Deriziotis P, Andre R, Smith DM, Goold R, Kinghorn KJ, Kristiansen M, Nathan JA, Rosenzweig R, Krutauz D, Glickman MH. et al. Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry. EMBO J. 2011;15:3065–3077. doi: 10.1038/emboj.2011.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Lindquist S. Conversion of PrP to a self-perpetuating PrPSc-like conformation in the cytosol. Science. 2002;15:1785–1788. doi: 10.1126/science.1073619. [DOI] [PubMed] [Google Scholar]
- Kang SC, Brown DR, Whiteman M, Li R, Pan T, Perry G, Wisniewski T, Sy MS, Wong BS. Prion protein is ubiquitinated after developing protease resistance in the brains of scrapie-infected mice. J Pathol. 2004;15:603–608. doi: 10.1002/path.1555. [DOI] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;15:863–873. doi: 10.1016/S0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- Broer L, Ikram MA, Schuur M, DeStefano AL, Bis JC, Liu F, Rivadeneira F, Uitterlinden AG, Beiser AS, Longstreth WT. et al. Association of HSP70 and its co-chaperones with Alzheimer’s disease. J Alzheimers Dis. 2011;15:93–102. doi: 10.3233/JAD-2011-101560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajou K, Maillard C, Jost M, Lijnen RH, Gils A, Declerck P, Carmeliet P, Foidart JM, Noel A. Host-derived plasminogen activator inhibitor-1 (PAI-1) concentration is critical for in vivo tumoral angiogenesis and growth. Oncogene. 2004;15:6986–6990. doi: 10.1038/sj.onc.1207859. [DOI] [PubMed] [Google Scholar]
- Hirakawa S, Kodama S, Kunstfeld R, Kajiya K, Brown LF, Detmar M. VEGF-A induces tumor and sentinel lymph node lymphangiogenesis and promotes lymphatic metastasis. J Exp Med. 2005;15:1089–1099. doi: 10.1084/jem.20041896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid AE, Hudson KF, Richey MW, Bartz JC. Rapid transepithelial transport of prions following inhalation. J Virol. 2012;15:12731–12740. doi: 10.1128/JVI.01930-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner DJ, Bartnikas TB, Gitlin JD. The role of copper in neurodegenerative disease. Neurobiol Dis. 1999;15:221–230. doi: 10.1006/nbdi.1999.0250. [DOI] [PubMed] [Google Scholar]
- Dexter DT, Jenner P, Schapira AH, Marsden CD. Alterations in levels of iron, ferritin, and other trace metals in neurodegenerative diseases affecting the basal ganglia: the Royal Kings and Queens Parkinson’s Disease Research Group. Ann Neurol. 1992;15(Suppl):S94–S100. doi: 10.1002/ana.410320716. [DOI] [PubMed] [Google Scholar]
- Gulcin I. Antioxidant and antiradical activities of L-carnitine. Life Sci. 2006;15:803–811. doi: 10.1016/j.lfs.2005.05.103. [DOI] [PubMed] [Google Scholar]