Abstract
Regulation of gene expression is mediated by diverse RNA binding proteins which play important roles in development and defense processes. Pumilio/FBF (Puf) protein in mammals functions as a posttranscriptional/translational repressor by binding to the 3′ UTR regions of its target mRNAs. Previous study reported that APUM5 provides protection against CMV infection by directly binding to CMV RNAs in Arabidopsis. CMV RNAs contain putative Pumilio-binding motifs and APUM5 bound to the 3′ UTR and some of its internal motifs both in vitro and in vivo. APUM5 works as a negative regulator of the 3′ UTR of CMV and it might regulate CMV replication. Our findings suggest that APUM5 acts as a defensive repressor in plants during CMV infection. However, functions of APUM5 and other APUM members are still not clear and more studies are needed to find out the interacting partners and target mRNAs in host plant.
Keywords: hunchback(hb), Cucumber mosaic virus (CMV), Pumilio homology domain (PHD), Pumilio/FBF (Puf) protein, RNA-binding proteins (RBPs)
Pumilio/FBF (Puf) proteins have an evolutionary conserved Pumilio homology domain (PHD) and are found in organisms as diverse as protozoa, fungi, mammals and plants.1-3 Puf is proposed to function as a posttranscriptional/translational repressor by interacting with sequence-specific motifs in the 3′ UTR of its target mRNAs4 or by associating with rRNA processing, possibly in a sequence non-specific manner in the nucleolus.5,6 The targets of Puf proteins are involved in differentiation, development, cell cycle, stem cell renewal and synaptic functions.7-9 In plant, Puf-like proteins are encoded by more copies of genes than mammalian Puf proteins. For example, Arabidopsis and rice genomes contain 25 and 19 putative Puf genes whereas human, Drosophila, C. elegans and yeast genomes contain two, one, nine and six Puf genes, respectively. In a bioinformatics analysis, the expression of some plant Puf genes increased in response to biotic and abiotic stresses and exhibited tissue-dependent differential expression.10This demonstrates that plant Puf proteins could play diverse roles in plant development, differentiation and biotic and abiotic stresses.
Recently, targets of the Arabidopsis Puf protein were selected by yeast three-hybrid (Y3H) screening using APUM2 as bait, and five target mRNA sequences were found.11Unexpectedly, these candidate genes were not associated with well-known target genes, such as those involved in development, stem cell renewal and differentiation. One of the candidate target mRNAs was RD19 (At4g39090), RESPONSIVE TO DEHYDRATION 19, which responds to desiccation as an abiotic stress.11 RD19 interacts with the Ralstonia solanacearum type III effector PopP2.12 In terms of biotic stress regulation, the direct binding of mammalian Puf proteins to target genes has not yet been reported. However, the expression of 13 genes that encode proteins involved in antibacterial and antifungal activity was upregulated in Drosophila Pum mutant, as determined by genome-wide analysis,13 although homologs of these genes do not exist in Arabidopsis.
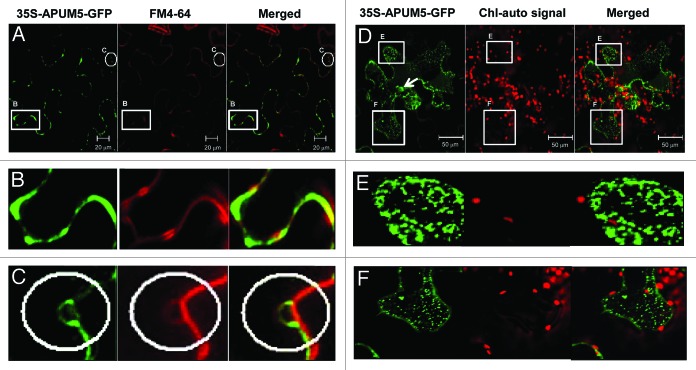
Previous studies indicated that some Pufs localize predominantly to the cytoplasm, but diverse Pufs are also known to localize to multiple subcellular organelles.14-16 CMV RNAs replicate in the vacuolar membranes and tonoplasts by means of replicase protein complexes.17,18 In our study, some co-localization events were detected between APUM5-GFP and FM4-64 in the plasma membrane (Fig. 1A and B) and tonoplast-like structures (Fig. 1A and C). However, some of the regions labeled with APUM5-GFP did not coincide with FM4-64 labeling (Fig. 1A). In addition, some of the APUM5-GFP signal was detected in vesicle-like structures and in parts of the nucleus (Fig. 1D-F). Thus, APUM5 might have other functions to regulate endogenous target mRNAs because APUM5 localizes to multiple subcellular organelles.
Figure 1. Subcellular Localization of APUM5-GFP in N. benthamiana. (A) C-terminal GFP-tagged APUM5 was transiently expressed in N. benthamiana. The FM4-64 stain was used as a plasma membrane marker. Merged image shows colocalized regions. Scale bars are indicated at the pictures. (B) APUM5-GFP signals were merged with FM4-64 in the plasma membrane. (C) APUM5-GFP signals were merged with FM4-64 in vacuolar-like membrane. (Band C) Images are enlarged images of those indicated as boxes and circles in the (A) image. (D) APUM5-GFP was localized to vesicle-like structures and nucleus. The arrow indicates the nucleus. Red signals are chloroplast-auto signals. Scale bars are indicated at the pictures. (EandF) Enlarged images of those indicated by boxes in (D).
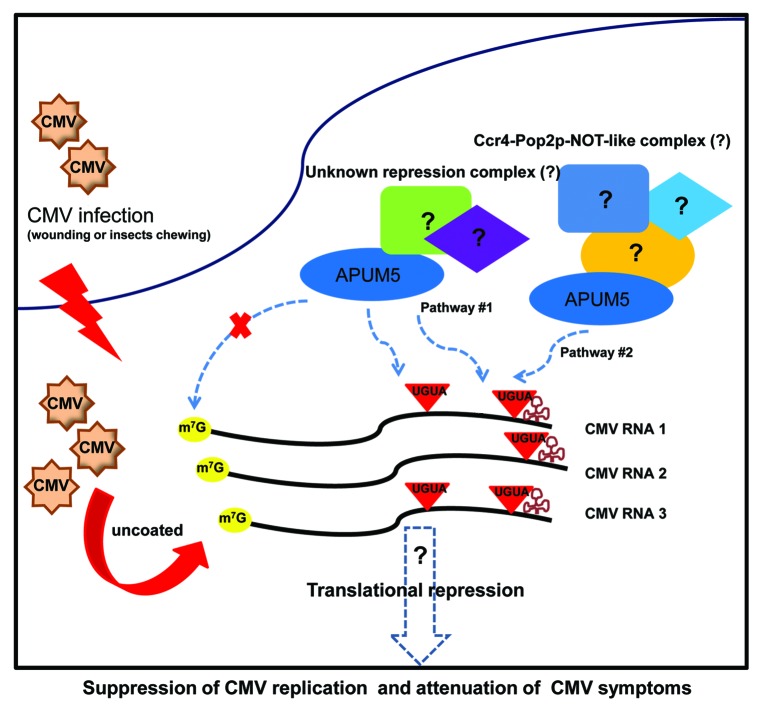
APUM5 bound to the putative Pumilio-binding motifs in the CMV RNAs and hbNRE2.19 Drosophila Pum binds to hbNRE2 mRNA and represses its ability to regulate embryo development via deadenlyase-dependent and -independent pathways.20,21 This indicates that APUM5 might be associated with deadenylase-dependent and -independent pathways to repress CMV RNAs, even though CMV has a tRNA-like structure (TLS) instead of a poly(A) tail at its 3′ end.22 Interestingly, only CMV group-I but not CMV group-II strains contained conserved Pumilio-binding motifs at the 3′ UTR. Furthermore, some internal, putative Pumilio-binding motifs of CMV RNAs could also serve as targets for the repression of CMV infection (Fig. 2).
Figure 2. Proposed working model for APUM5 suppression of CMV infection. First, CMV entry into the host cells occurs by mechanical wounding or insects chewing. Then CMV RNAs replicate and synthesize viral proteins. CMV RNAs are stabilized by a tRNA-like structure (TLS) which is recognized by tRNA-specific enzymes. APUM5 recognizes the CMV 3′ UTR and some internal Pumilio binding motifs in the CMV genome either with unknown repression complexes or in association with Ccr4-Pop2p-NOT-like complex. Finally, APUM5 suppresses CMV infection via the direct binding of viral RNAs and attenuates CMV symptoms.
mRNA repression by yeast Puf3, 4 and 5, human Pum, Drosophila Pum and C. elegans FBF seems to follow a general regulatory mechanism.4,23 For example, yeast Puf5 directly interacts with Pop2p and forms the Ccr4-Pop2p-NOT mRNA deadenylase complex that then attacks the 3′ UTR of target mRNA.4,24 We investigated the Arabidopsis homologs of mammalian Pop2p deadenylase. Pop2p is also known as carbon catabolite repressor 4-CCR4-associated factor1 (CCR4-CAF1) and catalyzes mRNA deadenylation.25 The CAF1 family in Arabidopsis has 11 members, some of which respond to environment and biotic stresses.26 AtCAF1a and AtCAF1b have active involvement in biotic and abiotic stresses.26 Thus, Arabidopsis APUM5 might interact with similar partners that are associated with deadenylation in planta through a more general model, as suggested by the Puf interaction with the Ccr4-Pop2p-NOT complex. Unlike Xenopus Pum2, APUM5 did not bind to the 5′ m7G cap structure for blocking the assembly of the initiation complex (Fig. 2).19,27 Thus, APUM5 might inhibit CMV infection together with unknown repression complexes.
Puf protein is very useful to regulate target RNA because of sequence-specific binding capacity. PHD of Puf could be engineered to specifically recognize different Pumilio binding site in animal system.28-30 However, plant Puf protein function is still not fully known compared with animal Pufs. Thus, more accurate analysis of the function of APUM5 and other APUM family needs to be done in plant. And further finding of its endogenous mRNA targets and de novo interacting partners could also be helpful in dissecting the function of plant Puf proteins.
Acknowledgment
This work was supported by the Wujangchoon Project (PJ007850) from the Rural Development Administration, Republic of Korea.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/23975
References
- 1.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3’UTR regulation as a way of life. Trends Genet. 2002;18:150–7. doi: 10.1016/S0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 2.Wharton RP, Aggarwal AK. mRNA regulation by Puf domain proteins. Sci STKE. 2006;2006:pe37. doi: 10.1126/stke.3542006pe37. [DOI] [PubMed] [Google Scholar]
- 3.Quenault T, Lithgow T, Traven A. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011;21:104–12. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–9. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 5.Droll D, Archer S, Fenn K, Delhi P, Matthews K, Clayton C. The trypanosome Pumilio-domain protein PUF7 associates with a nuclear cyclophilin and is involved in ribosomal RNA maturation. FEBS Lett. 2010;584:1156–62. doi: 10.1016/j.febslet.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abbasi N, Kim HB, Park NI, Kim HS, Kim YK, Park YI, et al. APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2010;64:960–76. doi: 10.1111/j.1365-313X.2010.04393.x. [DOI] [PubMed] [Google Scholar]
- 7.Szakmary A, Cox DN, Wang Z, Lin H. Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr Biol. 2005;15:171–8. doi: 10.1016/j.cub.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Vessey JP, Schoderboeck L, Gingl E, Luzi E, Riefler J, Di Leva F, et al. Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc Natl Acad Sci USA. 2010;107:3222–7. doi: 10.1073/pnas.0907128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Archer SK, Luu VD, de Queiroz RA, Brems S, Clayton C. Trypanosoma brucei PUF9 regulates mRNAs for proteins involved in replicative processes over the cell cycle. PLoS Pathog. 2009;5:e1000565. doi: 10.1371/journal.ppat.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tam PP, Barrette-Ng IH, Simon DM, Tam MW, Ang AL, Muench DG. The Puf family of RNA-binding proteins in plants: phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 2010;10:44. doi: 10.1186/1471-2229-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francischini CW, Quaggio RB. Molecular characterization of Arabidopsis thaliana PUF proteins--binding specificity and target candidates. FEBS J. 2009;276:5456–70. doi: 10.1111/j.1742-4658.2009.07230.x. [DOI] [PubMed] [Google Scholar]
- 12.Bernoux M, Timmers T, Jauneau A, Brière C, de Wit PJ, Marco Y, et al. RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell. 2008;20:2252–64. doi: 10.1105/tpc.108.058685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci USA. 2006;103:4487–92. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu W, Deng Y, Zenklusen D, Singer RH. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004;18:1452–65. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Rodríguez LJ, Gay AC, Pon LA. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J Cell Biol. 2007;176:197–207. doi: 10.1083/jcb.200606054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dallagiovanna B, Correa A, Probst CM, Holetz F, Smircich P, de Aguiar AM, et al. Functional genomic characterization of mRNAs associated with TcPUF6, a pumilio-like protein from Trypanosoma cruzi. J Biol Chem. 2008;283:8266–73. doi: 10.1074/jbc.M703097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huh SU, Kim MJ, Ham BK, Paek KH. A zinc finger protein Tsip1 controls Cucumber mosaic virus infection by interacting with the replication complex on vacuolar membranes of the tobacco plant. New Phytol. 2011;191:746–62. doi: 10.1111/j.1469-8137.2011.03717.x. [DOI] [PubMed] [Google Scholar]
- 18.Cillo F, Roberts IM, Palukaitis P. In situ localization and tissue distribution of the replication-associated proteins of Cucumber mosaic virus in tobacco and cucumber. J Virol. 2002;76:10654–64. doi: 10.1128/JVI.76.21.10654-10664.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huh SU, Kim MJ, Paek KH. Arabidopsis Pumilio protein APUM5 suppresses Cucumber mosaic virus infection via direct binding of viral RNAs. Proc Natl Acad Sci USA. 2013;110:779–84. doi: 10.1073/pnas.1214287110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chagnovich D, Lehmann R. Poly(A)-independent regulation of maternal hunchback translation in the Drosophila embryo. Proc Natl Acad Sci USA. 2001;98:11359–64. doi: 10.1073/pnas.201284398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wreden C, Verrotti AC, Schisa JA, Lieberfarb ME, Strickland S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997;124:3015–23. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- 22.Rao AL. Genome packaging by spherical plant RNA viruses. Annu Rev Phytopathol. 2006;44:61–87. doi: 10.1146/annurev.phyto.44.070505.143334. [DOI] [PubMed] [Google Scholar]
- 23.Campbell ZT, Menichelli E, Friend K, Wu J, Kimble J, Williamson JR, et al. Identification of a conserved interface between PUF and CPEB proteins. J Biol Chem. 2012;287:18854–62. doi: 10.1074/jbc.M112.352815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Etten J, Schagat TL, Hrit J, Weidmann CA, Brumbaugh J, Coon JJ, et al. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J Biol Chem. 2012;287:36370–83. doi: 10.1074/jbc.M112.373522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–86. doi: 10.1016/S0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 26.Walley JW, Kelley DR, Nestorova G, Hirschberg DL, Dehesh K. Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol. 2010;152:866–75. doi: 10.1104/pp.109.149005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao Q, Padmanabhan K, Richter JD. Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA. 2010;16:221–7. doi: 10.1261/rna.1884610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choudhury R, Tsai YS, Dominguez D, Wang Y, Wang Z. Engineering RNA endonucleases with customized sequence specificities. Nat Commun. 2012;3:1147. doi: 10.1038/ncomms2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filipovska A, Rackham O. Designer RNA-binding proteins: New tools for manipulating the transcriptome. RNA Biol. 2011;8:978–83. doi: 10.4161/rna.8.6.17907. [DOI] [PubMed] [Google Scholar]
- 30.Cooke A, Prigge A, Opperman L, Wickens M. Targeted translational regulation using the PUF protein family scaffold. Proc Natl Acad Sci USA. 2011;108:15870–5. doi: 10.1073/pnas.1105151108. [DOI] [PMC free article] [PubMed] [Google Scholar]