Purposeful genomic mutation mediates critical aspects of both adaptive and innate immunity. A physiological role for purposeful mutation is best illustrated by the AID/APOBEC family of enzymes, which can deaminate cytosine to introduce rogue uracil bases into DNA.1 In adaptive immunity, introduction of unnatural uracil into the immunoglobulin locus by activation-induced deaminase (AID) seeds the process of antibody diversification and maturation via somatic hypermutation (SHM) and class-switch recombination (CSR). In innate immunity, deaminases from the APOBEC3 subfamily can purposefully mutate foreign viral genomic intermediates to promote their degradation, prevent viral integration, or garble the coding sequences of the pathogens.
While critical to proper physiological immune function, purposeful genomic mutation is also fraught with risk, and a pathologic role for cytosine deamination is emerging in oncogenesis. It has long been known that off-target deamination by AID contributes to B-cell malignancy by generating mutations in oncogenes or driving aberrant chromosomal translocations.2 More recently, deamination from ectopically expressed APOBEC3 family members, pathologically targeting the human genome over foreign genomes, has emerged as a source of mutations in breast and other cancers.3,4 This observed pattern of mutagenesis involves clustered regions of hypermutation within the genome, known as kataegis. A notable feature of these mutations is their sequence specificity, where cytosine is preferentially mutated in certain sequence contexts over others, leaving a mutational signature characteristic of particular APOBEC family members.
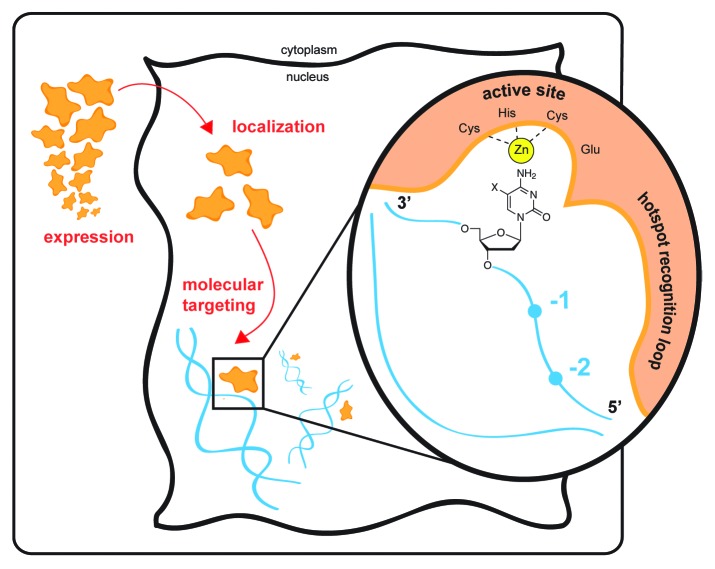
To understand the roles of the AID/APOBEC family in immunity and cancer more thoroughly, it is critical to elucidate how these enzymes target cytosine for mutagenic deamination. Targeting is achieved across many levels (Fig. 1), beginning with regulation of tissue-specific expression, and being further refined by both cellular localization and interactions with protein partners. The regulatory influences at these higher levels that help promote purposeful over pathologic mutation are starting to come to light. At a more molecular level, our understanding of targeting of particular genomic hotspots by AID/APOBEC enzymes is also rapidly evolving. Previous studies have demonstrated a “hotspot recognition” loop within the AID/APOBEC family that targets the enzymes to particular trinucleotide DNA patches spanning the target cytosine and the 2 upstream bases.5,6 Remarkably, this small protein loop can be grafted from a donor family member to change the mutational signature of an acceptor AID/APOBEC family member. More recent studies have also pointed to steric determinants, which may explain differential targeting of canonical cytosine base versus cytosine bases modified at the 5-position,7 with implications for DNA demethylation and epigenetic mechanisms linked to cancer.
Figure 1. Levels of targeting of AID/APOBEC deaminases. Physiological targeting of the purposeful mutator AID/APOBEC enzymes occurs at many levels. Expression of AID is generally confined to the activated B cell, while APOBEC3 family restriction factors are largely expressed in myeloid and lymphoid lineages and, in certain settings, may be induced by inflammation or cytokine signaling. Beyond tissue-specific localization, cellular localization is critical for protecting genomic DNA from deaminases, and protein partners may be involved in engaging with genomic targets. At the molecular level, the enzymes preferentially engage with single-stranded DNA. Each family member shows a distinctive sequence preference that can span several residues surrounding the target base (−2 to 0 positions shown). The mutational signatures are conferred via the enzyme’s “hotspot recognition” loop that engages the target sequence. The active site, depicted with bound zinc (Zn) and the active site acid–base residue (Glu), engages with the cytosine base. The enzyme shows discrimination against 5-position modified cytosines (shown with an X). The deoxyribosyl sugar is a key determinant of proper targeting of DNA over RNA, facilitating a conformation which permits targeting of cytosine for deamination. These multiple levels of targeting govern the proper physiological engagement of substrate and perturbations at any of these levels may contribute to pathological pro-oncogenic mutation by AID/APOBEC enzymes.
Focusing further on the molecular aspects of targeting, we recently sought to address a longstanding point of uncertainty: the mechanism of AID’s selective deamination of DNA over RNA.8 AID/APOBEC family members are known to be inhibited by tightly bound RNA, suggesting a common nucleic acid binding pocket, yet AID and its APOBEC3 homologs show selectivity for DNA deamination. Focusing on the B-cell mutator AID, we created an RNA-like substrate and sequentially introduced increasingly longer patches of DNA at the target cytosine and surrounding residues. Notably, in agreement with prior studies examining hotspot targeting with AID, we found that, in a sequence that otherwise mimics RNA, the minimal DNA patch necessary for efficient deamination spans the target cytosine and 2 nucleotides upstream. Thus, the major determinants for recognition of DNA over RNA and of the specific hotspot sequence within DNA appear to be largely confined to this trinucleotide region of the substrate. Focusing specifically on the contribution of nucleotide sugar toward targeting, we found that, within a substrate that is otherwise entirely composed of DNA, the introduction of a single ribocytidine at the target site, rather than deoxyribocytidine, was sufficient to disrupt deamination by at least 500-fold. A series of chimeric substrates containing alternative substitutions at the 2’ position of the ribosyl sugar suggested a mechanistic basis for this preference: ultimately, the target nucleotide’s rotational conformation, also known as sugar pucker, facilitates productive interactions between the enzyme and substrate that promote DNA over RNA deamination.
Mutation by AID/APOBEC enzymes is a double-edged sword. In vitro studies that reveal the molecular basis for targeting are key to understanding how purposeful mutation is achieved over pathologic mutation in vivo. The defining molecular signatures of AID/APOBEC enzymes—including the sequence contexts for mutation, clustering, and localization—are likely intricately linked to the biochemical attributes of the deaminases. Consolidating our findings with other biochemical studies on molecular targeting, we envision a mechanistic model in which AID/APOBEC3 enzymes use the “hotspot recognition” loop to engage with target DNA preferentially, and that the sugar pucker of the target nucleotides mediates important interactions that facilitate cytosine entering the active site for efficient deamination. Engagement of the DNA backbone is also likely to contribute to the mutational clustering through processive scanning of the target genome. While biochemical studies to date provide a foundation for understanding molecular targeting, structural insights into a deaminase bound to nucleic acid would likely aid in uncovering additional enzymatic and nucleic acid determinants for targeting. Beyond insights at a molecular level, the many higher layers of targeting—cellular expression, localization, and genomic targeting—remain important areas for continued study.
Nabel CS, et al. Proc Natl Acad Sci U S A. 2013;110:14225–30. doi: 10.1073/pnas.1306345110.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/27036
References
- 1.Conticello SG. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robbiani DF, et al. Annu Rev Pathol. 2013;8:79–103. doi: 10.1146/annurev-pathol-020712-164004. [DOI] [PubMed] [Google Scholar]
- 3.Burns MB, et al. Nature. 2013;494:366–70. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandrov LB, et al. Australian Pancreatic Cancer Genome Initiative. ICGC Breast Cancer Consortium. ICGC MMML-Seq Consortium. ICGC PedBrain Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohli RM, et al. J Biol Chem. 2010;285:40956–64. doi: 10.1074/jbc.M110.177402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M, et al. J Exp Med. 2010;207:141–53. doi: 10.1084/jem.20092238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nabel CS, et al. Nat Chem Biol. 2012;8:751–8. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nabel CS, et al. Proc Natl Acad Sci U S A. 2013;110:14225–30. doi: 10.1073/pnas.1306345110. [DOI] [PMC free article] [PubMed] [Google Scholar]