Abstract
Juvenile hormone (JH) controls key events in the honey bee life cycle, viz. caste development and age polyethism. We quantified transcript abundance of 24 genes involved in the JH biosynthetic pathway in the corpora allata-corpora cardiaca (CA-CC) complex. The expression of six of these genes showing relatively high transcript abundance was contrasted with CA size, hemolymph JH titer, as well as JH degradation rates and JH esterase (jhe) transcript levels. Gene expression did not match the contrasting JH titers in queen and worker fourth instar larvae, but jhe transcript abundance and JH degradation rates were significantly lower in queen larvae. Consequently, transcriptional control of JHE is of importance in regulating larval JH titers and caste development. In contrast, the same analyses applied to adult worker bees allowed us inferring that the high JH levels in foragers are due to increased JH synthesis. Upon RNAi-mediated silencing of the methyl farnesoate epoxidase gene (mfe) encoding the enzyme that catalyzes methyl farnesoate-to-JH conversion, the JH titer was decreased, thus corroborating that JH titer regulation in adult honey bees depends on this final JH biosynthesis step. The molecular pathway differences underlying JH titer regulation in larval caste development versus adult age polyethism lead us to propose that mfe and jhe genes be assayed when addressing questions on the role(s) of JH in social evolution.
Introduction
Juvenile Hormone (JH), synthesized by the corpora allata (CA), a small pair of glands located in the retrocerebral complex of the insect brain, is best known for its pleiotropic functions in insect metamorphosis and reproduction [1]. Building on these basic functions, different classes of insects have apparently co-opted this hormone and its downstream signaling pathways and regulatory modules into functions that permit specific adaptations in their life cycles (e.g., diapause, [2]) or complex life histories (e.g., seasonal or caste polyphenisms, [3]). In the honey bee Apis mellifera, JH plays essential functions in caste determination during larval phases of development and controls the age-related transition from within-nest to foraging tasks in adult workers [4].
During larval development, the JH titer reaches a peak in the third to fourth larval instar and then drops to low levels at the beginning of the fifth instar in both castes. This peak in the early larval stages is particularly pronounced in queens [5], [6] and is an important factor for the caste-specific organ differentiation, especially the larval ovaries [7]. In adult bees, JH does not appear to play a role as a gonadotropin, as is the case in many other insects [8]. Rather, after a small peak of JH that initiates vitellogenin synthesis in the late pharate adult stage of queens [9], the JH titer stays at low levels throughout a queen's adult life cycle. In contrast, it has taken on a role in pleiotropically setting the physiological conditions for age-specific tasks in adult workers. In young workers, the hemolymph JH levels are low, and these bees exert intranidal tasks, primarily feeding the brood with secretions from their well-developed hypopharyngeal glands [10]. As the bees grow older and switch to performing more hazardous extranidal task, viz. foraging for nectar, pollen and water, their JH titers are typically increased [4].
Understanding regulatory mechanisms that underlie the fluctuations in the hemolymph JH titers is, thus, a major issue for coming to terms with honey bee sociality. Such regulation can occur via two ways, in the CA, via modulation of enzyme levels and enzyme activity in the biosynthetic steps of the sesquiterpenoid JH molecule, and via degradation and clearance of secreted JH in the hemolymph. JH-precursor manipulation and pharmacological inhibition experiments have shown that the final steps in JH synthesis are critically regulated in the honey bee CA [11], with their activity being modulated by biogenic amines [12] and also by the insulin-signaling pathway [13]. RNAi and partition assay experiments provided evidence that the honey bee JH esterase (AmJHE), but not the JH epoxide hydrolase (AmJHEH), is capable of degrading circulating JH [14], [15].
What is largely lacking in this picture is functional information on honey bee genes encoding enzymes of the JH biosynthetic pathway in the CA. To provide such information we searched for homologs of genes known to be involved in the JH biosynthetic pathway in Drosophila melanogaster and Anopheles gambiae. The expression levels of these genes were quantified by RT-qPCR in two contrasting situations of the honey bee life cycle. The first one comprised the fourth larval instar, where the JH titers in prospective queens are much higher than those in prospective workers [6]. The second contrasting situation concerned young adult workers performing intranidal tasks (nurses) versus older workers undergoing foraging tasks, i.e., two stages in the adult life cycle where JH titers are widely different [4], [16]. So as to then correlate the respective gene expression levels with the JH titers in these stages we also measured the general diameter of the CA and the diameters of CA nuclei as a proxy for CA activity.
Results
In silico analyses of JH biosynthetic pathway genes
Juvenile hormone biosynthesis involves the production of farnesyl pyrophosphate (farnesyl-PP) from acetyl-CoA via the mevalonic acid pathway, followed by converting farnesyl-PP into JH-precursors (farnesoic acid and methyl farnesoate) (Figure S1). To characterize candidate genes encoding enzymes in the JH biosynthetic pathway of honey bees, we used EST data generated from CA of D. melanogaster and A. gambiae [17] as queries in BLASTP searches to find orthologous sequences in the honey bee genome (version 4.0). EST libraries gave us information on the JH biosynthetic pathway genes that were actually expressed in these glands, as deduced by the detection of their respective transcripts. This useful information was important for the characterization of the expression of these genes in the CA-CC complexes of the honeybee.
Reciprocal matches corresponding to 25 enzyme-encoding sequences were found and the respective conserved functional motifs were identified. The predicted sequences identified in the honey bee genome (GBs), their corresponding orthologs in A. gambiae and D. melanogaster, as well as the predicted enzymes with their respective functions in the JH biosynthetic pathway are listed in Table 1. In accordance with prior in silico analyses [18] and in contrast to other insects [19], [20], we retrieved seven genes, putatively paralogs, that encode enzymes with a farnesyl diphosphate synthase (prenyltransferase) (FPPS) function. The predicted structure of these 25 genes is represented in Figure S2.
Table 1. Genes encoding enzymes of the JH biosynthetic pathway in Apis mellifera. Prediction based on BLASTP comparisons using Anopheles gambiae and Drosophila melanogaster sequences against the honey bee genome (version 4.0).
| Enzyme | Symbol | Function | E.C. number | A. gambiae ortholog | D. melanogaster ortholog | A. mellifera ortholog | E-value |
| Methionine adenosyl transferase | MAT | Synthesis of S-adenosyl-L-methionine (AdoMet) | 2.5.1.6 | XM_307861 | NM_164362 | XM_623666 | e-132 |
| Adenosylhomocysteinase | AHC1 | Hydrolysis of S-adenosyl-L-homocysteine (AdoHcy) | 3.3.1.1 | XM_311257 | NM_078609 | XM_391917 | 0.0 |
| AHC2 | XM_624149 | e-124 | |||||
| Adenosine kinase | AK | Phosphorylates adenosine | 2.7.1.20 | XM_307001 | NM_168532 | XM_391988 | e-110 |
| Acetoacetyl CoA thiolase | ACT | Condenses 2 molecules of Acetyl-CoA | 2.3.1.9 | XP_321828 | NP_612094 | XM_391843 | 2e-77 |
| Hydroxymethylglutaryl-CoA redutase | HMGR | Reduces HMG-CoA to mevalonate | 1.1.1.34 | XP_307890 | - | XM_623115 | 0.0 |
| Hydroxymethylglutaryl-CoA synthase | HMGS | Condenses Acetoacetyl-CoA+acetyl-CoA | 2.3.3.10 | XP_315872 | NP_725570 | XM_397202 | e-171 |
| Mevalonate kinase | MK | Phosphorylates mevalonate | 2.7.1.36 | XM_319701 | NM_176158 | XM_624542 | 1e-30 |
| Phosphomevalonate kinase | PMK | Phosphorylates phosphomevalonate | 2.7.4.2 | XP_310779 | Q9VIT2 | XM_001120920 | 1e-34 |
| Isopentenyl-diphosphate delta-isomerase | IPPI | Isomerization of IPP to DMAPP | 5.3.3.2 | XP_321388 | NP_650962 | XM_001121223 | 4e-27 |
| Farnesyl diphosphate synthase (prenyltransferase) | FPPS1 | Sequential condensation of IPP with DMAPP and then GPP to form FPP | 2.5.1.1/10 | XP_308653 | NP_477380 | XM_001122575 | 2e-66 |
| FPPS2 | XM_623583 | 8e-79 | |||||
| FPPS3 | XM_326224 | 8e-76 | |||||
| FPPS4 | XM_001122685 | 5e-59 | |||||
| FPPS5 | XM_624295 | 2e-59 | |||||
| FPPS6 | XM_001123325 | 1e-37 | |||||
| Citrate (si)-synthase | CS | Synthesis of citrate in the mitochondria | 2.3.3.1 | XM_320478 | - | XM_393545 | 0.0 |
| ATP citrate lyase | ATPCL | Synthesis of cytosolic acetyl-CoA from citrate | 2.3.3.8 | XM_319323 | - | XM_623080 | 0.0 |
| Mitochondrial citrate transport protein | MCTP1 | Transports citrate from mitochondria to cytosol | - | XP_308964 | NP_727450 | NM_001010975 | e-128 |
| MCTP2 | XM_395934 | e-110 | |||||
| Short-chain dehydrogenase | SCD | Oxidation of farnesol to farnesal? | - | XM_556135 | NM_132467 | XM_624031 | 5e-57 |
| Crustacean “Farnesoic acid O-methyltransferase” homolog | FAMET | Function unclear | - | XP_318631 | NP_611544 | XM_623143 | e-101 |
| Bombyx JHA methyl transferase ortholog | MT | Transfers methyl group from AdoMet to farnesoic acid | - | XP_314173 | NP_609793 | XM_001119986 | 1e-40 |
| Methyl farnesoate epoxidase (CYP15) | MFE | Oxidation of MF into JH III | - | XP_315675 | NP_649151 | XM_623572 | 5e-80 |
Enzyme Commission (E.C.) classification and enzyme functions are shown.
Expression of JH biosynthetic pathway genes in different tissues of forager bees
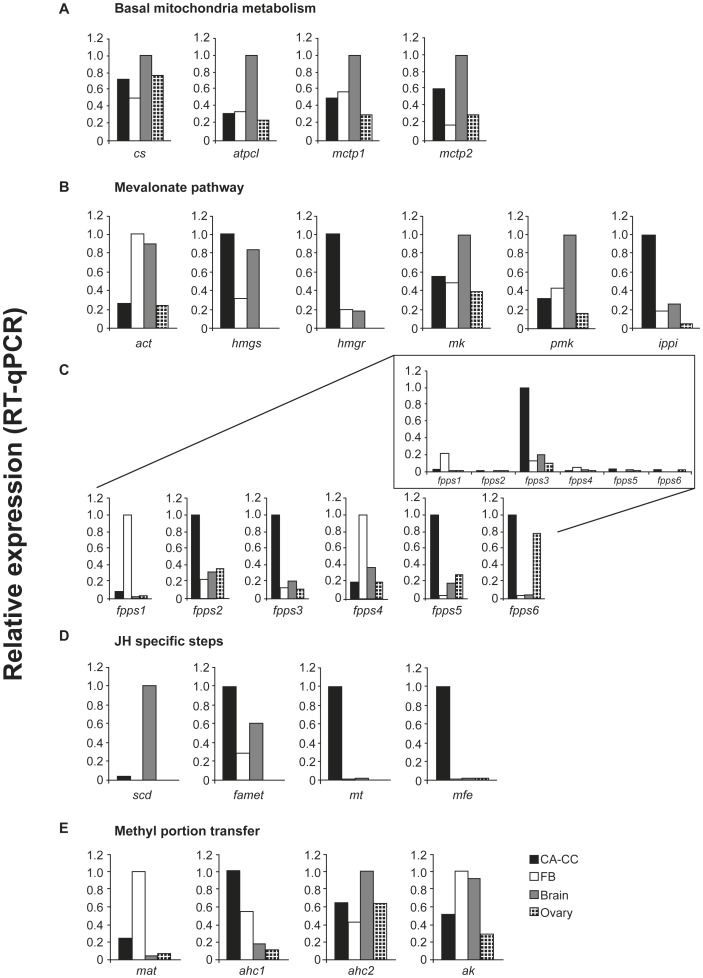
The expression of the genes involved in JH biosynthesis (Table 1), except fpps7, was investigated in the CA-CC complex, brain, fat body and ovaries of forager bees (Figure 1). These genes are potentially involved in basal mitochondrial metabolism leading to Acetyl-CoA production (Figure 1A), and in the mevalonate pathway (Figure 1B, 1C), or specific steps of JH biosynthesis (Figure 1D, 1E).
Figure 1. Expression of JH biosynthesis pathway genes in tissues from forager bees.
Genes are grouped according to the function of the corresponding enzymes: (A) basal mitochondrial metabolism; (B, C) mevalonate pathway; (D) JH-specific steps; (E) methyl moiety transfer. Total RNA extracted from corpora allata-corpora cardiaca complexes (CA-CC), brain, fat body (FB), and ovaries were used for transcript quantification by real time RT-PCR (RT-qPCR). Each column in the graphs represents transcript levels in a single sample of 20–25 pooled CA-CC complexes, and 10 pooled brains, fat bodies and ovaries. The highest expression value for each gene was converted to 1. In the insert (C) the highest expression level among fpps genes was converted to 1.
Transcripts of all these genes encoding enzymes of the JH biosynthetic pathway were detected in the CA-CC complex (Figure 1). This is consistent with the high rates of JH biosynthesis in the CA and high titers of JH in the hemolymph of foragers [4]. The expression of several of these genes was undetectable, or detected at only basal levels in fat body (fpps5, fpps6, scd, mt, mfe,), ovaries (hmgs, hmgr, ippi, fpps1, scd, famet, mt, mfe, mat), or in the brain (fpps1, fpps6, mt, mfe, mat). Expression profiles of the FPPS codifying genes in tissues and organs of forager bees were compared separately. As fpps3 turned out to be highly expressed in the CA-CC complex (Figure 1C - insert) this suggests that it is the bona fide farnesyl diphosphate synthase gene involved in JH biosynthesis in honey bees.
We then selected six genes, mfe, mt, hmgr, ippi, fpp3 and hmgs for in-depth studies. As shown in Figure 1, these genes are highly expressed in CA-CC complexes and can, thus, serve as markers for comparing JH biosynthesis in honey bee castes and during development.
JH biosynthesis gene expression in the CA-CC complex in relation to JH dynamics
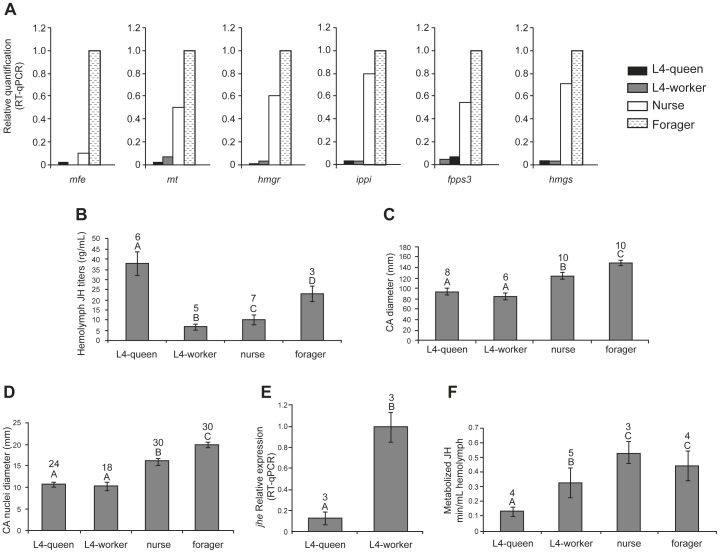
Transcript abundance of the six JH biosynthetic pathway genes, mfe, mt, hmgr, ippi, fpp3 and hmgs was contrasted to CA size and the hemolymph JH titers and metabolism in adult workers performing intranidal versus forager tasks, and also in fourth instar queen versus worker larvae (Figure 2).
Figure 2. Transcript levels of JH biosynthesis pathway genes, JH titer, corpora allata (CA) size, and JH metabolism in honey bee larvae and adults.
(A) relative expression levels of genes encoding enzymes of the JH biosynthesis pathway in CA-CC complexes; transcript levels were determined by RT-qPCR using a pool of 10 CA-CC pairs; rp49 expression was used for normalization. The highest expression value for each gene was converted to 1. (B) Hemolymph JH titers measured by RIA. (C and D) CA diameter and diameter of CA nuclei measured by confocal microscopy following DAPI staining. (E) relative expression of the jhe gene, the major gene involved in JH metabolism, in the larval whole body. (F) JH metabolism (JHE activity) assayed by partition assay. Data refer to fourth instar (L4) queen and worker larvae, and two stages in the life cycle of adult workers (nurse and forager). Means ± SEM are represented in all panels, with sample sizes (N) shown above bars, except for panel 2A, where transcripts levels were quantified in single pools of CA-CC complexes. Different letters represent statistical differences. Statistical analyses: (2B, C and D) One-way ANOVA, post-hoc Holm-Sidak test, p≤0.001; (2F) One-way ANOVA, post-hoc Tukey test, p≤0.001; (2E) t-test: p≤0.001.
In the case of adult workers, the RT-qPCR analysis of a sample consisting of 10 CA-CC complexes suggested that transcript abundance for all these genes was higher in foragers than in nurses (Figure 2A). Although this statement cannot be supported statistically because it is based on a single-sample analysis, we found that these differences correlated well with the high hemolymph JH titer in foragers (Figure 2B).
Furthermore, when comparing CA size (Figure 2C) and the size of CA nuclei (Figure 2D) these were also significantly larger in foragers. Therefore, the expression of these six genes in adult workers was correlated with JH hemolymph titer and CA size.
In contrast to our findings for adult workers, there was no such correlation in the fourth instar larvae. In fact, while the JH titer in queen larvae was considerably higher than in worker larvae (Figure 2B), in accordance with literature data [6], there were no apparent differences in transcript abundances for any of the six JH biosynthetic pathway genes (Figure 2A). Quite strikingly, transcript abundances for all these genes seemed rather low when compared to those seen in CA-CC complexes of adult bees (Figure 2A). Furthermore, we also did not observe any significant difference in general CA size (Figure 2C) and CA nuclei diameters in these fourth instar queen and worker larvae (Figure 2D), this suggesting that JH titer regulation in fourth instar queen and worker larvae does not depend on the differential expression of JH biosynthesis genes and is also independent of CA size. In more general terms, this would mean that the gene-regulatory network underlying JH synthesis in the CA-CC complex and the consequent modulation of the JH titer follow different rules in larval versus adult honey bees. To follow up on this question we next looked at JH degradation as a means of creating divergence in JH hemolymph titers.
JH esterase gene (jhe) expression and JH degradation activity in hemolymph
In a previous study [14] we had shown that the jhe gene, which is predicted to encode a putative JH esterase, has an expression pattern in the fat body that runs exactly opposite to the JH titer. We therefore investigated jhe transcript abundance in queen and worker larvae (Figure 2E) and contrasted these with the esterase-related JH degradation activity, measured by a JH partitioning assay, in the respective hemolymph samples (Figure 2F). In accordance with our predictions, jhe transcript abundance was considerably lower (p≤0.001) in fourth instar queen larvae when compared to same instar workers (Figure 2E), and JH degradation activity was also significantly lower in queen hemolymph (Figure 2F), this indicating that JH degradation is an important regulatory factor for the physiological hormone titer. It is also worthy of note that there was no significant difference in JH esterase activity in the hemolymph samples from adult workers (Figure 2F), lending further support to our hypothesis concerning differences in mechanisms underlying JH biosynthesis and titer regulation in larvae versus adult honey bees.
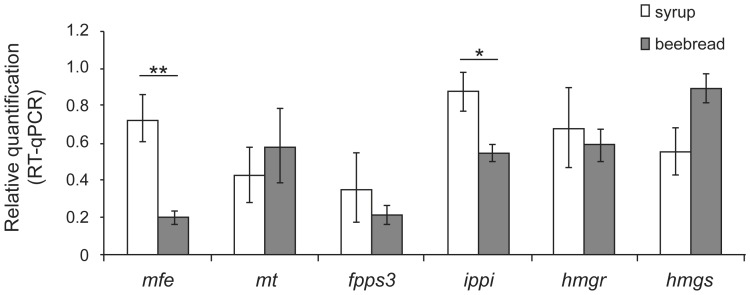
Genes encoding JH biosynthesis enzymes respond to differential feeding during adulthood
Hive bees mainly feed on a protein-rich diet, processed from the pollen stores in the hive, whereas foragers prefer an almost pure carbohydrate diet [21], [22]. As nutrition is linked to behavior and aging, we investigated whether and how diet affects the transcription levels of JH biosynthesis pathway genes. Newly emerged workers were kept for seven days in wooden cages and fed with a beebread/syrup diet (beebread) or with syrup only. For both groups we then assessed the transcript levels of six candidate genes (mfe, mt, fpps3, ippi, hmgr and hmgs) in the CA-CC complexes (Figure 3). We found that only ippi and mfe were differentially expressed under the two dietary conditions, both being more expressed in the syrup fed group (Figure 3). This is in agreement with the above reported findings on the transcript levels of the mfe and ippi genes (Figure 2A), suggesting that these genes are upregulated in foragers that feed on a carbohydrate-rich diet and typically have a high JH titer, when compared to nurse bees that feed on a protein-rich diet and have a low JH titer.
Figure 3. Effect of diet on transcript levels of genes encoding enzymes of the JH biosynthesis pathway and expressed in CA-CC complexes.
Newly-emerged workers were kept for 7 days in cages and fed on a protein-enriched diet (beebread plus syrup), or on a pure carbohydrate diet (syrup only). Each bar represents the mean ± SEM of three independent samples, each consisting of eight CA-CC pairs. (**) t-test; p≤0.001; (*) t-test; p≤0.05.
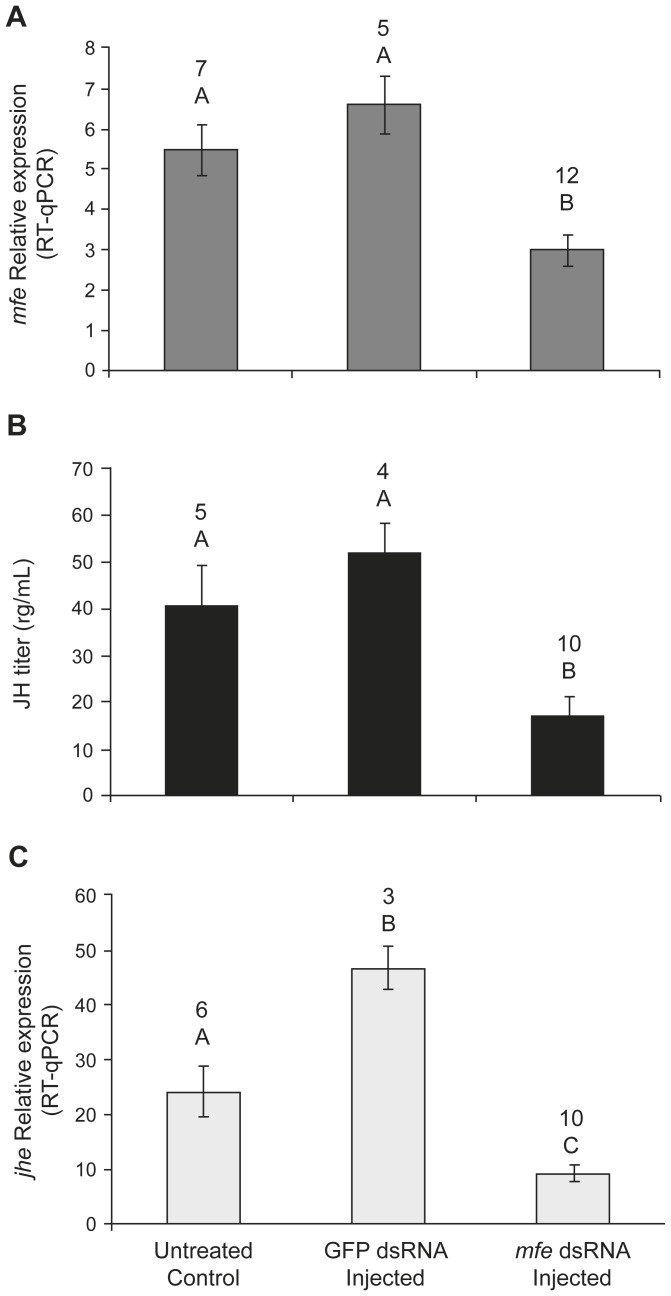
RNAi-mediated knockdown of methyl farnesoate epoxidase (mfe) gene function
The mfe gene, which putatively encodes an enzyme in the final JH biosynthesis steps, was chosen for a functional gene disruption assay. Three groups of newly emerged workers were set up in this experiment, one was injected with target mfe dsRNA, the second one received GFP dsRNA as an off-target dsRNA control, and the third group was left untreated (control). Seven days after the treatment the bees were collected, total RNA was individually extracted and used to check knockdown efficiency.
The mfe dsRNA-injected group showed an up to 50% reduction in mfe mRNA abundance when compared to the two other groups (Figure 4A). As this gene is primarily expressed in the CA-CC complexes (Figure 1D), this is a good indication of the silencing efficiency in the targeted organ. A similar level of reduction was also seen in the hemolymph JH titer (Figure 4B), thus reinforcing the prior evidence and directly linking this gene to the regulation of the JH titer in adult honey bees. Further support also came from the RT-qPCR assays run on jhe gene expression, which was significantly decreased in the mfe-knockdown group (Figure 4C). But here the link between the two genes probably goes through the regulation of the hemolymph JH titer, as in a previous study [14] it has been shown that a topical application of synthetic JH-III can induce jhe transcription, and that jhe knockdown bees had higher JH titers in their hemolymph. This RNAi assay on mfe, an apparently critical gene in the JH biosynthetic pathway, thus lends further support to the hypothesis that, in adult honey bees, the regulation of the JH titer associated with division of labor in workers involves the transcriptional regulation of JH biosynthesis-specific genes, this contrasting with the situation in the larval stages, where the regulation of a gene encoding a JH-degrading esterase (jhe) would be more crucial for the divergence in queen/worker phenotype development.
Figure 4. Effects of RNAi-mediated silencing of methyl farnesoate epoxidase (mfe) gene function on major control genes of JH biosynthesis and metabolism and on the hemolymph JH titer.
Newly-emerged adult workers were injected with 3 µg mfe dsRNA or GFP dsRNA or left untreated. The bees were collected 7 days after the treatment. (A) mfe transcript levels determined by RT-qPCR using whole body RNA. (B) JH hemolymph titers measured by RIA. (C) jhe transcript levels determined by RT-qPCR using whole body RNA. rp49 was used for normalization. Each bar represents the mean ± SEM of independent samples; sample size is shown above bars. Different letters represent statistical differences assessed by One-way ANOVA followed by Tukey's post hoc test; (p≤0.001 in A and C; p = 0.003 in B).
Discussion
Two hallmarks of the social organization of honey bees, caste development and age-related division of labor of adult workers, are controlled by the hemolymph JH titer. Whereas the dependence of these processes on JH received much attention and is worked out in descriptive and mathematical models [23], [24], the mechanisms underlying JH titer regulation are not yet completely known. The steady-state hemolymph JH titer is modulated by the activity of enzymes in the biosynthetic pathway in the CA (Figure S1), or by enzymes involved in its degradation which in honey bees occurs primarily via a JH esterase, or by a combination of both. Whilst the genes encoding JH degrading enzymes and their functionalities have been fairly well characterized in honey bees [14], [15], this is not so for the JH biosynthesis steps.
Annotation and expression of JH biosynthesis genes
Based on computational analysis of A. mellifera candidate genes in JH biosynthetic pathway [18] and using a database comprising ESTs from insect CA transcriptomes [17], [20], [25]–[27] 25 genes were found and annotated in the honey bee genome. Amongst these, the farnesyl diphosphate synthase (fpps) deserves special attention due to the high number of paralogs. While the fpps gene is a single-copy gene in all sequenced Diptera [17], it is represented by seven copies in the honey bee genome. The fpps genes are not a unique case of gene duplication or gene family expansion in the honey bee genome, as this also happened in another 60 genes, and these have been interpreted as possible signatures of solitary to social life transitions in bees [18]. Our RT-qPCR results now showed that all these fpps paralogs are expressed in the retrocerebral CA-CC complex, and fpps3 seemed to be the most expressed one (Figure 1C - insert), thus suggesting that it is a bona fide candidate for an FPPS function in the JH biosynthetic pathway.
The expression of 24 of the 25 annotated JH biosynthesis genes was assayed in RNA extracts from CA-CC complexes, remaining brain tissue, fat body, and ovary. All genes were expressed in the CA-CC complexes, placing in evidence their putative role in JH biosynthesis. Transcription for most of the genes was also evidenced in the other tissues, which is not surprising, considering that the mevalonate pathway enzymes are required not only for JH production but also for the production of other bioactive terpenoids and for the farnesylation of proteins [28]. In contrast, two genes involved in JH-specific steps, those encoding O-methyltransferase (mt) and methyl farnesoate epoxidase (mfe), were specifically expressed in the CA-CC complexes.
Methyl farnesoate is the immediate precursor of JH in insects [29] and has hormonal functions in crustaceans [30]. Farnesoic acid O-methyltransferase (FAMeT) and JH acid O-methyltransferase (JHAMT) (called here MT) are described as catalyzing the methylation of farnesoic acid to methyl farnesoate with S-adenosyl-L-methionine functioning as cofactor [25]. In the fruit fly, D. melanogaster, a recombinant FAMeT (rFAMeT) was cloned, expressed, and a specific antiserum was generated. Immunohistochemical analysis confirmed the presence of FAMeT in the CA portion of the ring gland, but rFAMeT presented no enzymatic activity in a radiochemical assay, inferring that D. melanogaster FAMeT has little, if any role in sesquiterpenoid biosynthesis [31]. Notwithstanding, the DmJHAMT gene is predominantly expressed in the fly CA. Moreover, the recombinant protein was capable of catalyzing the methylation/esterification of JH-III acid or farnesoic acid in the presence of S-adenosyl-L-methionine [32]. Subsequently, Marchal et al. [33], using the locust Schistocerca gregaria as a model, showed that JHAMT, and not FAMeT, is the enzyme involved in JH synthesis. In the honey bee genome we found orthologs for both, famet and mt genes, and when assaying their expression by means of RT-qPCR we found that mt transcripts were specifically detected in CA-CC complex, whereas famet was transcribed more ubiquitously, as is also the case in the highly eusocial stingless bee Melipona quadrifasciata [34]. This leads us to infer that mt and not famet is the gene encoding a functional methyltransferase activity in the honey bee CA.
The mt gene (AmJHAMT gene) was recently assayed in terms of expression patterns and functionality in honey bee larvae and pupae [35]. It was observed that in these whole body RNA extracts the AmJHAMT gene expression profile during the fourth and fifth larval instars does not follow the modulation in JH levels. For instance, low mt transcript levels coincided with high JH levels in the fourth larval instar, and this was inverted in the fifth instar. Furthermore, as revealed by our data, mt transcript abundance in fourth instar queens and workers stood in contrast with the respective JH levels. Therefore, neither of these studies could find a positive correlation between levels of AmJHAMT transcripts and JH in the larval stages. Our hypothesis is that the larval JH titer is primarily regulated by JHE activity, and this is supported by evidence showing an increase in jhe transcripts in fourth and fifty instar worker larvae [14].
Methyl farnesoate epoxidase (MFE) versus JH esterase (JHE) – different modes of JH titer control in caste development and age-related division of labor?
The steady-state JH titer in hemolymph is a snapshot picture resulting from a balance in JH synthesis and release from the CA, and degradation by metabolizing enzymes. The regulation of synthesis has been pointed out as an important step in this process. So as to understand these processes at the level of transcriptional control and functionality of certain key genes in honey bees we looked at two contrasting situations in the honey bee life cycle, i.e., caste development and age-related division of labor.
For caste development we put our focus on the fourth larval instar, which is the stage when the JH titer in queen and worker larvae shows maximal differences [5], [6]. Earlier results obtained by a radiochemical assay of JH biosynthesis in vitro showed that MFE activity is not only the final step of JH synthesis in honey bee CA, but is also a limiting factor for JH production in CA of honey bee larvae [36]. The honey bee MFE is clearly the ortholog of a P450 enzyme (CYP15A1), which catalyzes the production of JH from methyl farnesoate in D. punctata [26]. Nonetheless, mfe transcript abundance is basal, or undetectable in the CA of fourth instar queen and worker larvae (Figure 2A), this raising the question of how the hemolymph JH titer could be so much higher in queen larvae (Figure 2B). One possibility would be CA size, but these turned out to be similar for queen and worker larvae (Figure 2C, 2D), and in this respect, honey bee larvae are similar to Manduca sexta, where CA volume does not correlate with the hemolymph JH level [37]. In late fifth instar A. mellifera larvae, CA volume varies very little [36], and this is also the case for M. sexta, where CA volumes were found to be much more related to larval age/size than to JH synthesis activity [38].
With larval CA size out of the question, the factor left was JH degradation and, indeed, this was found to be the case, as revealed by a much lower rate of JH metabolism in hemolymph of queen larvae (Figure 2F). These low rates were furthermore in accordance with reduced expression levels of the jhe gene (Figure 2E). For JH titer regulation in the fourth larval instar, the transcriptional control of jhe, a gene driving JH degradation in honey bees [14] should, thus be the primary critical factor. This assumption is in accordance with the respective rates of JH biosynthesis measured in a radiochemical in vitro assay [39], showing that JH biosynthesis rates in this instar are still very similar in queen and worker larvae, whereas their JH hemolymph titers are maximally different [6]. In the fifth larval instar, this regulatory mode may, however, change, as JH biosynthesis rates drastically increase in queen larvae until the end of the feeding phase, but concomitantly, the JH titer in hemolymph drops to basal levels, thus indicating increased clearance of JH also in queen larvae.
In adult workers, the results of the JH titer measurements were consistent with previous results showing that foragers have higher hormone levels than the younger nurse bees [4], [16], [40], [41]. As we could show, these differences in steady-state JH levels were not due to differences in JH degradation, but seemed associated with the expression levels for all the six JH biosynthesis pathway genes studied herein (Figure 2). This suggests that all these genes are upregulated once workers switch from performing within-hive tasks to become foragers. Interestingly, the gene that showed a high relative expression was mfe, and it was also this gene that was significantly upregulated in caged workers fed on a carbohydrate-rich diet (Figure 3). The expression of this gene seems, thus, not only linked with the age-related behavioral switch to foraging, but also reflects the nutritional physiology of foragers [22], even when these bees are kept in cages and cannot fly. Among the other genes assayed with respect to the bees' nutritional conditions, only ippi was also significantly upregulated in syrup-fed bees. But whereas there is strong experimental evidence for MFE as a key factor in controlling the JH titer [36], this is not the case for ippi, which encodes an isopentenyl diphosphate isomerase in an alternative route of the mevalonate pathway (see Figure S1).
Bees fed on a pure carbohydrate diet did not accumulate vitellogenin in their hemolymph [42], and, rather, it is the depletion of vitellogenin which has been linked to the hive bee-to-forager transition and predicted in the double repressor hypothesis [43] as an activation of an allato-regulatory central nervous system pathway. This hypothesis has since been validated by manipulation of JH levels, as well as by knocking down vitellogenin transcript levels by dsRNA injection [44]–[46], all these experiments confirming that the increase in JH titer at the transition from hive bee to forager behavior is contingent on a reduction in vitellogenin levels. Herein we now showed that the expression of mfe, a key gene in JH biosynthesis, is also directly contingent on the nutritional state, thus adding an extra component to this double repressor circuitry.
As demonstrated by several authors [47], [48] the process of behavioral maturation in bees is complex and influenced by different intrinsic and extrinsic factors, with age and nutritional state being of prime importance. The interference of the nutritional status with the onset of foraging is a well-described fact [49], [50], and seemingly, this involves the insulin-signaling pathway. Ament et al. [51] showed that an insulin receptor encoding gene, InR isoform 1, is upregulated in the abdomen of honey bee workers in response to a sugar diet, thus connecting the insulin-signaling pathway with behavioral maturation.
So as to better understand the connection between mfe expression and JH titer, we performed an experiment silencing mfe gene function by dsRNA injection. This experiment showed that not only was the JH titer level reduced in mfe knockdown bees, as expected, but that this also led to a reduction in jhe transcript abundance, indicating that JH degradation would also be reduced. A decrease in jhe transcript levels was detected as early as five days after the injection, but was more consistent on day 7 (Figure 4). We hypothesize that this may represent part of a feedback circuitry, whereby circulating JH may directly control its main degrading enzyme [14], so as to prevent excessive variation in steady-state hormone levels, this further illustrating the complexity in transcriptional control within the JH biosynthetic/degradation pathways in relation to age-related division of labor in honey bees.
MFE activity is evidenced as a limiting factor for JH production in the CA of honey bee larvae [36], but regulation of JH biosynthesis may occur to some extent at each one of the sequential steps in the biosynthetic pathway. Interference in this pathway, as well as in JH degradation via JHE activity [14] may affect the temporal fluctuation of JH titer. But these may not be the only factors. Recent in silico analyses performed in our laboratory revealed target binding sites for several known microRNAs in genes encoding enzymes of the JH-biosynthetic pathway. Importantly, target binding sites for twelve of these microRNAs (among them ame-miR-375-3p, ame-miR-3794-5p and ame-miR-6059-3p) are shared by the mfe, jhe and jheh genes, suggesting that these may be co-regulated. We also observed that each of these genes may be targeted by exclusive microRNAs. This emerging array of noncoding small RNAs, which regulate gene expression at the post-transcriptional level, adds a novel interesting twist to the mechanism underlying JH titer regulation and clearly deserves further attention.
Conclusion
We annotated 25 genes of the JH biosynthetic pathway in the honey bee genome and assayed the expression levels of 24, showing that several of these are prominently expressed in CA-CC complexes, when compared to other tissues. Six genes comprising representatives of the mevalonate pathway, as well as of the JH-specific steps were selected for transcript abundance measurements in two contrasting situations of the honey bee life cycle, the fourth larval instar with its caste-specific differences in the JH titer, and in adult workers, which show task-related JH titer differences. This revealed that the transcriptional regulation of a methyl farnesoate epoxidase-encoding gene (mfe) involved in JH biosynthesis in the CA, and of a JH esterase encoding gene (jhe) involved in JH degradation, should be key events for the control of JH levels in the hemolymph. Interestingly, these genes appear to be regulated in a different fashion in the fourth larval instar, when compared to adult workers. Whereas in the fourth larval instar the JH titer was closely correlated with jhe transcript levels, in the adults this was the case for mfe. For adult workers we could furthermore show that mfe expression is also contingent on the nutritional status. This, together with the finding that jhe transcript levels were reduced in workers that had their mfe gene function diminished by RNAi, makes a case for the caste- and stage-specific complexity in the regulation of the hemolymph JH titer in honey bees. Probably this is what is expected of a pleiotropic hormone, and this could also be the center court for the evolvability of its functions in the diversity and plasticity of insect life cycles.
The molecular pathway differences underlying JH titer regulation in honey bee caste development versus age-related division of labor may reflect differences in selection pressures on key enzymes during the evolution of sociality. This may not only be reflected in the expansion of the fpps genes in the honey bee, as well in synonymous versus non-synonymous changes in the sequences of other genes involved in JH biosynthesis and metabolism. Both aspects could now be investigated by comparing these genes across bee genomes currently under study. In particular studies intending to investigate the co-option and changing roles of JH in social insects under a phylogenetic hypothesis should consider measuring the transcript levels of mfe and jhe orthologs in addition to the respective JH hemolymph titers.
Materials and Methods
Bees
Workers and queens of Africanized hybrids of the honey bee, A. mellifera, were collected from hives maintained at the Experimental Apiary of the University of São Paulo at Ribeirão Preto, Brazil. Queens were obtained by a standard apicultural queen rearing technique, whereby first instar female larvae were transferred to artificial queen cells containing a drop of royal jelly and then introduced into a queenless colony to be reared until the desired stage [9]. Newly emerged workers were paint-marked, introduced into queenright hives and subsequently collected according to the activity they were performing: nourishing larvae (nurse bees, between 7 and 10 days old), or foraging for pollen (older workers entering the hive carrying pollen).
DAPI staining of CA-CC complexes
To assess size variation in the CA-CC complex as a possible indicator of JH synthesis levels, the CA dissected from fourth instar queen and worker larvae and from adult workers (nurses and foragers) were stained with DAPI [37]. Briefly, CA-CC complexes were dissected in PBS (137 mM NaCl, 8.1 mM Na2HPO4, 1.47 mM KH2PO4, 2.68 mM KCl), fixed in 4% paraformaldehyde in PBS for 30 min at room temperature and washed several times in PBS. The complexes were incubated in DAPI dissolved in PBS (1∶4000 w/v) for 4 min at room temperature, washed several times in PBS and mounted in 80% glycerol in PBS for analysis by confocal microscopy (Leica TCS-SP5 Laser Microscope; Leica Microsystems, Wetzlar, Germany). Measurements were taken on CA from ten foragers, ten nurses, eight fourth instar queen-destined larvae and six fourth instar worker-destined larvae. In total we measured the diameters of 34 CA and of 102 CA nuclei. The data were analyzed by one-way ANOVA followed by a post-hoc Holm-Sidak test at a significance level of p≤0.05.
JH synthesis candidate genes - annotation and primer design
The honey bee genome database (http://www.hgsc.bcm.tmc.edu/projects/honeybee/) (version 4.0) downloaded to our local server (http://zulu.fmrp.usp.br/beelab) was searched for sequences encoding enzymes of the JH biosynthetic pathway, using Drosophila melanogaster and Anopheles gambiae orthologs as queries. The predicted protein sequences obtained by BLASTN analyses were confirmed by investigating the presence of specific motifs. Primer3 software [52] was used to design specific primers (Table 2) for quantitative PCR analysis. The structural architecture, including intron/exon boundaries of these genes, was manually annotated against the honey bee genome sequence using Artemis 7.0 software [53] implemented in a LINUX server.
Table 2. Primers for assaying JH synthesis pathway genes, and for a JH metabolism gene (jhe). rp49 was used as reference gene.
| Gene | Primer forward (Sequence 5′-3′) | Primer reverse (Sequence 5′-3′) | Amplicon length (nt) |
| rp49 | CGTCATATGTTGCCAACTGGT | TTGAGCACGTTCAACAATGG | 150 |
| jhe | GTTATCGCTTCTGATATGGCT | GATGGGAAATAGGTACCGAC | 121 |
| act | GGCCTCAAACGACTCTTCAA | CGTGCCAGTGGTGTAAGTTG | 163 |
| hmgs | CCGTATCTTGGGTAGAAAGC | GATGATCTCACTCCACGATC | 172 |
| hmgr | CCTGCACAGAATGTTGGAAG | GTGCAGGAAGAATAGTTCCG | 178 |
| mk | TTCCGCTTCTTTTGCAGTTT | TTCCAGATGGATTACCATGC | 155 |
| pmk | CTCATTGGGCCAAATCTCAT | TATCTAGAGCCGCACGACAA | 147 |
| ippi | GCTGAAGCTACTTTGCAGCTA | GCTCGATGAAGTAAAACTTGTCC | 137 |
| fpps1 | GAAGATCAGTACCAATTGTAATC | GTTGGTTGATTTCGTCGCATG | 183 |
| fpps2 | GAGATCAGTATCGCTTATAGTC | GTTGGTTGATTTCGTCGCATG | 174 |
| fpps3 | GGAAGACGAAGATCATATCACT | GTTGGTTGATTTCGTCGCATG | 175 |
| fpps4 | GCGCATGGTGAATGTTTAGA | CGAAACGCATAGCAAGACTG | 186 |
| fpps5 | CGAAAGGAGGAAAACGAAGA | CTGCCATTGTGTGAACTGCT | 140 |
| fpps6 | CATCGATAGATTCGGGAAAGA | ATCGATTATGCCAGCGAAAC | 185 |
| scd | AGAGCTATCGCTGCTCTTGC | TGTGGGAAATGATGCTAACG | 150 |
| famet | GTAAAGAAGGTGAGCCAAGTG | CGATTAACCATTCACCAGAAG | 111 |
| mt | CCTTCACTGGTGCCAAAACT | TGGCCTATATCGAGGATTCG | 140 |
| mfe | GGAATCATTTCTTGCGGAGA | GTTATGCGCGCTATGGAAAT | 145 |
| cs | AATGAATGGTTTGGCTGGAC | GAACGACTTGACCGCTCTTC | 152 |
| mctp1 | TTGATTGTTTTGTGCGCATT | TGTCAACACCACCAAGGAAA | 150 |
| mctp2 | GAAATTTGGCTTCTGGTGGA | CAATCACCCAGACCTTTGAA | 133 |
| atpcl | CTGGCGCTTATGTTCCTGAT | ACCAAGTTCCCTAGCCCAAT | 152 |
| mat | TGCAAAAGTGGCTTGTGAGA | GCCAATCGAATCCTTTTGAA | 152 |
| ahc1 | TGGATGAAGCATCACGAAAA | TGCAACATCAATTTCGCAGT | 143 |
| ahc2 | GCCGAAGATAAGCCATTGAA | CAGCAGCCACTTCATTTTGA | 154 |
| ak | GCAAATGATGCAATTCTTGCT | ATTTGGTTTCCCCAAGAACC | 141 |
Tissue dissection, RNA extraction and cDNA synthesis
For comparative analysis of the transcript levels of genes encoding enzymes putatively involved in the JH biosynthetic pathway, foragers were dissected in ice-cold Ringer solution to retrieve the CA-CC complex. Due to the difficulty of quickly dissecting individual CA, each gland pair was removed together with the adhering CC. The CA-CC total RNA from foragers was extract using the GenElute™ Mammalian Total RNA Kit (Sigma). After dissecting remnant brain tissue, fat body, and ovaries, these tissue samples were transferred to 1 ml of Trizol Reagent (Invitrogen) and stored at −80°C until RNA extraction following the manufacturer's instructions. CA-CC complexes from fourth instar queen and worker larvae were also dissected in Ringer solution and RNA was extracted by homogenization in Trizol reagent.
RNA quantity and quality was assessed spectrophotometrically using a Nanodrop-1000 system (Thermo Scientific, Wilmington, MA, USA). Aliquots of 1 µg total RNA were treated with DNase I (Promega) before first-strand cDNA synthesis by reverse transcription using SuperScript II Reverse Transcriptase (Invitrogen) and an oligo(dT)12–18 primer (Invitrogen) following the manufacturer's instructions.
Quantitative Real-Time RT-PCR (RT-qPCR)
By means of RT-qPCR assays we quantified the expression of a set of 24 candidate genes encoding enzymes putatively involved in the JH biosynthetic pathway. The assays were run in a 7500 Real Time PCR System (Applied Biosystems, Wilmington, MA, USA) under the following conditions: 50°C for 2 min, 95°C for 10 min and 40 cycles at 95°C for 15 s and 60°C for 1 min. The quantification was carried out in 20 µl reaction volumes containing 10 pmol of each primer, 10 µl SYBR Green Master Mix 2X (Applied Biosystems), 1 µl of cDNA, and water to complete the volume. The specificity of the primers was checked by sequencing of the amplicons. Primer efficiency (E) was calculated based on the slope of a standard curve (E = 10(−1/slope)) generated from serial 1∶10 dilutions of cDNA. The rp49 ribosomal protein gene (GenBank accession number AF441189), also known as rpl32 (GenBank accession number NM 001011587.1), was used as reference gene based on prior validation experiments [54]. Data were analyzed according to the comparative threshold cycle (Ct) method, in which the amount of the target transcript is normalized to the reference gene and relative to a calibrator and is given as 2−ΔΔCt fold change.
JH titer quantification by radioimmunoassay (RIA)
Hemolymph was collected from fourth instar larvae (queens and workers) and from adult workers (nurses and foragers) using a microcapillary, and 1 µl aliquots were immediately transferred to 0.5 ml of acetonitrile in a Teflon-lined screw cap vial and stored at −20°C until analysis. The JH extraction from hemolymph followed the liquid-phase separation protocol established by Huang et al. [55], and the antibody used was developed by Goodman et al. [56]. The protocol for the use with honey bee samples is described in full detail in Hartfelder et al. [57]. Based on standard curve values, JH titers were calculated by non-linear four-parameter regression (ImmunoAssay Calculations spreadsheet, Bachem, Bubendorf, Switzerland) and expressed as JH-III equivalents (pg/µl hemolymph). The data were analyzed by one-way ANOVA followed by a post-hoc Tukey test, at a significance level of p≤0.05.
Partition assay for JH esterase (JHE) activity
JHE activity was estimated by a partition assay developed by Share and Roe [58] which uses 3H-labeled JH as substrate. The potent and specific JH esterase inhibitor 3-octylthio-1,1,1-trifluoro-2-propanone (OTFP) was used to measure JH degrading activity in the honey bee hemolymph. Hemolymph samples were collected with a microcapillary from an incision in the integument of fourth instar larvae (queens and workers) and adult workers (nurses and foragers), and centrifuged at 5,000× g for 5 min at 4°C. Hemolymph (10 µl) was mixed with 10 µl PBS and incubated with 1 µl ethanol or 1 µl 2 mM OTFP diluted in ethanol for 10 min at 30°C, followed by incubation at 30°C for 45 min with 100 µl of 6.08 nmol radiolabeled JH-III ([10–3H(N)]-JH-III, specific activity 11.8 Ci/mmol; PerkinElmer Life Sciences) diluted in PBS, to give approximately 6200 cpm per assay. To stop JHE activity at the end of the assay, 50 ml of methanol, water and ammonia solution (10∶9∶1, v/v) were added and the mixture briefly stirred before adding 250 µl isooctane to partition non-hydrolyzed JH-III into the organic phase. A 75 µl aliquot of the aqueous phase containing the water-soluble JH degradation product was taken for liquid scintillation counting (LS 6500 Scintillation Counter Beckman, Ramsey, MN, USA) in Optiphase HiSafe3 cocktail (Packard). Each sample was assayed in triplicate. JHE activity was expressed as nmol JH acid produced per min per ml hemolymph.
Differential feeding experiment of adult workers
This experiment was done to investigate the effects of food intake on the transcript levels of JH biosynthesis genes. Newly-emerged workers (n = 20) were kept for seven days in 8×11×13 cm screened wooden cages at 34°C and 80% humidity, where they received either syrup (50% sugar in water), or beebread (pollen collected from cell combs) plus syrup (30% beebread in syrup) diets. In both treatments, the bees received water and the respective diet ad libitum. Three pools of eight CA-CC complexes were dissected from bees submitted to each treatment and used for total RNA extraction with GenElute™ Mammalian Total RNA Kit (Sigma); cDNA synthesis was done as described above.
RNA interference for silencing mfe gene in adult workers
The function of the methyl farnesoate epoxidase-encoding gene, mfe (Official Gene Set 3.2, GB15634), in the JH biosynthetic pathway was investigated by an RNAi experiment performed in adult workers based on a protocol established by Amdam et al. [59]. Briefly, dsRNA was synthesized using a RiboMax Large Scale RNA Production System – T7 (Promega), following the manufacturer's protocol and using gene-specific primers (dsMFE-f: TAATACGACTCACTATAGGGCGAGATCAATGTGGTCTCAGTG and dsMFE-r: TAATACGACTCACTATAGGGCGATCGTGATCTACCTACTACG). The dsRNA was purified using Trizol Reagent and maintained at −80°C until use.
For injection, cold anesthetized newly-emerged workers were immobilized on a piece of styrofoam where they were kept at 8°C during the procedure. Two groups of these bees were injected with double-stranded mfe RNA (mfe dsRNA) or with a dsRNA for Green Fluorescent Protein (control) (GFP dsRNA). One µl of the dsRNA solution (3 µg dsRNA per µl Ringer saline) was injected laterally into the abdomen between the 5th and 6th tergites. A third group served as untreated control. All groups were kept in 8×11×13 cm screened wooden cages at 34°C and 80% humidity where they were fed with syrup and water ad libitum. After 7 days, 2 µl of hemolymph were collected from an incision in the thorax of cold anesthetized bees, to be used for JH titer analysis by RIA. The remaining whole body was stored in Trizol Reagent (Invitrogen) and maintained at −80°C until use. After RNA extraction, 3 µg of DNase I-treated total RNA was used for cDNA synthesis. A total of 12 bees injected with mfe dsRNA, 5 GFP dsRNA injected bees and 7 untreated bees were analyzed for mfe expression by means of RT-qPCR assays.
After validating the knockdown, hemolymph samples of those bees showing a significant decrease in mfe transcript levels (one-way ANOVA post-hoc Tukey test and significance at p≤0.05) were used for measuring JH titers.
Supporting Information
JH-III biosynthetic pathway (modified from Bellés et al. 2005 Annu Rev Entomol 50: 181–99, and Noriega et al. 2006 Insect Biochem Mol Biol 36:11 366–374). The underlined enzymes correspond to the genes whose expression is shown in Figure 1B–E .
(TIF)
(A) Representation of the A. mellifera genes encoding enzymes of the JH biosynthetic pathway. Exons and introns are represented by non-scaled boxes and lines, respectively. Nucleotide numbers are indicated to provide a general estimation of gene size. The direction of transcription is indicated by an arrow. (B) Architecture of the genes encoding Farnesyl diphosphate synthase (FPPS). Exons are named by their respective lengths. Lines between exons indicate intron and their respective length is indicated. The direction of transcription is indicated by a black arrow. The dashed arrows indicate that part of the sequence is not available in the honey bee genome database (version 4). Arrowheads indicate primers position.
(EPS)
Acknowledgments
We are very grateful to Marcela A. F. B. Laure for her help with corpora allata dissections. We also thank L.R. Aguiar for his valuable technical assistance in the apiary.
Funding Statement
This research was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo [research grant numbers 05/03926-5, 11/03171-5], which also provided a fellowship [09/05675-0] to ADB. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Nijhout HF (1994) Insect hormones. Princeton: Princeton University Press. 267 p. [Google Scholar]
- 2.Denlinger DL, Yocum GF, Rinehart JP (2012) Hormonal control of diapause. In: Gilbert JL editor. Insect Endocrinology. London: Academic Press. pp. 430–463. [Google Scholar]
- 3.Hartfelder K, Emlen DJ (2012) Endocrine control of insect polyphenism. In: Gilbert JL editor. Insect Endocrinology. London: Academic Press. pp. 464–522. [Google Scholar]
- 4. Huang Z, Robinson GE, Tobe S, Yagi K, Strambi C, et al. (1991) Hormonal regulation of behavioral development in the honey bee is based on changes in the rate of juvenile hormone biosynthesis. J Insect Physiol 37: 733–741. [Google Scholar]
- 5. Rembold H (1987) Caste specific modulation of juvenile-hormone titers in Apis mellifera . Insect Biochem 17: 1003–1006. [Google Scholar]
- 6. Rachinsky A, Strambi C, Strambi A, Hartfelder K (1990) Caste and metamorphosis: hemolymph titres of juvenile hormone and ecdysteroids in last instar honeybee larvae. Gen Comp Endocrinol 79: 31–38. [DOI] [PubMed] [Google Scholar]
- 7. Schmidt Capella IC, Hartfelder K (1998) Juvenile hormone effect on DNA synthesis and apoptosis in caste-specific differentiation of the larval honey bee (Apis mellifera L.) ovary. J Insect Physiol 44: 385–391. [DOI] [PubMed] [Google Scholar]
- 8. Robinson GE, Vargo EL (1997) Juvenile hormone in adult eusocial Hymenoptera: gonadotropin and behavioral pacemaker. Arch Insect Biochem Physiol 35: 559–583. [DOI] [PubMed] [Google Scholar]
- 9. Barchuk AR, Bitondi MM, Simões ZL (2002) Effects of juvenile hormone and ecdysone on the timing of vitellogenin appearance in hemolymph of queen and worker pupae of Apis mellifera . J Insect Sci 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winston ML (1987) The Biology of the Honey Bee. Cambridge, MA: Harvard University Press. pp. 1–281. [Google Scholar]
- 11. Rachinsky A, Tobe S, Feldlaufer M (2000) Terminal steps in JH biosynthesis in the honey bee (Apis mellifera L.): developmental changes in sensitivity to JH precursor and allatotropin. Insect Biochem Mol Biol 30: 729–737. [DOI] [PubMed] [Google Scholar]
- 12. Rachinsky A, Feldlaufer M (2000) Responsiveness of honey bee (Apis mellifera L.) corpora allata to allato regulatory peptides from four insect species. J Insect Physiol 46: 41–46. [DOI] [PubMed] [Google Scholar]
- 13. Corona M, Velarde R, Remolina S, Moran-Lauter A, Wang Y, et al. (2007) Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Proc Natl Acad Sci USA 104: 7128–7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mackert A, do Nascimento AM, Bitondi MMG, Hartfelder K, Simões ZLP (2008) Identification of a juvenile hormone esterase-like gene in the honey bee, Apis mellifera L. - expression analysis and functional assays. Comp Biochem Physiol B Biochem Mol Biol 150: 33–44. [DOI] [PubMed] [Google Scholar]
- 15. Mackert A, Hartfelder K, Bitondi MMG, Simões ZLP (2010) The juvenile hormone (JH) epoxide hydrolase gene in the honey bee (Apis mellifera) genome encodes a protein which has negligible participation in JH degradation. J Insect Physiol 56: 1139–1146. [DOI] [PubMed] [Google Scholar]
- 16. Amdam GV, Norberg K, Omholt SW, Kryger P, Lourenço AP, et al. (2005) Higher vitellogenin concentrations in honey bee workers may be an adaptation to life in temperate climates. Insectes Soc 52: 316–319. [Google Scholar]
- 17. Noriega F, Ribeiro J, Koener J, Valenzuela J, Hernandez-Martinez S, et al. (2006) Comparative genomics of insect juvenile hormone biosynthesis. Insect Biochem Mol Biol 36: 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. The Honey Bee Genome Sequencing Consortium (2006) Insights into social insects from the genome of the honeybee Apis mellifera . Nature 443: 931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cusson M, Beliveau C, Sen S, Vandermoten S, Rutledge R, et al. (2006) Characterization and tissue-specific expression of two lepidopteran farnesyl diphosphate synthase homologs: implications for the biosynthesis of ethyl-substituted juvenile hormones. Proteins 65: 742–758. [DOI] [PubMed] [Google Scholar]
- 20. Kinjoh T, Kaneko Y, Itoyama K, Mita K, Hiruma K, et al. (2007) Control of juvenile hormone biosynthesis in Bombyx mori: cloning of the enzymes in the mevalonate pathway and assessment of their developmental expression in the corpora allata . Insect Biochem Mol Biol 37: 808–818. [DOI] [PubMed] [Google Scholar]
- 21. Haydak MH (1970) Honey bee nutrition. Annu Rev Entomol 15: 143–156. [Google Scholar]
- 22. Crailsheim K, Schneider LHW, Hrassnigg N, Buhlmann G, Brosch U, et al. (1992) Pollen consumption and utilization in worker honeybees (Apis mellifera carnica) - dependence on individual age and function. J Insect Physiol 38: 409–419. [Google Scholar]
- 23. Amdam GV, Omholt SW (2002) The regulatory anatomy of honeybee lifespan. J Theor Biol 216: 209–228. [DOI] [PubMed] [Google Scholar]
- 24. Leimar O, Hartfelder K, Laubichler MD, Page RE Jr (2012) Development and evolution of caste dimorphism in honeybees – a modeling approach. Ecol Evol 2: 3098–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shinoda T, Itoyama K (2003) Juvenile hormone acid methyltransferase: a key regulatory enzyme for insect metamorphosis. Proc Natl Acad Sci USA 100: 11986–11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Helvig C, Koener J, Unnithan G, Feyereisen R (2004) CYP15A1, the cytochrome P450 that catalyzes epoxidation of methyl farnesoate to juvenile hormone III in cockroach corpora allata . Proc Natl Acad Sci USA 101: 4024–4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ollivier M, Legeai F, Rispe C (2010) Comparative analysis of the Acyrthosiphon pisum genome and expressed sequence tag-based gene sets from other aphid species. Insect Mol Biol 19: 33–45. [DOI] [PubMed] [Google Scholar]
- 28. Goldstein JL, Brown MS (1990) Regulation of the mevalonate pathway. Nature 343: 425–430. [DOI] [PubMed] [Google Scholar]
- 29.Goodman WG, Cusson M (2012) The juvenile hormones. In: Gilbert JL editor. Insect Endocrinology. London: Academic Press. pp. 310–365. [Google Scholar]
- 30. Laufer H, Landau M, Homola E, Borst D (1987) Methyl farnesoate - its site of synthesis and regulation of secretion in a juvenile crustacean. Insect Biochem 17: 1129–1131. [Google Scholar]
- 31. Burtenshaw S, Su P, Zhang J, Tobe S, Dayton L, et al. (2008) A putative farnesoic acid O-methyltransferase (FAMeT) orthologue in Drosophila melanogaster (CG10527): relationship to juvenile hormone biosynthesis? Peptides 29: 242–251. [DOI] [PubMed] [Google Scholar]
- 32. Niwa R, Niimi T, Honda N, Yoshiyama M, Itoyama K, et al. (2008) Juvenile hormone acid O-methyltransferase in Drosophila melanogaster . Insect Biochem Mol Biol 38: 714–720. [DOI] [PubMed] [Google Scholar]
- 33. Marchal E, Zhang J, Badisco L, Verlinden H, Hult EF, et al. (2011) Final step in juvenile hormone biosynthesis in the desert locust, Schistocerca gregaria . Insect Biochem Mol Biol 41: 219–227. [DOI] [PubMed] [Google Scholar]
- 34. Sousa CS, Serrão JE, Bonetti AM, Marques I, Amaral R, et al. (2013) Insights into the Melipona scutellaris fat body transcriptome (Hymenoptera, Apidae, Meliponini). Genet Mol Biol 36: 292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li W, Huang ZY, Liu F, Li Z, Yan L, et al. (2013) Molecular cloning and characterization of juvenile hormone acid methyltransferase in the honey bee, Apis mellifera, and its differential expression during caste differentiation. PLoS One 8: e68544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rachinsky A, Hartfelder K (1991) Differential production of juvenile hormone and its desoxy precursor by corpora allata of honeybees during a critical period of caste development. Naturwissenschaften 78: 270–272. [Google Scholar]
- 37. Ueda H, Shinoda T, Hiruma K (2009) Spatial expression of the mevalonate enzymes involved in juvenile hormone biosynthesis in the corpora allata in Bombyx mori . J Insect Physiol 55: 798–804. [DOI] [PubMed] [Google Scholar]
- 38. Granger NA, Bollenbacher WE, Vince R, Gilbert LI, Baehr JC, et al. (1979) In vitro biosynthesis of juvenile hormone by the larval corpora allata of Manduca sexta . Mol Cell Endocrinol 16: 1–17. [DOI] [PubMed] [Google Scholar]
- 39. Rachinsky A, Hartfelder K (1990) Corpora allata activity, a prime regulating element for caste-specific juvenile-hormone titer in honey bee larvae (Apis mellifera carnica). J Insect Physiol 36: 189–194. [Google Scholar]
- 40. Robinson GE, Strambi C, Strambi A, Feldlaufer MF (1991) Comparison of juvenile hormone and ecdysteroid titers in adult worker and queen honey bees (Apis mellifera). J Insect Physiol 37: 929–935. [Google Scholar]
- 41. Amdam GV, Nilsen KA, Norberg K, Fondrk MK, Hartfelder K (2007) Variation in endocrine signaling underlies variation in social life history. Am Nat 170: 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bitondi MMG, Simões ZLP (2006) The relationship between level of pollen in the diet, vitellogenin and juvenile hormone titres in Africanized Apis mellifera workers. J Apic Res 35: 27–36. [Google Scholar]
- 43. Amdam G, Omholt S (2003) The hive bee to forager transition in honey bee colonies: the double repressor hypothesis. J Theor Biol 223: 451–464. [DOI] [PubMed] [Google Scholar]
- 44. Guidugli KR, Nascimento AM, Amdam GV, Barchuk AR, Omholt S, et al. (2005) Vitellogenin regulates hormonal dynamics in the worker caste of a eusocial insect. FEBS Lett 579: 4961–4965. [DOI] [PubMed] [Google Scholar]
- 45. Nelson C, Ihle K, Fondrk M, Page R, Amdam G (2007) The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biology 5: 673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marco Antonio DS, Guidugli-Lazzarini KR, do Nascimento AM, Simoes ZLP, Hartfelder K (2008) RNAi-mediated silencing of vitellogenin gene function turns honeybee (Apis mellifera) workers into extremely precocious foragers. Naturwissenschaften 95: 953–961. [DOI] [PubMed] [Google Scholar]
- 47. Chandrasekaran S, Ament SA, Eddy JA, Rodriguez-Zas SL, Schatz BR, et al. (2011) Behavior-specific changes in transcriptional modules lead to distinct and predictable neurogenomic states. Proc Natl Acad Sci USA 108: 18020–18025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ament SA, Wang Y, Chen CC, Blatti CA, Hong F, et al. (2012) Bee social behavior and behavior-related gene expression. PLoS Genetics 8: e1002596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schulz D, Huang Z, Robinson G (1998) Effects of colony food shortage on behavioral development in honey bees. Behav Ecol Sociobiol 42: 295–303. [Google Scholar]
- 50. Toth A, Robinson GE (2005) Worker nutrition and division of labour in honeybees. Anim Behav 69: 427–435. [Google Scholar]
- 51. Ament SA, Corona M, Pollock HS, Robinson GE (2008) Insulin signaling is involved in the regulation of worker division of labor in honey bee colonies. Proc Natl Acad Sci USA 105: 4226–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–86. [DOI] [PubMed] [Google Scholar]
- 53. Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, et al. (2000) Artemis: sequence visualization and annotation. Bioinformatics 16: 944–945. [DOI] [PubMed] [Google Scholar]
- 54. Lourenço AP, Mackert A, Cristino AD, Simões ZLP (2008) Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie 39: 372–385. [Google Scholar]
- 55. Huang ZY, Robinson GE, Borst DW (1994) Physiological correlates of division of labor among similarly aged honey bees. J Comp Physiol A 174: 731–739. [DOI] [PubMed] [Google Scholar]
- 56. Goodman WG, Coy DC, Baker FC, Xu L, Toong YC (1990) Development and application of a radioimmunoassay for the juvenile hormones. Insect Biochem 20: 357–364. [Google Scholar]
- 57. Hartfelder K, Bitondi MMG, Brent CS, Guidugli-Lazzarini KR, Simões ZLP, et al. (2013) Standard methods for physiology and biochemistry research in Apis mellifera . J Apic Res 52 1: 47. [Google Scholar]
- 59. Amdam GV, Simões ZL, Guidugli KR, Norberg K, Omholt SW (2003) Disruption of vitellogenin gene function in adult honeybees by intra-abdominal injection of double-stranded RNA. BMC Biotechnology 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
JH-III biosynthetic pathway (modified from Bellés et al. 2005 Annu Rev Entomol 50: 181–99, and Noriega et al. 2006 Insect Biochem Mol Biol 36:11 366–374). The underlined enzymes correspond to the genes whose expression is shown in Figure 1B–E .
(TIF)
(A) Representation of the A. mellifera genes encoding enzymes of the JH biosynthetic pathway. Exons and introns are represented by non-scaled boxes and lines, respectively. Nucleotide numbers are indicated to provide a general estimation of gene size. The direction of transcription is indicated by an arrow. (B) Architecture of the genes encoding Farnesyl diphosphate synthase (FPPS). Exons are named by their respective lengths. Lines between exons indicate intron and their respective length is indicated. The direction of transcription is indicated by a black arrow. The dashed arrows indicate that part of the sequence is not available in the honey bee genome database (version 4). Arrowheads indicate primers position.
(EPS)