β-barrel membrane proteins are composed of multiple antiparallel β-strands and form pores in the outer membranes of endosymbiotic organelles like mitochondria and the evolutionary related Gram-negative bacteria. These β-barrel channels are crucial for signal transduction, metabolite transport, and protein translocation.
How are β-barrel membrane proteins assembled into the outer membrane? After synthesis, β-barrel proteins have to be transported to the inner face of the outer membrane, where specific machineries assemble the β-barrel proteins into the membrane. In bacteria the β-barrel precursor proteins are synthesized in the cytosol, secreted across the inner membrane by the Sec translocase, and subsequently transferred by periplasmic chaperones to the β-barrel assembly machinery (BAM) of the outer membrane (Fig. 1).1
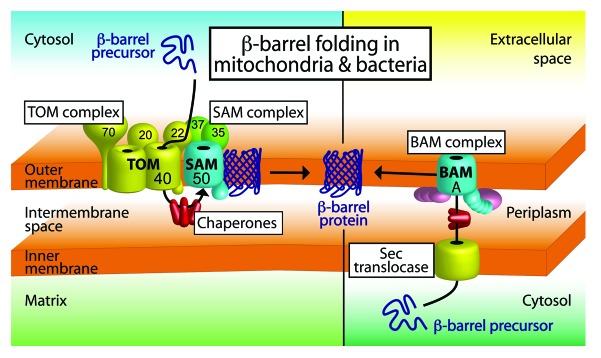
Figure 1. Model of β-barrel membrane protein biogenesis in mitochondria and bacteria. β-barrel precursor proteins are synthesized in the cytosol of bacteria and eukaryotic cells. Mitochondrial β-barrel precursors are imported by the TOM–SAM supercomplex composed of the translocase of the outer membrane and the sorting and assembly machinery. Chaperones help to shield the precursors on the intermembrane space side of the supercomplex. In bacteria, β-barrel proteins are transported across the inner membrane by the Sec translocase. Periplasmic chaperones are required to transfer the precursors to the bacterial β-barrel assembly machinery (BAM) in the outer membrane. The SAM and BAM complexes mediate β-barrel folding and membrane insertion into the outer membranes. Figure modified from reference 2 and from video related to reference 6.
In eukaryotic cells, the vast majority of mitochondrial proteins, including β-barrel precursors, are encoded in the nucleus and synthesized outside of the organelle. The translocase of the outer mitochondrial membrane (TOM) facilitates the import of these proteins into mitochondria. The receptor proteins Tom20, Tom22, and Tom70 recognize the precursors, and the β-barrel protein Tom40 forms the essential protein import channel in the mitochondrial outer membrane.2 After translocation across the outer membrane hydrophobic precursors engage with chaperones in the intermembrane space. β-barrel precursors contain a C-terminal β-signal, which is recognized by the sorting and assembly machinery (SAM/TOB) of the outer mitochondrial membrane (Fig. 1).3 The SAM complex is comprised of 3 main subunits. The core subunit Sam50 is a β-barrel membrane protein itself. Sam50 is homologous to the core subunit of the bacterial β-barrel assembly machinery, called BamA, and both proteins are members of the Omp85 protein family.2 Sam37 and Sam35 are located on the cytosolic leaflet of the mitochondrial outer membrane and are not related to other bacterial BAM lipoprotein subunits.1,2
Mitochondrial β-barrel proteins have retained the ability to assemble into the bacterial outer membrane and vice versa.4 Therefore, it is assumed that the mechanism of β-barrel formation and membrane insertion by the bacterial BAM complex is similar to the mitochondrial SAM complex. Studies with the reconstituted BAM complex imply that the SAM/BAM complexes are sufficient for the folding and insertion of β-barrel proteins into membranes.5 By introduction of cysteine residues at defined positions of the precursor and monitoring of formed disulfides, the formation of mature β-barrels could be demonstrated in mitochondria. The β-barrel was folded and inserted into the outer membrane, while the precursor was in contact with the SAM complex.6 The essential components Sam50 and BamA play a crucial role for β-barrel formation in vivo.1,2 Structural data of BamA indicate that a lateral release of the substrate β-strands from the SAM/BAM channel into the membrane might be feasible.2,3,7 Alternatively, Sam50/BamA might act as scaffolds for β-barrel formation in the outer membrane.
Surprisingly efficient β-barrel folding in mitochondria also depends on Tom22, the central import receptor of the TOM complex. How can the import of β-barrel precursors by the TOM complex affect the folding of the barrel at the SAM complex? Qiu et al. show by affinity purification and native gel electrophoresis that TOM and SAM form a supercomplex at a 1:1 ratio, and that the cytosolic receptor domain of Tom22 is crucial for supercomplex formation.6 These results support a new model for the biogenesis of β-barrel proteins in mitochondria. Upon its translocation through the TOM complex the C-terminal β-signal is transferred to the adjacent SAM complex. β-signal binding to SAM initiates β-barrel assembly, and the consecutive β-strand association during barrel formation might pull the precursor through the TOM complex. Thus, protein import and β-barrel folding in mitochondria are coupled by the TOM–SAM supercomplex (Fig. 1).6 However, it is assumed that TOM complexes are more abundant than SAM complexes, and therefore only some of the TOM complexes are part of a TOM–SAM supercomplex. This raises the question of how β-barrel precursors in the cytosol selectively recognize the TOM–SAM supercomplexes. One possibility is that the cytosolically exposed subunits of the SAM complex target the β-barrel precursors specifically to TOM-SAM supercomplexes. Supporting this notion, Sam35 binds the β-signal of the precursor, and Sam37 was initially identified as protein import receptor.3,8
Qiu J, et al. Cell. 2013;154:596–608. doi: 10.1016/j.cell.2013.06.033.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/27035
References
- 1.Rigel NW, et al. Curr Opin Microbiol. 2012;15:189–93. doi: 10.1016/j.mib.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfanner N, et al. Nat Struct Mol Biol. 2004;11:1044–8. doi: 10.1038/nsmb852. [DOI] [PubMed] [Google Scholar]
- 3.Kutik S, et al. Cell. 2008;132:1011–24. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 4.Walther DM, et al. Mol Biol Evol. 2010;27:887–95. doi: 10.1093/molbev/msp294. [DOI] [PubMed] [Google Scholar]
- 5.Hagan CL, et al. Science. 2010;328:890–2. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu J, et al. Cell. 2013;154:596–608. doi: 10.1016/j.cell.2013.06.033. [DOI] [PubMed] [Google Scholar]
- 7.Noinaj N, et al. Nature. 2013;501:385–90. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gratzer S, et al. J Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]


