Abstract
The glycosylphosphatidylinositol (GPI) anchor is an essential glycolipid that tethers certain eukaryotic proteins to the cell surface. The core structure of the GPI anchor is remarkably well conserved across evolution and consists of NH2-CH2-CH2-PO4-6Manα1,2Manα1,6Manα1,4-GlcNα1,6-myo-inositol-PO4-lipid. The glycan portion of this structure may be modified with various side-branching sugars or other compounds that are heterogeneous and differ from organism to organism. One such modification is an α(1,2)-linked fourth mannose (Man-IV) that is side-branched to the third mannose (Man-III) of the trimannosyl core. In fungi and mammals, addition of Man-III and Man-IV occurs by two distinct Family 22 α(1,2)-mannosyltransferases, Gpi10/PigB and Smp3/PigZ, respectively. However, in the five protozoan parasite genomes we examined, no genes encoding Smp3/PigZ proteins were observed, despite reports of tetramannosyl-GPI structures (Man4-GPIs) being produced by some parasites. In this study, we tested the hypothesis that the Gpi10/PigB proteins produced by protozoan parasites have the ability to add both Man-III and Man-IV to GPI precursors. We used yeast genetics to test the in vivo specificity of Gpi10/PigB proteins from several Plasmodium and Trypanosoma species by examining their ability to restore viability to Saccharomyces cerevisiae strains harboring lethal defects in Man-III (gpi10Δ) or Man-IV (smp3Δ) addition to GPI precursor lipids. We demonstrate that genes encoding PigB enzymes from T. cruzi, T. congolense and P. falciparum are each capable of separately complementing essential gpi10Δ and smp3Δ mutations, while PIGB genes from T. vivax and T. brucei only complement gpi10Δ. Additionally, we show the ability of T. cruzi PIGB to robustly complement a gpi10Δ/smp3Δ double mutant. Our data suggest that certain Plasmodium and Trypanosoma PigB mannosyltransferases can transfer more than one mannose to GPI precursors in vivo, and suggest a novel biosynthetic mechanism by which Man4-GPIs may be synthesized in these organisms.
Introduction
The glycosylphosphatidylinositol (GPI) anchor serves to attach many eukaryotic proteins to the cell surface. GPIs are essential glycolipids that are assembled stepwise on a phosphatidylinositol backbone within the endoplasmic reticulum (ER) and become attached to the carboxy-termini of certain secretory proteins prior to exiting the ER and trafficking to the cell surface via the secretory pathway. In mammals, GPI anchored proteins serve a variety of functions on the cell surface and GPI biosynthesis is essential for normal development [1]. In fungi, GPI biosynthesis is essential for viability as GPI anchored proteins are critical components of the yeast cell wall [2]. In protozoan parasites, GPIs are important in host-parasite interaction and also play a role in the pro-inflammatory immune response to parasitic infection [3], [4], [5].
Most known GPIs contain a common core structure of NH2-CH2-CH2-PO4-6Manα1,2Manα1,6Manα1,4-GlcNα1,6-myo-inositol-PO4-lipid (Figure 1) [6], [7], [8]. However, mature protein-bound GPI structure varies significantly across the whole of eukaryotic biology due to the presence of numerous moieties that become side branched to the GPI glycan and variations in lipid composition from organism to organism (Table 1) [2], [6], [7], [8], [9], [10]. Additionally, GPI structure is highly dynamic within any given organism with both additions and subtractions being made to the GPI molecule throughout its transport to the cell surface.
Figure 1. Schematic of the core GPI anchor glycan and its side chain modifications.
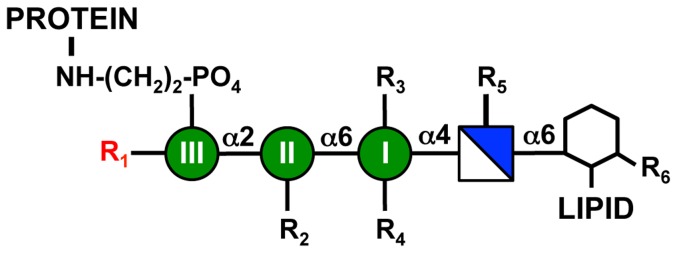
GPI anchors consist of a highly conserved carbohydrate sequence that is linked to the inositol head group (hexagon) of phosphatidylinositol (hexagon + LIPID). The core GPI glycan contains glucosamine (blue/white square) and three mannoses (Man I-III, green circles). A protein is linked to the third mannose via an ethanolamine phosphate. R indicates positions where modification of the core GPI structure can occur, and possible R modifications in various species are described in Table 1. The red R1 indicates the position of fourth mannose addition.
Table 1. Modifications of the GPI anchor core structure observed in various organisms1.
| Organism | R1 | R2 | R3 | R4 | R5 | R6 | Lipid |
| S. cerevisiae | Man(α1,2)Man(α1,2) or Man(α1,3)Man(α1,2) | ±EthNP | ±EthNP | OH | OH | OH | DAG |
| H. sapiens | ±Man(α1,2) | ±EthNP | EthNP | (±NANA)(±Gal(β1,3))(±GalNAc(β1,4)) | OH | ±palmitate | AAG or DAG |
| T. brucei | OH | OH | OH | ±Galα1−2(Galα1−2Galα1−6)Galα1−3 | OH | OH | DMG |
| T. cruzi | Man(α1,2) | OH | OH | OH | AEP | OH | AAG/Cer |
| P. falciparum | Man(α1,2) | OH | OH | OH | OH | ±myristic acid | DAG |
Chart describing known side chain modifications of the GPI core in various species. R1-R6 and Lipid positions on the GPI core are indicated in Figure 1. DAG: diacylglycerol; AAG: alkylacylglycerol; DMG: dimyristylglycerol; Man: mannose; EthNP: ethanolamine phosphate; GalNAc: N-acetylgalactosamine; AEP: aminoethylphosphonate; Cer: ceramide.
See references 7, 10, 16, 17, 20
One interesting modification of the core GPI structure is the presence of a fourth mannose (Man-IV) α1,2-linked to the third mannose (Man-III) of the GPI. The Man-IV modification has been observed on GPIs isolated from various proteins in mammals [11], [12], [13], [14], fungi [15], and the protozoan parasites Trypanosoma cruzi [16] and Plasmodium falciparum [17]. The addition of Man-IV has been well defined in yeast and mammals [18], [19]. In both groups, Man-IV is added to GPI precursors during their biosynthesis by a conserved family of GPI mannosyltransferases (Smp3p/PigZp family). However, the two taxa differ in their relative requirement for Man-IV on the GPI glycan. In the yeast Saccharomyces cerevisiae, Man-IV is added by the essential Smp3 protein in a mandatory step of biosynthesis that precedes addition of EthN-P to Man-III and GPI transfer to protein [18]. Thus, it is believed that all yeast GPIs bear four mannoses. Conversely, in mammalian cells, trimannosyl-GPIs (Man3-GPIs) predominate with addition of Man-IV to GPIs by the PigZ protein occurring less frequently, perhaps in a tissue or cell-specific manner [19]. However, the physiological significance of this modification in mammals is still unclear.
The presence of Man-IV has also been observed in some protozoan parasites, but not others. For example, while the GPIs of Trypanosoma brucei do not contain Man-IV [20], the P. falciparum MSP-1 and MSP-2 proteins each bear GPIs with four mannoses [17]. Additionally, a glycoproteomic study of GPIs in T. cruzi showed that Man-IV was present on each of over 90 observed GPI structures suggesting that this organism exclusively synthesizes Man4-GPIs [16]. Interestingly, the genomes of both P. falciparum and T. cruzi lack genes encoding obvious orthologs of the Smp3/PigZ fourth mannosyltransferase family [21], suggesting that Man-IV addition to GPIs in these organisms likely occurs via a different mechanism.
In the present study, we have explored a hypothesis that the PigB family of GPI mannosyltransferases (GPI-MT-III) that add an α1,2-linked third mannose to GPIs in certain protozoan parasites may also be capable of adding the α1,2-linked fourth mannose. We have used yeast genetics to explore the in vivo specificity of various parasite PigB GPI mannosyltransferases by determining their ability to restore viability to yeast mutants harboring lethal defects in either Man-III or Man-IV addition to GPI precursor lipids. We report that the GPI-MT-III (PigB) proteins from T. cruzi, T. congolense and P. falciparum are capable of complementing both gpi10Δ and smp3Δ mutations. Additionally, we show that expression of T. cruzi PIGB can restore viability of a yeast double mutant defective in both Man-III and Man-IV addition to GPI precursors. This study represents the first evidence that a single GPI mannosyltransferase is capable of performing multiple sugar additions to the GPI anchor and provides insight into a possible mechanism for Man4-GPI formation in certain protozoan parasites.
Materials and Methods
Sequence database searching
The algorithms BLASTP and TBLASTN [22], [23] were used with default settings to identify putative Smp3p and Gpi10p sequences in various eukaryotic genomes present in GenBank. Searches were performed using the BLAST server at the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov) with Smp3 and Gpi10 protein sequences from S. cerevisiae and mammals as search queries. In certain cases, SMP3 or GPI10 genes were deemed to be absent from a genome if no hits with an e-value less than 1×10−2 were observed using both BLASTP and TBLASTN and the genomic data searched represented at least 7X genome sequence coverage. All identified genes are listed in Table S1.
Yeast strains and culture media
A haploid S. cerevisiae strain (YCT138) containing a lethal smp3::KanR null allele and the rescuing vector pGAL-ScSMP3 (URA3, CEN) was previously described [19]. A similar haploid strain (YCT684) having a gpi10::KanR allele and a rescuing pGAL-ScGPI10 vector (URA3, CEN) was constructed by introduction of pGAL-ScGPI10 into a heterozygous GPI10/gpi10::KanR diploid (Life Technologies) and dissection of tetrads to isolate a viable gpi10::KanR haploid segregant harboring pGAL-ScGPI10. These haploid stains (YCT138 and YCT684) were used as the base strains for the transformation of pPGK-PIGB expression plasmids and subsequent plasmid shuffling experiments.
To generate the gpi10Δ/smp3Δ double mutant strain, a haploid gpi10::KanR strain containing pGAL-TcrPIGB was mated with a haploid smp3::KanR strain containing pGAL-TcrPIGB. The resulting diploid was induced to sporulate and asci (“tetrads”) containing four haploid spores, the meiotic progeny of the cross, were dissected and assessed for their ability to grow on YPGal agar. For tetrads yielding four viable progeny, each haploid was scored for its resistance to kanamycin (G418). Tetrads showing the non-parental ditype segregation pattern were screened by PCR to identify haploids that contained both null gpi10 and smp3 loci as well as the pGAL-TcrPIGB plasmid.
Compositions of yeast growth media SGal, YPGal and SD have been previously described [19], [24]. Amino acids were routinely supplemented in growth media to complement strain auxotrophies as previously described [24] and Geneticin (G418, Gibco) was used at a final concentration of 200 µg/mL. SD 5FOA medium contained 0.1% (w/v) 5-fluoroorotic acid (Toronto Research Chemicals) and 50 µg uracil per mL of SD medium.
Expression of GPI10, SMP3 and PIGB genes
S. cerevisiae GPI10, SMP3 and T.cruzi GPI10 were cloned into the pMW20 (pGAL) expression vector using a previously described scheme [19]. The coding regions of S. cerevisiae GPI10 and SMP3 were each amplified by PCR from S. cerevisiae genomic DNA and cloned into the PstI-BamHI sites (for GPI10) or NotI-AscI sites (for SMP3) of the yeast expression vector pPGK-415. Vector pPGK-415 was constructed in pRS-415 [25], [26] (CEN, LEU2) and contained 500 bp of the PGK1 promoter and 249 bp of the PGK1 terminator cloned between the SacI-XhoI sites. Three unique sites (NotI, PacI and AscI) are situated between the promoter and terminator to facilitate cloning of cDNA sequences for expression. Wild-type cDNA sequences of the T. brucei PIGB (AB033824.1), T. cruzi PIGB (XM_801669.1), T. vivax PIGB (HE573026.1, locus tag TVY486_1005620), and T. congolense PIGB (HE575323.1, locus tag TCIL3000_10_4690) genes were each synthesized and cloned into pUC57 (Genscript). Additionally, P. falciparum PIGB (XM_001350101.1) was synthesized with codon optimization for expression in yeast (Genscript). Each gene was amplified using Phusion High-Fidelity DNA Polymerase (New England Biolabs) with primers containing a C-terminal HA tag sequence and NotI or AscI restriction sites at the 5′ or 3′ end of the cDNA, respectively (See Table S2 for primers used in cloning). Amplified DNA fragments were digested and cloned into the NotI-AscI sites of pPGK-415. Chimera 1, consisting of nucleotides 1-306 of TbPIGB (which includes the ‘long motif’ region) fused to nucleotides 457-1827 of TcrPIGB, was synthesized (Genscript) and subcloned into pPGK-415 as described above. Chimera 2 was generated by ‘PCR knitting’ using primer sets 1, 16 and 2, 15 (Table S2) to first amplify fragments of TcrPIGB from a pPGK-415-TcrPIGB template. These fragments contained complementary overlapping regions corresponding to the TbPIGB ‘short motif’ region, and were fused together by a second round of PCR using primers 1 and 2. The resulting DNA fragment (chimera 2) contained the TcrPIGB cDNA with its short motif replaced with that of TbPIGB (i.e. a fusion consisting of TcrPIGB nt 1-1146, TbPIGB nt 1006-1053 and TcrPIGB nt 1195-1827). Chimera 2 was cloned into the NotI-AscI sites of pPGK-415 for expression as described above. Chimera 3 was generated using the same scheme as for the generation of chimera 2, but using chimera 1 as the template for the initial PCRs. This chimera therefore consisted of a fusion of nucleotides 1-306 of TbPIGB, nucleotides 457-1146 of TcrPIGB, nucleotides 1006-1053 of TbPIGB and nucleotides 1195-1827 of TcrPIGB. Typical Phusion PCR conditions were as follows: 98°C for 3 min, 10 cycles of 98°C for 15 s, 54°C for 15 s and 72°C for 1 min 15 s (20–30 s per kb), followed by 25 cycles of 98°C for 15 s, 63°C for 15 s and 72°C for 1 min 15 s (20–30 s per kb), and completed by a final extension at 72°C for 5 min. All primers used for the generation of expression constructs are listed in Table S2. Vectors were introduced into YCT684 and YCT138 using lithium acetate as described in [27] with the carrier DNA and PEG/TE/LiOAc replaced by NEB Yeast Transformation Reagent (New England Biolabs).
Complementation of gpi10 or smp3 null mutations
Complementation of gpi10 or smp3 lethal null mutations was assessed by ‘plasmid shuffling’. This was performed by streaking isolated colonies of strains transformed with a pPGK-PIGB expression vector from selective medium (SGal) to non-selective medium (YPGal) to permit plasmid loss. These strains were subsequently subjected to two rounds of counter-selection on SD 5FOA agar plates at 25°C for 3–7 days to identify strains capable of growing in the absence of either of the parental URA3-containing vectors (pGAL-ScSMP3 or pGAL-ScGPI10). Following the second round of 5FOA counter-selection, viable strains were assessed for complete loss of vectors pGAL-ScSMP3 or pGAL-ScGPI10 by PCR using primers that anneal to URA3 (Primers P1 and P2, Table S3) and amplify a diagnostic amplicon if either vector is still present. Strains were also tested for the presence of a null gpi10 or smp3 locus (Primers P3–P5, Table S3) and the pPGK plasmid (Primers P6–P14, Table S3). See below for DNA extraction and PCR reaction conditions. All plasmid shuffling experiments were repeated at least twice.
PCR confirmation of yeast strain genotypes
Total DNA was extracted from yeast strains as previously described [28]. PCR was performed using the Phusion High Fidelity Master Mix with HF Buffer (New England Biolabs), the primers listed in Table S3 and 25 ng of isolated DNA. Typical PCR cycling conditions were 95°C for 5 min, followed by 35 cycles of 95°C for 15 s, 53°C for 15 s and 72°C for 20 s and a final 5 min extension at 72°C.
Serial drop cultures of yeast strains
Serial dilutions of yeast cultures were used to assess the robustness of growth of engineered strains compared to control strains. Yeast cells were suspended in 5 mL of appropriate medium after which 1 mL was used to determine cell density via light scattering (OD600) in a spectrophotometer (Amersham). Cells in the remaining 4 mL were pelleted by centrifugation for 15 min at 1850×g and resuspended in appropriate medium to a concentration of 2×107 cells per mL. Serial 10-fold dilutions were performed and 5 µL of each dilution was plated onto appropriate agar medium. Plates were incubated for 3–5 days at 25°C.
Results
Identification of GPI α(1,2)-mannosyltransferase families
Synthesis of the mannose core of the GPI glycan occurs via sequential action of three distinct GPI mannosyltransferases that transfer mannose from the polyisoprenoid sugar donor dolichol phosphomannose (Dol-P-Man) to the GPI precursor. These three enzymes (GPI-MT-I, GPI-MT-II and GPI-MT-III) add α(1,4)-, α(1,6)- and α(1,2)-linked mannose, respectively (For a review see reference [6]). The structures of known GPIs across eukaryotic biology almost invariably contain this arrangement of mannoses, and as such, these three protein families (Gpi14p/PigMp, Gpi18p/PigVp, and Gpi10p/PigBp, respectively) are each highly conserved and their genes are straightforward to identify in genomic sequences. It is worth noting that some apicomplexan parasites such as Babesia bovis, Babesia microti, Theileria annulata, and Theileria parva lack a Gpi10/PigB family protein and B. bovis has been further shown to contain GPI anchor structures containing only two mannoses [29], [30]. In mammals and yeast, a fourth mannose (Man-IV) that is α(1,2)-linked to Man-III may be added to varying degrees by the Dol-P-Man utilizing Smp3/PigZ mannosyltransferase family (GPI-MT-IV). While this enzyme functions separately from GPI-MT-III, the two protein families both belong to glycosyltransferase family 22 [31], harbor similar domain organization [32] and share some isolated protein sequence homology. To date, the only known mechanism for addition of Man-IV to GPIs is via the Smp3/PigZ family of mannosyltransferases.
Precise GPI structural data is available for a relatively small number of GPI anchored proteins from few classes of organisms. Therefore, to better understand what organisms might be able to add Man-III and/or Man-IV to the GPI core structure, we examined the distribution of the Gpi10/PigB and Smp3/PigZ protein families across a wide array of eukaryotic organisms (Table S1). The BLASTP algorithm was used to identify both Gpi10/PigB and Smp3/PigZ protein sequences in the NCBI database. Candidate Gpi10p/PigBp sequences were encoded by every genome we examined, consistent with this enzyme's essential role in formation of the conserved trimannosyl core GPI glycan. In contrast, Smp3p/PigZp sequences were not as widely present in animal genomes (Table S1). In vertebrates, probable Smp3/PigZ proteins were encoded by each of 46 mammalian genomes and 11 avian genomes, but were absent from all 6 fish genomes we searched. In invertebrates, putative Smp3/PigZ proteins were encoded by each of the 31 arthropod genomes we examined and were also found in several primitive multicellular animals. However, several genomes lacked Smp3/PigZ proteins including those of nematodes and sea urchins. Outside of Animalia, possible Smp3/PigZ proteins were found encoded by the genomes of most fungi, several green algae and phytoplankton, but were absent from each of the 10 plant genomes we analyzed.
Curiously, Smp3p/PigZp sequences were absent from the genomes of all protozoan parasites we examined (Table S1), despite prior observations that Man-IV is present on GPIs on certain proteins from P. falciparum [17] and is abundantly present on GPI structures from T. cruzi [3]. This suggested that these organisms might employ a different mechanism for addition of Man-IV to GPIs. We hypothesized that the GPI-MT-III (PigB) proteins from these organisms might be able to add two consecutive α(1,2)-linked mannoses (Man-III and Man-IV) to GPIs.
Expression of parasite GPI mannosyltransferase genes in yeast gpi mutants
Yeast GPI biosynthesis involves the action of two independent α1,2-mannosyltransferases that transfer Man-III and Man-IV to the GPI precursor. The genes encoding these enzymes (GPI10 and SMP3, respectively) are both essential for cell viability, and conditional mutants of each gene accumulate GPI lipid intermediates with glycan head groups no larger than two mannoses [33] or three mannoses [18], respectively. Furthermore, overexpressed Gpi10 and Smp3 proteins are not capable of substituting for each other in vivo [18]. The latter observation suggests that while both genes encode yeast α1,2-mannosyltransferases, each protein exhibits strict substrate specificity, with Gpi10p adding mannose to Man2-GPI precursors and Smp3p adding mannose to Man3-GPI precursors. Together, these prior observations support the interpretation that the Gpi10 and Smp3 proteins each perform distinct, mandatory mannose transfer events in the yeast GPI assembly pathway. In this study, we exploited this unique feature of yeast GPI synthesis to query the ability of various parasite PIGB mannosyltransferase genes to complement lethal gpi10Δ or smp3Δ mutations as a way to interrogate the specificity of PigB proteins in vivo.
Complementation of yeast lethal gpi10 and smp3 null mutations by heterologously expressed parasite PIGB genes was assessed using a plasmid shuffling approach (Figure 2A). We used two haploid yeast strains that each contained a chromosomal deletion of either the gpi10 or smp3 gene and a rescuing URA3-containing plasmid that directed expression of wild-type S. cerevisiae GPI10 or SMP3 from the yeast GAL10 promoter (strains YCT684 and YCT138, respectively). A second expression vector (pPGK) was used to express candidate parasite PIGB genes from T. brucei (TbPIGB), T. cruzi (TcrPIGB), T. congolense (TcoPIGB), T. vivax (TvPIGB) and P. falciparum (PfPIGB) via the PGK1 promoter.
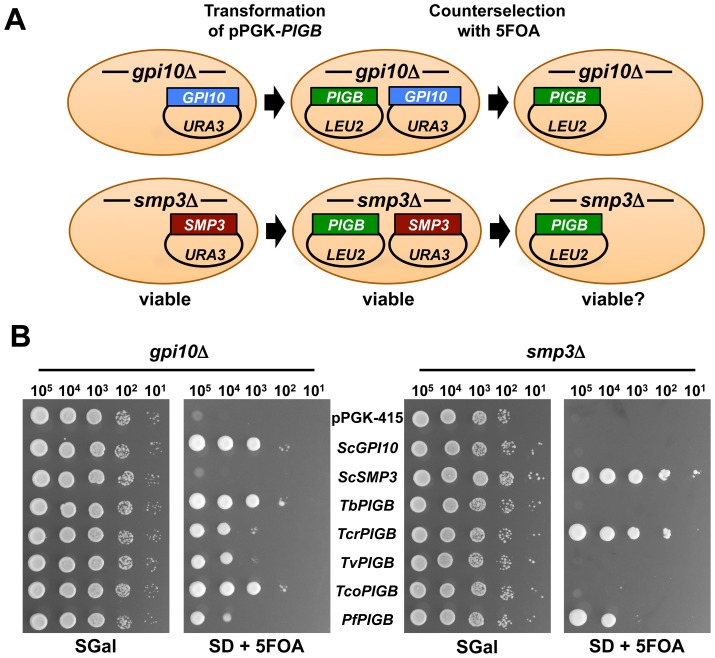
Figure 2. Complementation of yeast gpi10 and smp3 null mutants by protozoan PIGB genes.
A. Schematic of yeast plasmid shuffling experiments. Haploid gpi10::KanR containing pGAL-ScGPI10 (top row) or smp3::KanR containing pGAL-ScSMP3 (bottom row) strains were transformed with protozoan pPGK-PIGB expression plasmids. Yeasts carrying both plasmids were then induced to lose one plasmid via growth on rich medium, and then strains containing only the pPGK plasmids were selected for by growth on 5FOA. B. Serial drop cultures of yeast on SGal and SD + 5FOA media. Note that all strains grow on SGal medium as they contain both the pGAL rescuing plasmid and the pPGK expression plasmid. Following selection on 5FOA only the strains containing PIGB genes that can complement the indicated null mutation are able to grow.
To assess the ability of protozoan PIGB genes to complement either gpi10Δ or smp3Δ, each pPGK-PIGB expression vector was separately introduced into S. cerevisiae YCT684 and YCT138. Plasmid shuffling was performed through counterselection for the loss of the URA3-containing vector by growth on medium containing 5FOA (Figure 2A). Each candidate protozoan PIGB gene permitted growth of S. cerevisiae harboring the lethal gpi10Δ mutation on 5FOA-containing medium (Figure 2B), indicating that each of the protozoan PIGB genes selected for this study was functional in yeast and was capable of substituting for the yeast GPI-MT-III in vivo. Furthermore, in the smp3Δ background, TcrPIGB and PfPIGB expression permitted robust growth on 5FOA-containing medium (Figure 2B). TcoPIGB in the smp3Δ background also demonstrated reproducible growth on 5FOA-containing medium, although its growth was significantly slower than other strains and was just above the pPGK-415 negative control at the 105 cell dilution (Figure 2B, far right panel). Thus, the TcrPigB, TcoPigB and PfPigB proteins were able to overcome a GPI-MT-IV deficiency in vivo.
Verification of viable strains
To verify complementation of the lethal gpi10Δ and smp3Δ mutations by expression of protozoan PIGB genes, we isolated genomic DNA from all strains both before and after plasmid shuffling and performed a series of diagnostic PCR tests to confirm strain genotypes and the presence or absence of pGAL- and pPGK-derived plasmids (see Figure 3A–C for schematic). PCR with primers specific for URA3 was used to confirm the presence or absence of pGAL-SMP3 or pGAL-GPI10 in strains before and after plasmid shuffling (Figure 3A). PCR was then used to verify the presence of chromosomal gpi10::KanR or smp3::KanR alleles in the gpi10Δ or smp3Δ backgrounds, respectively (Figure 3B). Another PCR reaction was performed to confirm the presence of a pPGK-PIGB expression vector using individual forward primers that anneal to the 3′ end of each PIGB gene and a reverse primer specific for the pPGK backbone (Figure 3C).
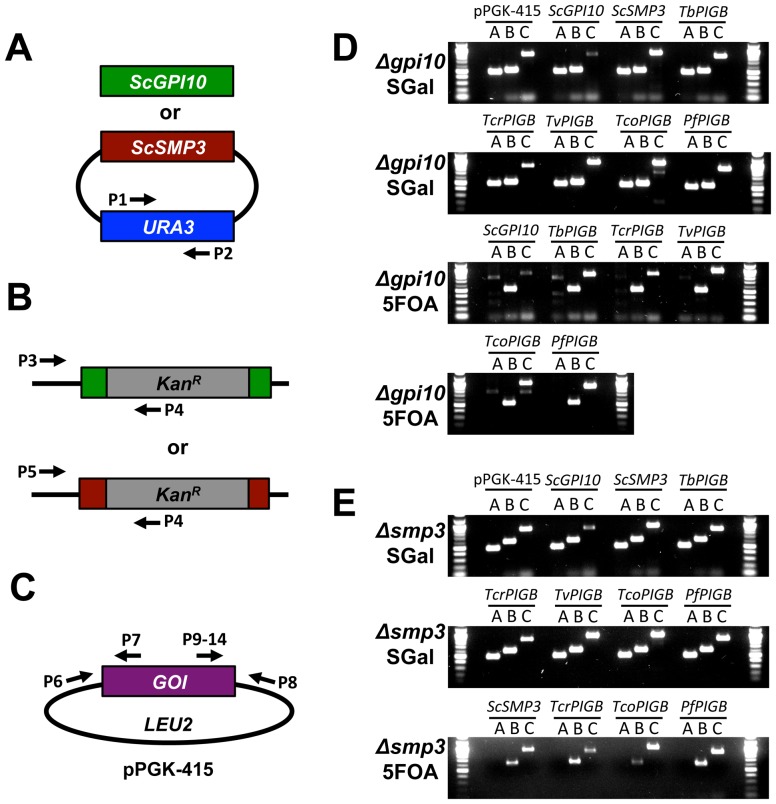
Figure 3. PCR confirmation of yeast strains.
A–C. Schematics of PCR reactions used to confirm strains. See Table S3 for sequences of the indicated primers. A. To confirm the presence (on SGal) or absence (on 5FOA) of the pGAL rescuing plasmid, the URA3 locus within this plasmid was amplified. B. To confirm that the endogenous gpi10 or smp3 locus was indeed null in all strains, forward primers residing in the 5′UTR of the genes were coupled with a reverse primer that sits within the kanamycin cassette that was inserted into the gene locus upon null strain generation. C. To confirm the presence of the pPGK plasmid, PCR was performed using a primer within the pPGK vector, coupled with a primer in the 3′end of the coding region of the PIGB gene of interest. D–E. Results of PCR experiments to confirm all yeast strains shown in Figure 2. DNA was isolated from all strains and the PCRs described in A–C were performed. D. gpi10Δ strains before (SGal) and after (5FOA) plasmid shuffling. E. smp3Δ strains before (SGal) and after (5FOA) plasmid shuffling. Reactions A–C correspond to the schematics A–C at left. Amplicon sizes are: Reaction A: 500 bp; Reaction B: 534 bp for gpi10, 616 bp for smp3; Reaction C: 902 bp for pPGK-415, 891 bp for pPGK-ScGPI10, 922 bp for pPGK-ScSMP3, 800 bp for pPGK-TbPIGB, 820 bp for pPGK-TcrPIGB, 879 bp for pPGK-TvPIGB, 932 bp for pPGK-TcoPIGB and 908 bp for pPGK-PfPIGB.
We confirmed that all strains shown in Figure 2B contained the expected arrangements of plasmids and knockout loci (Figure 3D and E). In particular, the URA3-plasmid containing the wild-type ScGPI10 or ScSMP3 complementing gene was present in all strains prior to plasmid shuffling (Figure 3D), but had been lost in all surviving strains after 5FOA counterselection (Figure 3E). Of note, even though the TcoPIGB expressing strain had shown minimal growth on 5FOA-containing medium in the drop culture experiment, we were able to isolate DNA and confirm that it harbored an smp3::KanR locus, lacked the wild-type S. cerevisiae SMP3 rescuing plasmid, and contained the pPGK-TcoPIGB plasmid (Figure 3E). From these data, we show that TcrPIGB and PfPIGB, and to a lessor extent TcoPIGB, are able to compensate for deficiencies in both Man-III and Man-IV addition to yeast GPI precursors in vivo.
Complementation of a gpi10/smp3 double null mutant
We sought further genetic evidence that some parasite PIGB genes are capable of performing the dual role of Man-III and Man-IV addition to GPI anchors. In a second complementation experiment, we investigated if TcrPIGB could restore viability to a haploid gpi10::KanR/smp3::KanR double mutant. We elected to use TcrPIGB expression for this experiment because it showed the most robust complementation of smp3Δ.
We generated a double knockout strain by mating a haploid gpi10Δ strain containing pGAL-TcrPIGB with a haploid smp3Δ strain containing pGAL-TcrPIGB and inducing sporulation. Following dissection of ascospores representing the meiotic progeny of the cross, we readily obtained tetrads showing a non-parental ditype pattern of kanamycin marker segregation, with two KanR segregants (double mutants) and two KanS segregants (wild-type) (Figure 4A). PCR was used to show that the two viable KanR segregants each harbored both the gpi10::KanR and smp3::KanR alleles (Figure 4B). Finally, a representative viable double null strain was scored for its ability to grow on medium containing galactose or glucose to induce or repress TcrPIGB expression, respectively (Figure 4C). The double mutant grew robustly in the presence of galactose, but not in glucose, further indicating that its viability was entirely dependent on expression of TcrPIGB from the complementing vector pGAL-TcrPIGB.
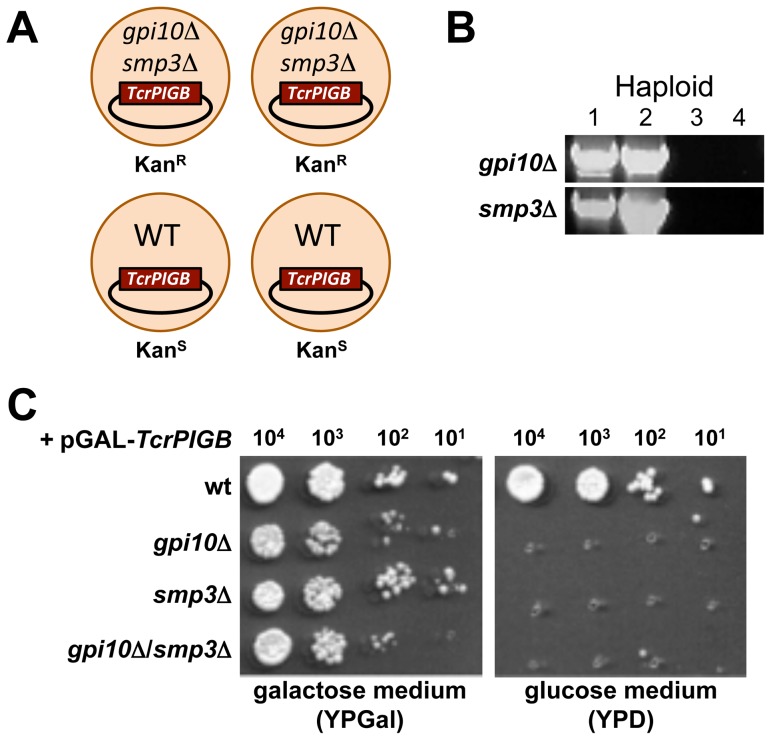
Figure 4. T. cruzi PIGB can complement a yeast strain deficient in both gpi10 and smp3.
A. Schematic of a tetrad showing the desired non-parental ditype pattern of kanamycin marker segregation generated from the mating of a haploid gpi10::KanR strain containing pGAL-TcrPIGB with a haploid smp3::KanR strain containing pGAL-TcrPIGB. B. PCR was used to test each haploid meiotic segregant from a non-parental ditype tetrad for the presence of gpi10Δ and smp3Δ alleles (see Figure 3B for PCR schematic). Haploids 1 and 2 are each double mutants that contain both null alleles. C. Serial drop cultures of both single gpi10Δ and smp3Δ mutant strains as well as the double mutant gpi10Δ/smp3Δ strain. Note that the strains grow on the galactose-containing medium that induces pGAL-TcrPIGB expression but not on the glucose medium, which represses pGAL-TcrPIGB expression.
These data unambiguously demonstrate that TcrPIGB is capable of restoring viability to cells defective in both Man-III and Man-IV addition to GPI precursors. The likeliest interpretation of these observations is that TcrPigB is capable of efficiently adding both the third and fourth mannoses to yeast GPI precursors during their biosynthesis in the ER.
Use of chimeric constructs to evaluate TcrPIGB substrate recognition in vivo
Little is known about the structure and function of Gpi10p/PigBp α(1,2)-mannosyltransferases. Several factors have precluded the use of classic in vitro biochemical characterization of these enzymes. First, members of this protein family are multi-spanning transmembrane proteins and are difficult to abundantly express and purify. Second, these enzymes transfer mannose from Dol-P-Man to low abundance intermediate lipids that cannot be purified in sufficient quantities for in vitro transferase assays. Therefore, we elected to use our system of complementation of yeast gpi10Δ and smp3Δ mutations to explore any relationship between highly conserved domains in Gpi10/PigB proteins and their ability to transfer Man-III or Man-IV to GPI precursors.
Each of the sequences of the parasite PigB proteins used in this study contained a conserved N-terminal “long motif” and a C-terminal HKEXRF “short motif” (Figure 5A) that were previously defined in bioinformatics studies that examined GPI mannosyltransferase sequences [32], [34]. These domains reside within putative loop regions of the protein and are hypothesized to be a part of the active site (long motif) or substrate acceptor site (short motif) of the enzyme [32]. We wished to determine whether either of these motifs was important for the ability of some parasite PigB proteins to add both Man-III and Man-IV to GPI precursors. Chimeric proteins were designed where the region of the protein containing the long motif and/or the short motif of the broader specificity TcrPigB protein was replaced by the protein domain containing the appropriate long (chimera 1) and/or short (chimeras 2 and 3) motifs from TbPigBp (Figure 5B). We selected TbPIGB and TcrPIGB for these experiments because they were from closely related organisms and they both gave robust complementation of gpi10Δ (TbPIGB and TcrPIGB) and/or smp3Δ (only TcrPIGB).
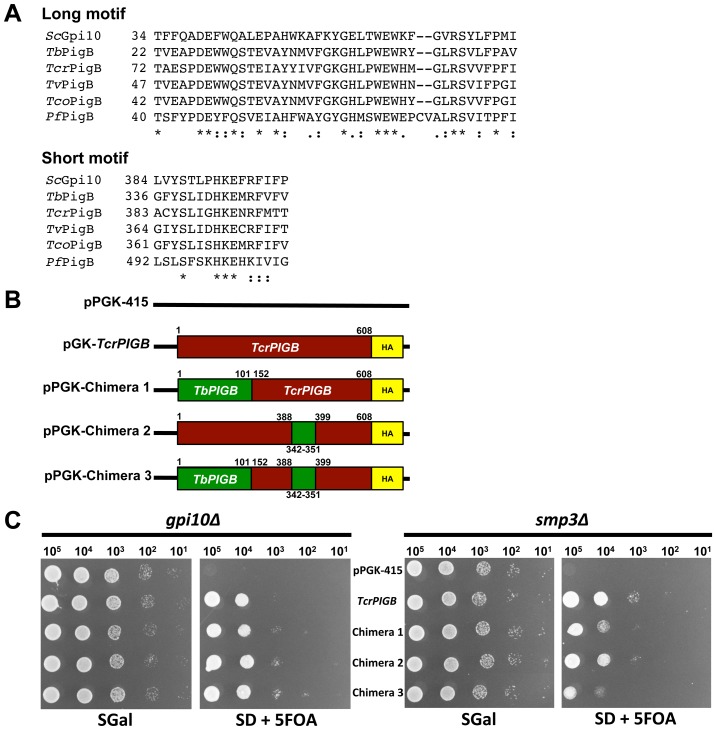
Figure 5. The long and short conserved motifs do not confer broad specificity to TcrPigB.
A. Alignment of the N-terminal long and C-terminal short motifs from yeast and protozoa. B. Schematics of the generated chimera constructs. The N-terminal long (chimera 1), C-terminal short (chimera 2) or both conserved motifs (chimera 3) of TcrPIGB (red) were replaced by those from TbPIGB (green). Numbers correspond to the amino acid positions of each domain within its native protein sequence. C. Serial drop cultures of control and chimeric constructs in both gpi10Δ and smp3Δ strains before (SGal) and after (SD + 5FOA) plasmid shuffling. Note that all three of the chimeric PigB proteins are able to complement the gpi10Δ and smp3Δ mutant strains.
Vectors directing expression of each chimera were introduced into haploid gpi10Δ and smp3Δ strains and were subjected to plasmid shuffling as described above. All three chimeric proteins were able to compensate for defects in Man-III addition to GPI precursors, indicating they were functional proteins (Figure 5C). Interestingly, despite containing the long and/or short motif sequences from the exclusively Man-III mannosyltransferase TbPIGB, each of the TcrPIGB chimeras was also still able to compensate for a deficiency in Man-IV addition to GPI precursors (Figure 5C). These data suggest that the two most highly conserved regions of PigB proteins are not alone or in combination responsible for the ability of some PigB proteins to discriminate between Man2-GPI and Man3-GPIs substrates for mannose transfer.
Discussion
In this study, after an exhaustive bioinformatics search, we found no SMP3/PIGZ sequences in the genomes of various protozoan parasites, despite prior reports that P. falciparum and T. cruzi harbor Man-IV on the GPI anchors of several GPI anchored proteins [16], [17]. We hypothesized that the α(1,2)-mannosyltransferase responsible for Man-III addition (PigB) might also be capable of adding Man-IV in these organisms. Because addition of Man-III and Man-IV to GPI precursors is performed by two separate essential mannosyltransferases in S. cerevisiae, we utilized complementation of yeast null mutants to explore the in vivo specificities of proteins encoded by various parasite PigB proteins. We showed that all parasite PIGB genes we examined could be functionally expressed in yeast and restored viability to cells defective in essential GPI-MT-III activity due to deletion of gpi10. Additionally, we found that expression of the PIGB genes from T. cruzi, T. congolense and P. falciparum restored viability to cells defective in essential GPI-MT-IV activity (smp3Δ. Finally, in a definitive experiment, we showed that T. cruzi PIGB was able to robustly complement a yeast gpi10Δ/smp3Δ double mutant strain defective in addition of both essential α(1,2)-linked mannoses (Man-III and Man-IV) to GPI precursors.
The likeliest interpretation of our complementation data is that certain parasite PIGB genes encode GPI mannosyltransferases that are capable of adding two α(1,2)-linked mannoses (Man-III and Man-IV) to GPI intermediate lipids during their biosynthesis in vivo. Interestingly, the pattern of complementation we observed correlated to the known presence or absence of Man-IV on GPI anchored proteins isolated from individual parasites. For example, TcrPIGB and PfPIGB each showed broad specificity by complementing both gpi10Δ and smp3Δ in yeast and Man4-GPI structures have been reported on GPI anchored proteins in both T. cruzi and P. falciparum [16], [17]. In contrast, TbPIGB complemented only gpi10Δ and Man-IV is not present on the two characterized GPIs from the VSG [20] and procyclin [35] proteins of T. brucei. Thus, broad specificity PigB proteins represent a possible mechanism by which Man4-GPIs are formed by protozoan parasites lacking SMP3/PIGZ. Furthermore, the absence of an SMP3/PIGZ sequence encoded in any eukaryotic genome likely does not exclusively predict the organism's inability to modify its GPIs with Man-IV.
In conclusion, our study represents the first report implicating a GPI mannosyltransferase in performing multiple sugar additions to the glycan of GPI intermediate lipids. Furthermore, the difference in specificity of some protozoan parasite PigB proteins and those of higher eukaryotes suggests that selective inhibition of PigBp-mediated mannose transfer to GPIs may be a plausible strategy for development of novel anti-parasitic chemotherapeutic agents.
Supporting Information
Putative Gpi10/PigB and Smp3/PigZ proteins present in Kingdom Animalia.
(DOC)
Primers used in the generation of plasmids.
(DOC)
Primers used to confirm yeast strains.
(DOC)
Acknowledgments
The authors would like to acknowledge Drs. Jeremy Foster, Andy Gardner, Barbara Taron and Saulius Vainauskas for critical reading of the manuscript. We are grateful to Dr. Donald Comb and New England Biolabs for the supportive research environment.
Funding Statement
This research was privately funded by New England Biolabs, Inc. The funders had no role in experimental design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1. Kinoshita T, Fujita M, Maeda Y (2008) Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: recent progress. J Biochem 144: 287–294. [DOI] [PubMed] [Google Scholar]
- 2. Pittet M, Conzelmann A (2007) Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1771: 405–420. [DOI] [PubMed] [Google Scholar]
- 3. Almeida IC, Gazzinelli RT (2001) Proinflammatory activity of glycosylphosphatidylinositol anchors derived from Trypanosoma cruzi: structural and functional analyses. J Leukoc Biol 70: 467–477. [PubMed] [Google Scholar]
- 4. Boutlis CS, Riley EM, Anstey NM, de Souza JB (2005) Glycosylphosphatidylinositols in malaria pathogenesis and immunity: potential for therapeutic inhibition and vaccination. Curr Top Microbiol Immunol 297: 145–185. [DOI] [PubMed] [Google Scholar]
- 5. Debierre-Grockiego F, Schwarz RT (2010) Immunological reactions in response to apicomplexan glycosylphosphatidylinositols. Glycobiology 20: 801–811. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson MAJ, Kinoshita T, Hart GW (2009) Glycosylphosphatidylinositol Anchors. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, et al.., editors. Essentials of Glycobiology. 2nd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. pp. 143–161. [PubMed]
- 7. Paulick MG, Bertozzi CR (2008) The glycosylphosphatidylinositol anchor: a complex membrane-anchoring structure for proteins. Biochemistry 47: 6991–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Orlean P, Menon AK (2007) Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J Lipid Res 48: 993–1011. [DOI] [PubMed] [Google Scholar]
- 9. Ferguson MA (1999) The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J Cell Sci 112 (Pt 17): 2799–2809. [DOI] [PubMed] [Google Scholar]
- 10. Serrano AA, Schenkman S, Yoshida N, Mehlert A, Richardson JM, et al. (1995) The lipid structure of the glycosylphosphatidylinositol-anchored mucin-like sialic acid acceptors of Trypanosoma cruzi changes during parasite differentiation from epimastigotes to infective metacyclic trypomastigote forms. J Biol Chem 270: 27244–27253. [DOI] [PubMed] [Google Scholar]
- 11. Rudd PM, Morgan BP, Wormald MR, Harvey DJ, van den Berg CW, et al. (1997) The glycosylation of the complement regulatory protein, human erythrocyte CD59. J Biol Chem 272: 7229–7244. [DOI] [PubMed] [Google Scholar]
- 12. Taguchi R, Hamakawa N, Harada-Nishida M, Fukui T, Nojima K, et al. (1994) Microheterogeneity in glycosylphosphatidylinositol anchor structures of bovine liver 5′-nucleotidase. Biochemistry 33: 1017–1022. [DOI] [PubMed] [Google Scholar]
- 13. Homans SW, Ferguson MA, Dwek RA, Rademacher TW, Anand R, et al. (1988) Complete structure of the glycosyl phosphatidylinositol membrane anchor of rat brain Thy-1 glycoprotein. Nature 333: 269–272. [DOI] [PubMed] [Google Scholar]
- 14. Brewis IA, Ferguson MA, Mehlert A, Turner AJ, Hooper NM (1995) Structures of the glycosyl-phosphatidylinositol anchors of porcine and human renal membrane dipeptidase. Comprehensive structural studies on the porcine anchor and interspecies comparison of the glycan core structures. J Biol Chem 270: 22946–22956. [DOI] [PubMed] [Google Scholar]
- 15. Fankhauser C, Homans SW, Thomas-Oates JE, McConville MJ, Desponds C, et al. (1993) Structures of glycosylphosphatidylinositol membrane anchors from Saccharomyces cerevisiae. J Biol Chem 268: 26365–26374. [PubMed] [Google Scholar]
- 16. Nakayasu ES, Yashunsky DV, Nohara LL, Torrecilhas AC, Nikolaev AV, et al. (2009) GPIomics: global analysis of glycosylphosphatidylinositol-anchored molecules of Trypanosoma cruzi. Mol Sys Biol 5: 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gerold P, Schofield L, Blackman MJ, Holder AA, Schwarz RT (1996) Structural analysis of the glycosyl-phosphatidylinositol membrane anchor of the merozoite surface proteins-1 and -2 of Plasmodium falciparum. Mol Biochem Parasitol 75: 131–143. [DOI] [PubMed] [Google Scholar]
- 18. Grimme SJ, Westfall BA, Wiedman JM, Taron CH, Orlean P (2001) The essential Smp3 protein is required for addition of the side-branching fourth mannose during assembly of yeast glycosylphosphatidylinositols. J Biol Chem 276: 27731–27739. [DOI] [PubMed] [Google Scholar]
- 19. Taron BW, Colussi PA, Wiedman JM, Orlean P, Taron CH (2004) Human Smp3p adds a fourth mannose to yeast and human glycosylphosphatidylinositol precursors in vivo. J Biol Chem 279: 36083–36092. [DOI] [PubMed] [Google Scholar]
- 20. Ferguson MA, Homans SW, Dwek RA, Rademacher TW (1988) Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science 239: 753–759. [DOI] [PubMed] [Google Scholar]
- 21. Cardoso MS, Junqueira C, Trigueiro RC, Shams-Eldin H, Macedo CS, et al. (2013) Identification and Functional Analysis of Trypanosoma cruzi Genes That Encode Proteins of the Glycosylphosphatidylinositol Biosynthetic Pathway. PLoS Negl Trop Dis 7: e2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gish W, States DJ (1993) Identification of protein coding regions by database similarity search. Nat Genet 3: 266–272. [DOI] [PubMed] [Google Scholar]
- 23. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 24. Sherman F (1991) Getting started with yeast. Methods Enzymol 194: 3–21. [DOI] [PubMed] [Google Scholar]
- 25. Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. [DOI] [PubMed] [Google Scholar]
- 26. Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gietz D, St Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Looke M, Kristjuhan K, Kristjuhan A (2011) Extraction of genomic DNA from yeasts for PCR-based applications. Biotechniques 50: 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cornillot E, Hadj-Kaddour K, Dassouli A, Noel B, Ranwez V, et al. (2012) Sequencing of the smallest Apicomplexan genome from the human pathogen Babesia microti. Nucleic Acids Res 40: 9102–9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rodriguez AE, Couto A, Echaide I, Schnittger L, Florin-Christensen M (2010) Babesia bovis contains an abundant parasite-specific protein-free glycerophosphatidylinositol and the genes predicted for its assembly. Vet Parasitol 167: 227–235. [DOI] [PubMed] [Google Scholar]
- 31. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, et al. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37: D233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oriol R, Martinez-Duncker I, Chantret I, Mollicone R, Codogno P (2002) Common origin and evolution of glycosyltransferases using Dol-P-monosaccharides as donor substrate. Mol Biol Evol 19: 1451–1463. [DOI] [PubMed] [Google Scholar]
- 33. Sutterlin C, Escribano MV, Gerold P, Maeda Y, Mazon MJ, et al. (1998) Saccharomyces cerevisiae GPI10, the functional homologue of human PIG-B, is required for glycosylphosphatidylinositol-anchor synthesis. Biochem J 332 (Pt 1): 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Delorenzi M, Sexton A, Shams-Eldin H, Schwarz RT, Speed T, et al. (2002) Genes for glycosylphosphatidylinositol toxin biosynthesis in Plasmodium falciparum. Infect Immun 70: 4510–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Treumann A, Zitzmann N, Hulsmeier A, Prescott AR, Almond A, et al. (1997) Structural characterisation of two forms of procyclic acidic repetitive protein expressed by procyclic forms of Trypanosoma brucei. J Mol Biol 269: 529–547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Putative Gpi10/PigB and Smp3/PigZ proteins present in Kingdom Animalia.
(DOC)
Primers used in the generation of plasmids.
(DOC)
Primers used to confirm yeast strains.
(DOC)