Abstract
Scaffold proteins are known to regulate important cellular processes by interacting with multiple proteins to modulate molecular responses. RACK1 (Receptor for Activated C Kinase 1) is a WD-40 type scaffold protein, conserved in eukaryotes, from Chlamydymonas to plants and humans, expresses ubiquitously and plays regulatory roles in diverse signal transduction and stress response pathways. Here we present the use of Arabidopsis RACK1A, the predominant isoform of a 3-member family, as a bait to screen a split-ubiquitin based cDNA library. In total 97 proteins from dehydration, salt stress, ribosomal and photosynthesis pathways are found to potentially interact with RACK1A. False positive interactions were eliminated following extensive selection based growth potentials. Confirmation of a sub-set of selected interactions is demonstrated through the co-transformation with individual plasmid containing cDNA and the respective bait. Interaction of diverse proteins points to a regulatory role of RACK1A in the cross-talk between signaling pathways. Promoter analysis of the stress and photosynthetic pathway genes revealed conserved transcription factor binding sites. RACK1A is known to be a multifunctional protein and the current identification of potential interacting proteins and future in vivo elucidations of the physiological basis of such interactions will shed light on the possible molecular mechanisms that RACK1A uses to regulate diverse signaling pathways.
Keywords: Arabidopsis, RACK1A, drought, photosynthesis, split-ubiquitin, transcription factor
Introduction
The widely conserved eukaryotic scaffold protein RACK1 (Receptor for Activating C Kinase1) regulates different signal transduction pathways ranging from cell division to environmental stress responses by interacting with diverse proteins.1-5 The protein was identified through its ability to function as a scaffold protein, stabilizing signaling complexes involving protein kinase C.6 RACK1 proteins with seven WD-40 repeats are highly conserved (70–80% at the protein level) in a wide range of species, including plants, humans, rats, chickens, flies, nematodes, algae and yeasts. Though plant RACK1 has been implicated in regulating diverse signaling pathways that include but are not limited to, drought and salt stress, growth hormones, innate immunity and rare sugars,4,7,8 compared with the metazoan domain, virtually no information is available about its interacting proteins. So far the Biomolecular Interaction Network Database9 reports that metazoan RACK1 interacts with 87 different proteins ranging from ion channels to diverse ribosomal proteins. However RACK1 has not been reported to be used as bait in any conventional yeast two hybrid (Y-2-H) based library screens where baits are required to transactivate reporter gene expression only after interacting with the prey. The reported RACK1 interacting proteins were identified, in most cases, when the respective protein was used as bait to detect RACK1 as the prey partner (BIND database). As Y-2-H based protein-protein interaction requires reconstitution of a split transcription factor in the yeast nucleus, frequently proteins with transactivation domains active in yeast are excluded as bait. The yeast split-ubiquitin system has been developed to overcome the limitations of the classical Y-2-H system.10,11 The split-ubiquitin method is based on the ability of Nub and Cub, the N- and C-terminal halves of ubiquitin fused to a bait or to a prey separately, to assemble into a quasi-native ubiquitin (Ub).10,11 Ub specific proteases recognize the reconstituted Ub, though not its halves, and cleave the Ub moiety off a reporter protein, which has been linked to the C-terminus of Cub. The release of the reporter serves as readout indicating interactions. As a consequence, the yeast cells lose their capability to grow in the absence of uracil, and thus permits selection in the presence of the otherwise toxic URA3p specific antimetabolite 5-fluoroorotic acid (5-FOA). This technique offers major advantages over classical yeast two-hybrid screen in that it can be modified to identify both cytosolic and membrane proteins, and to identify transient interactions.10,11
Due to the inability of using RACK1 as bait in a traditional yeast two hybrid screen, the repertoire of the RACK1 interacting proteins may actually be much larger than the subset currently identified. To ascertain for a potentially comprehensive functional role of RACK1 protein, it is imperative that we use a system where RACK1 could be used as bait to screen a random cDNA library. Here we report the use of Arabidopsis RACK1A as bait to screen for potential interacting proteins. As opposed to a single gene in metazoan, Arabidopsis genome maintains three different RACK1 genes—termed as RACK1A, RACK1B and RACK1C. RACK1A—the predominant member with an unequal redundancy effect has been extensively studied.4,7,8 We have employed the split-ubiquitin based system that does not require the nuclear transactivation process, a central impediment to use RACK1 as bait in the traditional Y-2-H library screens.10,11 The identification of more than 90 Arabidopsis RACK1A interacting proteins from the screen indicates the efficacy of the split-ubiquitin based library screen.
Results and Discussion
As a scaffold protein, RACK1 connects distinct signaling pathways to regulate important cellular activities including cell growth and proliferation, transcription and protein synthesis.5 Unlike animal RACK1, which is encoded by a single copy gene in the respective genomes, the presence of more than one copy of RACK1 in most plant species provides multiple opportunities for RACK1 based protein-protein interaction signaling modules. In this study, a split-ubiquitin based Arabidopsis inflorescence cDNA library was used to systematically screen for RACK1A interacting proteins. The split-ubiquitin based cDNA library screen allows to overcome problems associated with the auto-activation properties of RACK1 when used as bait in conventional Y-2-H screens as this screen does not depend on the transactivation capabilities of the bait.10 Any protein interacting with RACK1A would be recognized as the interaction mode would reconstitute a functional ubiquitin protein tagged with the URA3 reporter protein. Proteolytic degradation of the URA3 reporter protein renders the yeast cells URA-auxotroph and resistant to the counter selection agent 5-FOA induced toxicity. Non-interacting clones with the functional URA3 will not be able to survive the FOA toxicity. Following the scheme, several rounds of screens were performed using RACK1A as bait to identify potential interacting proteins. RACK1 Y248 residue phosphorylation has been implicated in many cellular functions (4). In order to discern whether RACK1A Y248 phosphorylation is required for any potential interaction, we have also used a site-directed mutagenesis construct Y248F-RACK1A, as bait. As can be seen from Figure 1, the RACK1A bait appeared to potentially interact with diverse proteins as FOA resistant colonies were found to grow on FOA selection plates (Fig. 1, upper panel right), whereas, when Y248F-RACK1A was used as bait, no FOA resistant colonies were found (Fig. 1, lower panel right) indicating that tyrosine phosphorylation on the Y248 residue may dictate any potential interaction under the conditions used. The successful yeast transformation process is evident from the growth of yeast colonies harboring both the bait (WT and mutant) and prey constructs on selection plates lacking both His and Trp (Fig. 1, upper and lower panel left). We are cognizant that bait stability is a concern for this kind of library screen. URA-prototrophy was examined by allowing the bait clone to grow on plates lacking uracil (data not shown). As the yeast colonies harboring the baits were able to grow on plates lacking uracil, we used the baits in the respective screening process.
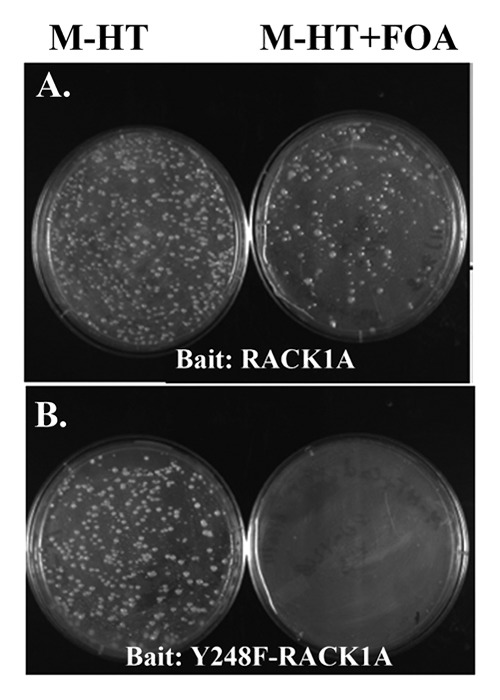
Figure 1.Arabidopsis cDNA library screen using the Split Ubiquitin Assay. Arabidopsis cDNA library screen was done with split ubiquitin assay using Arabidopsis RACK1A (upper panel) or Y248F-RACK1A (lower panel) as a bait. The left sides of the each panels show the yeast colonies transformed with the bait (-His) and the prey library (-trp). One tenths of the transformed colonies are grown on yeast minimal media lacking histidine and tryptophan. (Left sides of each panels) and the rest of the transformed colonies are plated on minimal media lacking histidine and tryptophan but with 0.5 mg/ml of FOA added (right sides of both panels). The image was taken after 48 h of growth at 30°C.
Like in other genetic and conventional Y-2-H screens, the split-ubiquitin system has a tendency to result in false positives that may arise from several distinct mechanisms during the initial growth selection process. Promoter mutations to activate the reporter gene even in the absence of any potential interaction have been cited to give rise to false positives.11 In addition, overexpression of certain proteins in yeast may non-specifically activate the reporter genes. However, these false positives can be partly eliminated using a set of genetic criteria that can be rapidly tested. Therefore, based on the published optimized procedure to screen a split ubiquitin library,11 a methodological scheme was undertaken to eliminate potential false positives. The FOA-resistant clones were re-streaked on minimal plate lacking His, Trp and Ura (M-HTU). Only the true interactors where the reporter URA3 clone is proteolytically degraded were not able to grow on the M-HTU plates (data not shown). The URA3 DNA sequence from a few of the selected clones was also checked for any mutations that may render such clones FOA-resistant. The low rate of mutations in the URA3 provided confirmation for the elimination of certain false positives by this scheme.
To identify the interacting partners, specific PCR primers targeting the prey protein constructs were used to rescue the plasmid. A typical result from the PCR reactions to identify the interacting proteins is shown in Figure 2. The amplified PCR products were sequenced to reveal the identity of the encoded interacting proteins. Some of the FOA resistant colonies did not produce any PCR bands; most likely being either false positives or due to failure of the PCR reaction. However, out of almost 150 PCR bands, positive identifications of 97 interacting proteins were completed by DNA sequencing (Table1; Table S1). Many of the key interacting proteins from the photosynthetic pathways were identified more than once, indicating that the library screen has been successful in eliminating potential false positives.
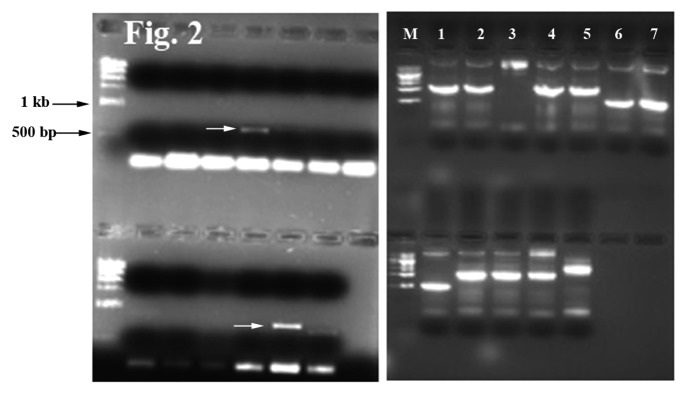
Figure 2. Amplified plasmid DNA from the prey colonies. DNA from FOA resistant colonies were PCR amplified using prey specific primers as described in the Methods section. The arrows in the left panel indicate amplification from interacting clones harboring CP12 (arrow on the upper half lanes) and Rubisco small subunit 1A (arrow on the lower half lanes) sequences. The right panel shows PCR bands from reactions using yeast colonies harboring interacting bait and prey plasmids. The PCR was done in duplicates or in triplicates. Upper half: lane 1 and 2, LHCA3 bands; lanes 4 and 5, LHCA2; lanes 6 and 7, ribosomal protein L29-1. Lower half: lane 1, ribosomal protein L29-1; lanes 4–6, aquaporin (later discarded as con-transformation failed to result in any FOA resistant colonies); lane 7, LHCA2. Different sizes of the PCR products indicate different prey constructs containing DNA inserts for distinct proteins. Overall the PCR band sizes ranged from 400–1,400 bp.
Table 1. List of multifunctional interacting partners of RACK1A.
| Locus name | Name of the protein | Function | Reference |
|---|---|---|---|
| AT1G67090 | Rubisco small subunit 1A | Oxidative and drought stress, photosynthesis | 17 |
| AT3G62030 | Peptidyl-prolyl cis-trans isomerase CYP20-3 | Stress response | 25 |
| AT3G13520 | Arabinogalactan protein 21 | Salt stress | 15 |
| AT1G08830 | Superoxide dismutase [Cu-Zn], CSD1 | Oxidative stress response | 14 |
| AT2G02850 | Plantacyanin | Stress response, defense | 26 |
| AT3G01500 | Carbonic anhydrase 1,CA1 | Stress response | 27 |
| AT5G04140 | Fd-GOGAT | Photorespiration | 18 |
| AT1G20340 | Plastocyanin major isoform, DRT112 | Photosynthesis | 28 |
| AT1G29930 | Chlorophyll a-b binding protein 1CAB1 | Photosynthesis | 29 |
| AT2G34420 | Light-harvesting complex II chlorophyll a/b binding protein 1 | Photosynthesis | 20 |
| AT5G20010 | GTP-binding nuclear protein Ran-1 | Protein import into the nucleus | 30 |
| AT1G06190 | Rho termination factor | Transcription termination | 31 |
| AT4G21660 | Splicing factor 3B subunit 4 | RNA splicing | 32 |
Selected proteins from the stress and photosynthetic pathways are presented. The functional categorization was ascertained by the entry of the references on the PubMed database. The complete list of identified interacting proteins is presented as a supplementary file.
Nine of the potential interacting proteins representing different classes of the identified proteins were tested for their ability as prey to interact with the bait RACK1A in a one-to-one split ubiquitin based interaction assay. As the Y248F-RACK1A bait failed to show any potential interactions (Fig. 1B), we used the bait as a negative control in the co-transformation assay. Figure 3A–C show the results from the representative selection plates with seven of the nine tested interactions. Both the individual bait (WT RACK1A or mutant Y248F-RACK1A) and the prey constructs: Chlorophyll a/b binding protein1 (CAB1) (lane 1); Plantacyanin (lane 2); Copper/Zinc Superoxide Dismutase 1, CSD1 (lane 3); Plastocyanin (lane 4); Rubisco small subunit 1A (lane 5); Carbonic anhydrase1 (lane 6); and Prolyl cis-trans isomerase (CYP20-3) (lane 7), were being expressed on the M-HT selection plate (Fig. 3A). When tested on M-HT+FOA plate, only the WT-RACK1A bait and individual prey harboring yeast colonies were able to grow, confirming their interaction (Fig. 3B). However, no colony was able to grow to a significant level on the same plate, indicating lack of or very weak interaction with the mutant bait (Fig. 3B). To further confirm that the growth from the M-HT+FOA plates resulted from the interaction based degradation of the reporter protein URA3, the same colonies from the FOA plates were tested on the selection plate lacking any exogenously supplemented uracil (Fig. 3C). As expected, the non-interacting bait and prey harboring yeast colonies were able to grow on the uracil lacking plates-indicating the maintenance of a functional URA3 protein; whereas the FOA resistant colonies were not able to survive on the plate. Similar results from additional plates were obtained with the Photosystem I Light Harvesting Complex Gene 2 (LHCA2) and Photosystem I Light Harvesting Complex Gene 3 (LHCA3) proteins. By considering both the positive and negative selection results from the represented interactions, it can be concluded that the screen employed to identify RACK1A protein interactors has been quite successful.
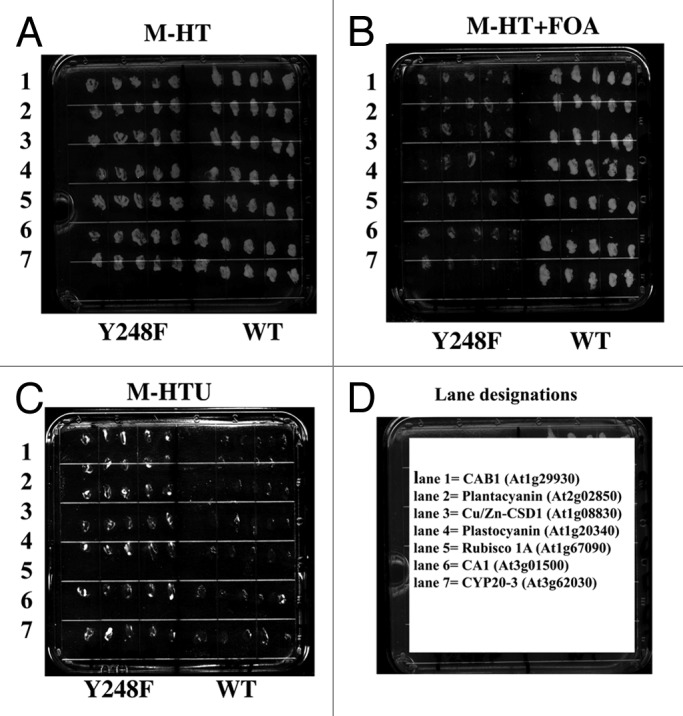
Figure 3. Interaction of a sub-set of proteins is reconfirmed by co-transformation of the individual bait and prey vectors. Individual split ubiquitin prey vector harboring DNA sequences from respective interacting proteins representing different classes of the total interacting protein sets and RACK1A bait (pMKZ-RACK1A) were co-transformed and five colonies are plated on minimal plates lacking histidine and tryptophan (M-HT) (A). As a negative control Y248F-RACK1A DNA harboring bait (pMKZ-Y248F-RACK1A) was used with the respective prey vectors. The prey vectors shown here are: 1, CAB1 (AT1G29930); 2, Plantacyanin (AT2G02850); 3, Cu/Zn-CSD1 (AT1G08830); 4, Plastocyanin (AT1G20340); 5, Rubisco small subunit 1A (AT1G67090); 6, Carbonic anhydrase (AT3G01500); and 7, Peptidyl-prolyl cis-trans isomerase, CYP20-3 (AT3G62030). Positive interactions with WT but not with Y248F baits are evident from FOA resistant growth (B). Lack of any growth on uracil lacking selection plate (M-HTU) from the FOA resistant colonies of (B) but growth of FOA sensitive colonies from the same panel reinforces results (C). The images were taken after 48 h of growth at 30°C. The lane designations are shown in (D).
Conforming to RACK1A’s role in the environmental stress response pathways,4 26% of the interacting partners are found to be directly or indirectly involved with environmental stress signaling pathways (Fig. 4). Also 14% of interacting proteins were found to be ribosomal proteins, supporting the reported localization of RACK1 in the ribosome to regulate global translation.12,13 Moreover, 18% of RACK1A interacting proteins have coordinated roles in regulating photosynthesis and light regulated physiological processes (Table 1). In addition, 30% of interacting proteins were found from different cellular functional categories, including transcriptional regulation, protein folding and RNA splicing. Several interacting partners appear to be unknown or novel (8%) and 4% of the interacting proteins are related to pathogen defense.
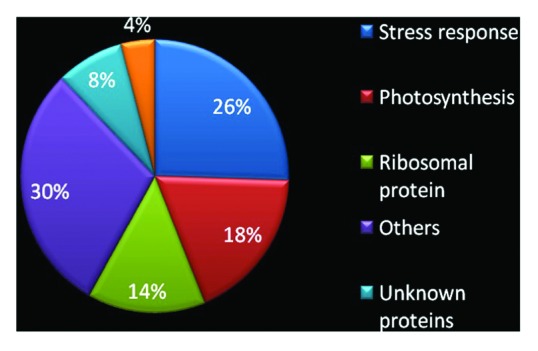
Figure 4. Pie chart representing functional categories of RACK1A interacting proteins. The total of 97 interacting proteins is categorized according to their functional attributes. The color coded pie-chart shows the percentage of proteins that interact with RACK1A from each category.
Interestingly, one of the interacting partners found in the cDNA library screen using RACK1A as bait was a Cu-Zn SOD an important enzyme to regulate the reactive oxygen species within the cell. In plants, Cu-Zn SOD is found in the cytosol and in the extracellular space as well as in the chloroplast. Cu-Zn SODs are important regulators of many of the environmental stress responses including drought, salt, flooding and pathogen infection pathways.14 It is conceivable that by potentially interacting with Cu-Zn SOD, RACK1A can negatively regulate the specific SOD function and, under stress conditions, this negative regulation can be relieved to allow the specific SOD enzyme to detoxify generated reactive oxygen species. Another important stress responsive protein found from the library screen was Arabinogalactan-protein9 (AGP9). AGPs play a crucial role in salt-stressed cells as massive upregulation of AGPs has been reported under such conditions.15 Photorespiration is one of the important plant metabolic processes that is activated significantly during drought and high temperature. Ribulose-1, 5-bisphosphate carboxylase oxygenase (Rubisco) is a key enzyme of the photorespiration process.16 The putative interaction of RACK1A with Rubisco subunits suggests a role for RACK1A in photorespiration, thus indirectly implicated in the drought response. It is possible that inactivation of RACK1A may provide plant with avenues to channel its excess absorbed energy during drought/high light stress condition by channeling the excess energy load toward the photorespiration pathway.4,17 On the other hand, ferredoxin-dependent glutamate synthase (Fd-GOGAT) also plays a major role in photorespiration in Arabidopsis.18 Thus, the putative interaction of RACK1A with Fd-GOGAT also implies that RACK1A functions in photorespiration and drought response. Another potential interacting protein identified during the library screen was peptidyl-prolyl cis-trans isomerase, which is also implicated in regulating drought responses.19 In drought-stressed rice plant, Rubisco were shown to be downregulated whereas peptidyl-prolyl cis-trans isomerase, which might alleviate the damage on Rubisco by drought stress, was upregulated.19 Interaction of RACK1A with many components of the drought stress pathways indicates that the protein may play a key role in coordinating Rubisco, Fd-GOGAT, Cu-Zn SOD and peptidyl-prolyl cis-trans isomerase function under these conditions. The chlorophyll a/b light-harvesting complex of photosystem II (LHC-II) is a conserved family of membrane proteins that has a major role in plant photosynthesis. Additionally, LHC-II also takes part in building the structure of the photosynthetic apparatus and regulates energy flow between the two photosystems.20 On the other hand, Photosystem I Light Harvesting Complex Gene 2 (LHCA2) is linked with photosystem II (PS II). Both LHCA2 and LHCA3 (Photosystem I Light Harvesting Complex Gene 3) are potential interacting partners of RACK1A, suggesting a possible role of RACK1A in the switching of PSI and PSII. The identification of diverse photosynthesis related genes: LHCA2, LHCA3, chlorophyll a/b binding protein 1 (CAB1); photosystem I reaction center subunit XI; photolyase/blue-light receptor 2, PHR2; photosystem I reaction center subunit III; photosystem I reaction center subunit N as potential interacting partner suggests a major role for RACK1A during photosynthesis (Table 1; Table S1). The potential interaction with carbonic anhydrase 1 (CA1) also indicates a role of RACK1A in the transition between C3 and C4 types of photosynthetic processes. In a related study, we have shown that RACK1A homo- and hetero-dimerizes and the tyrosine phosphorylation at Y248 residue is an important post-translational modification that dictates the dimerization event (Kundu N, 2012, PhD dissertation, Howard University). In this study, use of a mutant RACK1A (Y248F) as bait failed to the isolation of any interacting proteins (Fig. 1B). It remains to be seen whether dimerization of RACK1A is a pre-condition for interaction with other proteins.
The promoter regions of the stress and photosynthetic genes were analyzed to uncover conserved transcription factor binding sites (Fig. S1). The Athena database that allows rapid visualization and systematic analysis of Arabidopsis promoter sequences21 reveals three distinct TF sites within the 27 analyzed gene promoters. The ABRE-like binding site motif (C/G/T)ACGTG(G/T)(A/C) known to occur in genes encoding proteins that respond to dehydration and low temperature22 was found in 15 promoters with a p-value of < 10−4. The Ibox promoter motif (GATAAG),23 which is found in the promoters of light regulated genes was found in all of the 16 RACK1A interacting photosynthetic genes. The low statistical p-value of < 0.0040 indicates the significance of this finding. The MYB2AT site (TAACTG), known to be involved in the regulation of genes that are responsive to water-stress,24 is present in 11 of the stress related proteins that interact with RACK1A protein. Considering that the cDNA library was developed from inflorescence tissues, it is quite intriguing that many of the photosynthetic proteins appear to interact with RACK1A. Arabidopsis genome wide studies showed that the green tissues like sepals, carpels, siliques in the inflorescence tissue maintain many of the photosynthetic proteins33
Protein-protein interactions are the dominant modes to virtually regulate every cellular process ranging from DNA replication and translations to cellular stress responses. As a scaffold protein RACK1 brings other proteins together and is therefore directly and/or indirectly involved in many signaling pathways. Here we present a screen for the identification of Arabidopsis RACK1A interacting proteins. Conforming to its published role in the environmental stress response, translational control and developmental pathways, the screen revealed that the majority of RACK1A interacting proteins are related with these pathways. RACK1A was found to be a common interacting partner of many stress responsive proteins indicating a major role in crosstalk among these signaling pathways. Identification of the conserved TF sites within the promoters of genes encoding these interacting proteins should help to elucidation the regulatory mechanisms involved in the RACK1A-mediated environmental stress and photosynthetic pathways. Such knowledge will eventually help in developing biotechnological strategies to develop suitable environmental stress tolerant transgenic crops.
Materials and methods
E. coli and yeast strains
All plasmid constructions and amplifications were performed using the E. coli strain XL1-Blue and DH5α (Stratagene).The genotype of wild type yeast strain used for the split-ubiquitin assays was JD53 (Ura-, Trp-, Leu-, Lys-).
Plasmid Construct
The split-ubiquitin assay was performed in S. cerevisiae strain JD53. Gateway Entry clones were made by insertion of full length RACK1A cDNA into pCR8/GW/TOPO vector (Invitrogen). Entry clones were maintained in the E.coli strain DH5α in LB medium supplemented with spectinomycin (50 μg ml−1). Following recombination, the RACK1A from the Entry clone was transferred to the destination bait vector pMKZ [pMet-KZ::GWY Cassette-Cub-URA3-CYC1 (His)]. Similarly, Arabidopsis cDNAs were maintained in the prey vector pCUP-CGK-pCup-NuI::AttB1-EcoRI-cDNA-polyA-XhoI-AttB2-CYC1 (Trp). The prey vector was selected with ampicillin (100 μg ml−1).
cDNA Library construction
To construct the library, total RNA was isolated from frozen Col-0 floral material using the RNA extraction kit from RNAwiz (Ambion). PolyA+ RNA was purified with the Dynabead Kit and subsequently quantified. For the synthesis of the cDNA library, the cDNA synthesis kit from Statagene was used. Seven mg of polyA+ RNA was used for synthesis following the protocol of the manufacturer. Based on analyzing a random number of individual clones, the average size of the inserts was 1.3 kb. The cDNA library was contained in the pCup-CGK vector. Modified prey vector containing two bacterial selection markers, AmpR, KanR were used. This vector was compatible with the GATEWAY system.
Electroporation
Isolated bait plasmid RACK1A DNA was electroporated into Saccharomyces cerevisiae strain JD53. Transformants were selected on minimal medium plate with yeast nitrogen base without amino acids (Sigma) and glucose, supplemented with lysine, leucine, uracil, tryptophan (M-H) plates was used to select transfected colonies. pMKZ-RACK1 expressing yeast cells were transfected with Arabidopsis cDNA library plasmid DNA (2 μg). All transformants were selected on 5-fluoroorotic acid (5-FOA) plates containing minimal medium with yeast nitrogen base without amino acids (Sigma) and glucose, supplemented with leucine, lysine, uracil (M-HT), copper chloride (100 μM) and 0.5 mg/ml 5-FOA.
PCR amplification
Yeast DNA from FOA-resistant colonies was isolated after treating the cells with 40 U of lyticase (Sigma) for 10 min at 37°C. PCR-based amplification of the isolated DNA was performed in a 50 μl reaction mixture. Amplification of the DNA was performed by using NUI.F: 5′GATTTTCGTCAAGACTTTGACCGGTA3′ as forward and CYC.R: 5′TTTCGGTTAGAGCGGATGTG3′ as reverse primers in a thermocycler with an initial denaturation at 95°C for 2 min, followed by 35 cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec and extension at 72°C for 1 min, with a final extension at 72°C for 15 min. The amplified PCR products were gel purified and were sequenced using the NUI.F as sequencing primer. The PCR products were sequenced by a commercial vendor (Genewiz Inc.). BLAST search was used to reveal the identity of the DNA sequences (GenBank).
Split ubiquitin assay with identified interacting proteins
Nine RACK1A interacting proteins, LHCA2 (At3g61470), LHCA3 (At1g61520) CAB1 (At1g29930), Cu/Zn-CSD1 (At1g08830), Plantacyanin (At2g02850), Rubisco small subunit 1A (At1g67090), Carbonic anhydrase (At3g01500), Plastocyanin (At1g20340) and Peptidyl-prolyl cis-trans isomerase, CYP20-3 (At3g62030) identified in the cDNA library screen were reconfirmed by co-transformation. For this, the full length cDNAs from the respective clones were amplified by PCR and the PCR products were cloned in the pCR8/GW/TOPO entry vector (Invitrogen). Later through recombination, the cDNA is transferred to the prey destination vector, NuI [pCup-NuI-GWY-CYC1 (Trp)]. Plasmid DNA from pMKZ-RACK1A or pMKZ-Y248F-RACK1A bait and from NuI prey vector with the cloned cDNAs were electroporated into the JD53 yeast strain. The ability of the transfected yeast colonies to grow on plates lacking histidine and tryptophan supplemented with 0.5 mg/ml FOA and 100 μM CuCl2 (M-HT+ FOA) was indicative of protein interaction.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Science Foundation, USA to H.U. (MCB 0542312). We acknowledge the service of Ms Aniqa Tasnim for her proof reading the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/psb/article/24012
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/24012
References
- 1.McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharmacol. 2002;62:1261–73. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- 2.Sklan EH, Podoly E, Soreq H. RACK1 has the nerve to act: structure meets function in the nervous system. Prog Neurobiol. 2006;78:117–34. doi: 10.1016/j.pneurobio.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Chen S, Lin F, Shin ME, Wang F, Shen L, Hamm HE. RACK1 regulates directional cell migration by acting on G betagamma at the interface with its effectors PLC beta and PI3K gamma. Mol Biol Cell. 2008;19:3909–22. doi: 10.1091/mbc.E08-04-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ullah H, Scappini EL, Moon AF, Williams LV, Armstrong DL, Pedersen LC. Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana. Protein Sci. 2008;17:1771–80. doi: 10.1110/ps.035121.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams DR, Ron D, Kiely PA. RACK1, A multifaceted scaffolding protein: Structure and function. Cell Commun Signal. 2011;9:22–46. doi: 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mochly-Rosen D, Khaner H, Lopez J. Identification of intracellular receptor proteins for activated protein kinase C. Proc Natl Acad Sci USA. 1991;88:3997–4000. doi: 10.1073/pnas.88.9.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen JG, Ullah H, Temple B, Liang J, Guo J, Alonso JM, et al. RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J Exp Bot. 2006;57:2697–708. doi: 10.1093/jxb/erl035. [DOI] [PubMed] [Google Scholar]
- 8.Fennell H, Olawin A, Mizanur R, Ken I, Chen JG, Ullah H. Arabidopsis scaffold protein RACK1A modulates rare sugar D-allose regulated gibberellin signaling. Plant Signal Behav. 2012;7:1771–80. doi: 10.4161/psb.21995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alfarano C, Andrade CE, Anthony K, Bahroos N, Bajec M, Bantoft K, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33(Database issue):D418–24. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehming N. Analysis of protein-protein proximities using the split-ubiquitin system. Brief Funct Genomic Proteomic. 2002;1:230–8. doi: 10.1093/bfgp/1.3.230. [DOI] [PubMed] [Google Scholar]
- 11.Dirnberger D, Messerschmid M, Baumeister R. An optimized split-ubiquitin cDNA-library screening system to identify novel interactors of the human Frizzled 1 receptor. Nucleic Acids Res. 2008;36:e37. doi: 10.1093/nar/gkm1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shor B, Calaycay J, Rushbrook J, McLeod M. Cpc2/RACK1 is a ribosome-associated protein that promotes efficient translation in Schizosaccharomyces pombe. J Biol Chem. 2003;278:49119–28. doi: 10.1074/jbc.M303968200. [DOI] [PubMed] [Google Scholar]
- 13.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–6. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 14.Kar RK. Plant responses to water stress: role of reactive oxygen species. Plant Signal Behav. 2011;6:1741–5. doi: 10.4161/psb.6.11.17729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamport DT, Kieliszewski MJ, Showalter AM. Salt stress upregulates periplasmic arabinogalactan proteins: using salt stress to analyse AGP function. New Phytol. 2006;169:479–92. doi: 10.1111/j.1469-8137.2005.01591.x. [DOI] [PubMed] [Google Scholar]
- 16.Ramachandra Reddy A, Chaitanya KV, Vivekanandan M. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J Plant Physiol. 2004;161:1189–202. doi: 10.1016/j.jplph.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 17.Marraccini P, Freire LP, Alves GS, Vieira NG, Vinecky F, Elbelt S, et al. RBCS1 expression in coffee Coffea orthologs, Coffea arabica homeologs, and expression variability between genotypes and under drought stress, BioMed. Central. Plant. Biology. 2011;11:85–108. doi: 10.1186/1471-2229-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coschigano KT, Melo-Oliveira R, Lim J, Coruzzi GM. Arabidopsis gls mutants and distinct Fd-GOGAT genes. Implications for photorespiration and primary nitrogen assimilation. Plant Cell. 1998;10:741–52. doi: 10.1105/tpc.10.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji K, Wang Y, Sun W, Lou Q, Mei H, Shen S, et al. Drought-responsive mechanisms in rice genotypes with contrasting drought tolerance during reproductive stage. J Plant Physiol. 2012;169:336–44. doi: 10.1016/j.jplph.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Barros T, Kühlbrandt W. Crystallisation, structure and function of plant light-harvesting Complex II. Biochim Biophys Acta. 2009;1787:753–72. doi: 10.1016/j.bbabio.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 21.O’Connor TR, Dyreson C, Wyrick JJ. Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics. 2005;21:4411–3. doi: 10.1093/bioinformatics/bti714. [DOI] [PubMed] [Google Scholar]
- 22.Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6:410–7. doi: 10.1016/S1369-5266(03)00092-X. [DOI] [PubMed] [Google Scholar]
- 23.Giuliano KA, Khatib FA, Hayden SM, Daoud EW, Adams ME, Amorese DA, et al. Properties of purified actin depolymerizing factor from chick brain. Biochemistry. 1988;27:8931–8. doi: 10.1021/bi00425a009. [DOI] [PubMed] [Google Scholar]
- 24.Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 1993;5:1529–39. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laxa M, König J, Dietz KJ, Kandlbinder A. Role of the cysteine residues in Arabidopsis thaliana cyclophilin CYP20-3 in peptidyl-prolyl cis-trans isomerase and redox-related functions. Biochem J. 2007;401:287–97. doi: 10.1042/BJ20061092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong J, Kim ST, Lord EM. Plantacyanin plays a role in reproduction in Arabidopsis. Plant Physiol. 2005;138:778–89. doi: 10.1104/pp.105.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alver A, Şentürk A, Çakirbay H, Menteşe A, Gökmen F, Keha EE, et al. Carbonic anhydrase II autoantibody and oxidative stress in rheumatoid arthritis. Clin Biochem. 2011;44:1385–9. doi: 10.1016/j.clinbiochem.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Redinbo MR, Yeates TO, Merchant S. Plastocyanin: structural and functional analysis. J Bioenerg Biomembr. 1994;26:49–66. doi: 10.1007/BF00763219. [DOI] [PubMed] [Google Scholar]
- 29.Kargul J, Barber J. Photosynthetic acclimation: structural reorganisation of light harvesting antenna-role of redox-dependent phosphorylation of major and minor chlorophyll a/b binding proteins. FEBS. 2008;275:1056–68. doi: 10.1111/j.1742-4658.2008.06262.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Xu Y, Han Y, Bao S, Du J, Yuan M, et al. Overexpression of RAN1 in rice and Arabidopsis alters primordial meristem, mitotic progress, and sensitivity to auxin. Plant Physiol. 2006;140:91–101. doi: 10.1104/pp.105.071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciampi MS. Rho-dependent terminators and transcription termination. Microbiology. 2006;152:2515–28. doi: 10.1099/mic.0.28982-0. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe H, Shionyu M, Kimura T, Kimata K, Watanabe H. Splicing factor 3b subunit 4 binds BMPR-IA and inhibits osteochondral cell differentiation. J Biol Chem. 2007;282:20728–38. doi: 10.1074/jbc.M703292200. [DOI] [PubMed] [Google Scholar]
- 33.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–32. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


