Abstract
Adult adipose tissue contains a large supply of progenitors that can renew fat cells for homeostatic tissue maintenance and adaptive growth or regeneration in response to external challenges. However, the in vivo mechanisms that control adipocyte progenitor behavior are poorly characterized. We recently demonstrated that recruitment of adipocyte progenitors by macrophages is a central feature of adipose tissue remodeling under various adipogenic conditions. Catabolic remodeling of white adipose tissue by β3-adrenergic receptor stimulation requires anti-inflammatory M2-polarized macrophages to clear dying adipocytes and to recruit new brown adipocytes from progenitors. In this Extra Views article, we discuss in greater detail the cellular elements of adipogenic niches and report a strategy to isolate and characterize the subpopulations of macrophages and adipocyte progenitors that actively participate in adrenergic tissue remodeling. Further characterization of these subpopulations may facilitate identification of new cellular targets to improve metabolic and immune function of adipose tissue.
Keywords: adipocyte progenitors, macrophages, proliferation, adipogenesis, adipose tissue remodeling
Introduction
Adipocytes are a specialized cell type that stores excess energy as triglycerides and rapidly mobilizes free fatty acids (FFA) in response to systemic demands. Adult adipose tissue contains a large supply of progenitors that can renew fat cells for homeostatic turnover, adaptive hyperplastic expansion, and regeneration after injury.1 Highly committed adipocyte progenitors can be isolated from the stromovascular cell (SVC) fraction of adipose tissue using a combination of negative and positive selection of cell surface markers.2-4 More recently, genetic lineage tracing studies have identified specific subpopulations of SVC that become adipocytes in intact tissue.3,5-8
While adipocyte progenitors have been identified, the mechanisms that control progenitor behavior in vivo remain enigmatic. In many tissues, adult stem cells/progenitors reside in microenvironments that control the balance between the quiescent and activated state.9-11 These niches are defined in part by the surrounding cell types, which are recognized as important regulators of stem cell/progenitor behavior.11 Adipose tissue contains multiple cell types, such as immune cells, endothelial cells, and fibroblasts, all of which may be viewed as potential progenitor niche components.12
Macrophages are predominant immune cells in adipose tissue and can constitute up to 40% of total SVC fraction under certain physiological and pathological conditions.13-19 It is well documented that hypertrophied adipose tissues of obese individuals accumulate macrophages that release proinflammatory cytokines, which can contribute to insulin resistance.16,20,21 This inflamed adipose tissue is characterized by dying/dead adipocytes surrounded by macrophages, forming crown-like structures (CLS).22 However, a regulatory role of macrophages during in vivo adipogenesis from progenitors has not been established.
Recently, we have demonstrated interplay between macrophages and progenitors under various adipogenic conditions,19 including high-fat diet (HFD) feeding, β-adrenergic stimulation, and local injury. Use of β3 adrenergic receptor (ADRB3) agonists, in particular, offers a highly selective means of brown adipocyte recruitment in white adipose tissue (WAT) by activating protein kinase A (PKA)-mediated signals specifically in fat cells.23 We found that newly recruited brown adipocytes are derived from stromal cells that express the stem cell markers CD34, Sca1, and platelet-derived growth factor receptor α (PDGFRα+ progenitors). Genetic tagging of these progenitors with inducible Cre recombinase allowed us to trace their interactions with other cell types over the course of cell activation, proliferation, and differentiation.19 We reported that ADRB3 stimulation triggers an immune response that depends on FFA mobilization19,23 and have identified CLS containing anti-inflammatory macrophages as a transient adipogenic tissue niche.19 In this model (Fig. 1), ADRB3 stimulation provokes the death of vulnerable white adipocytes. Signals generated by dead/dying fat cells lead to the recruitment of noninflammatory macrophages that surround the dead fat cell and clear the large remnant triglyceride core. PDGFRα+ progenitors are recruited to sites of fat cell efferocytosis, where they proliferate as an intimate part of the CLS. The tight spatial and temporal associations between macrophage and progenitor recruitment suggested a cause–effect relationship, and we have identified osteopontin (OPN) as one of the molecular players released from macrophages to recruit progenitors to the site of adipocyte clearance.19 We found that OPN is chemotactic for PDGFRα+ cells in vitro, and genetic ablation of OPN diminished CLS formation and progenitor proliferation and differentiation in vivo. While we propose that adipocyte death is a key cellular event that initiates adipose tissue remodeling, OPN is required to promote auto-amplified recruitment of macrophages and enhance local interaction of macrophages with progenitors. Finally, rapid resolution is accompanied by increased oxidative metabolism of WAT and new brown adipocyte formation in adipose tissue.
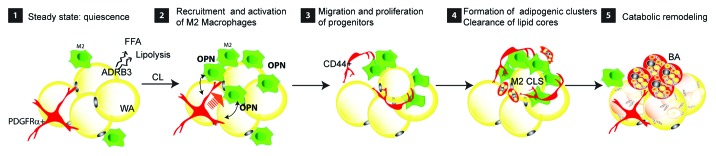
Figure 1. ADRB3 stimulation-induced adult adipose tissue remodeling. (1) During steady-state, adipocyte progenitors (PDGFRα+ cells) and tissue-resident macrophages (M2) are evenly distributed in white adipose tissue. (2) ADRB3 stimulation leads to adipocyte distress/death, potentially by excessive lipolysis, which recruits M2 macrophages that form CLS. Activated macrophages within the CLS release OPN that recruits adipocyte progenitors. (3) CD44+ PDGFRα+ progenitors proliferate and become an intimate part of CLS. (4) Macrophages clear lipid remnants from dying adipocytes and PDGFRα+ progenitors undergo adipogenesis. (5) As M2 macrophages regress, clusters of new brown adipocytes (BA) appear which contribute to catabolic remodeling of adipose tissue.
In this Extra Views article, we present an advancement of our previous work on the adipogenic niche by developing a strategy to isolate and characterize subpopulations of macrophages and progenitors that are actively participating in this remodeling process. We identified CD44 as a marker that can distinguish between activated progenitors and quiescent progenitors. In addition, we found that local proliferation of F4/80hi macrophages is a major source of M2 macrophage recruitment during CL-induced WAT remodeling.
Results
In vivo adipogenesis is restricted to CD44+ PDGFRα+progenitors
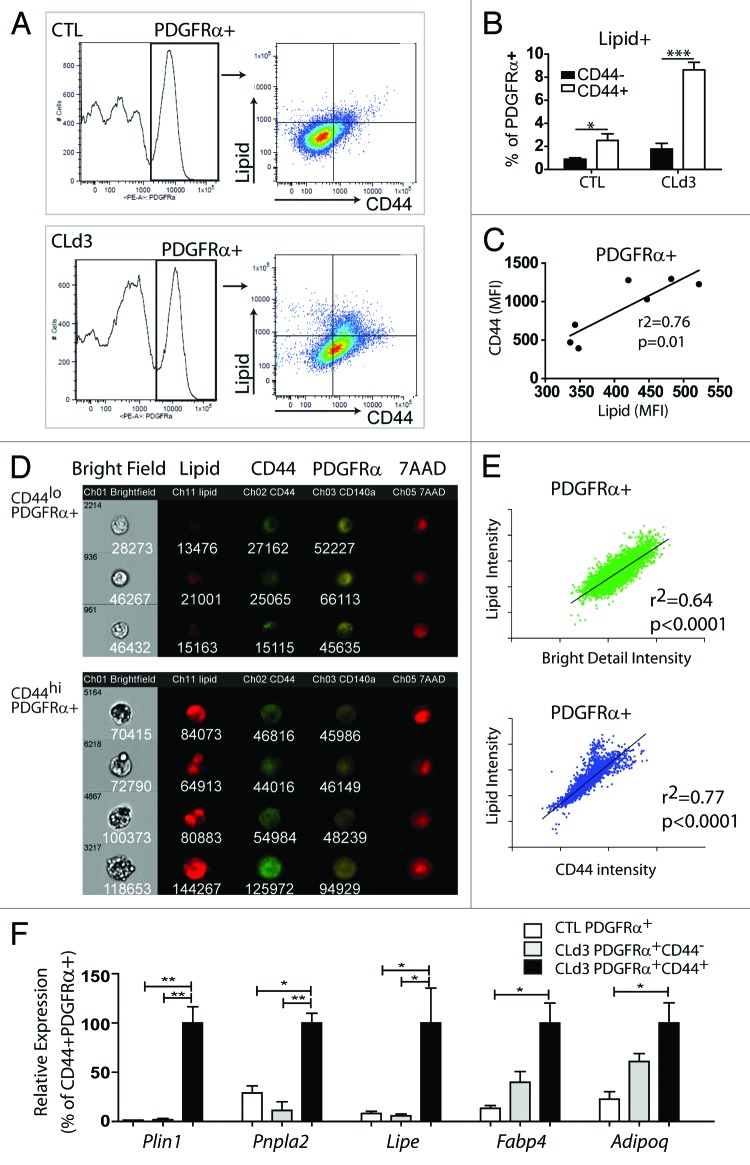
We previously demonstrated that ADRB3 stimulation recruits a subpopulation of PDGFRα+ progenitors that expressed an OPN receptor, CD44.19 Interestingly, cell proliferation is restricted to CD44+PDGFRα+progenitors, as indicated by expression of various proliferation markers (ki67, cyclin A2, and cyclin B1) and EdU incorporation.19 This suggested that CD44 expression may define progenitors that are primed to become adipocytes. To test if CD44 expression identifies a subpopulation of progenitors undergoing adipogenic differentiation, we examined lipid accumulation in CD44+ and CD44− subgroups of PDGFRα+ cells by flow cytometry (Fig. 2). Three days of CL treatment increased the lipid content in a subpopulation of PDGFRα+ cells in abdominal WAT, and this increase was restricted almost exclusively to CD44+PDGFRα+ cells (Fig. 2A and B). As expected, levels of CD44 expression were strongly correlated with lipid content in PDGFRα+ cells (Fig. 2C). Imaging flow cytometry confirmed the higher lipid content in PDGFRα+CD44hi cells compared with PDGFRα+CD44lo cells (Fig. 2D and E). Bright field images clearly indicated lipid droplet formation in individual CD44+PDGFRα+ cells (Fig. 2D). The Bright Detail Intensity tool of the IDEAS software captures the multilocular morphology seen in bright field images, and this parameter strongly correlated with cellular lipid content of CD44+PDGFRα+ cells (Fig. 2E). For further demonstration, we isolated CD44+PDGFRα+ fraction by FACS and examined adipogenic gene expression by quantitative PCR. Adipogenic marker expression (perilipin1 [Plin1], fatty acid binding protein 4 [Fabp4], patatin-like phospholipase domain containing 2 [Pnpla2], and adiponectin [Adipoq]) was highly enriched in CD44+ PDGFRα+ cells compared with PDGFRα+ cells from control conditions and CD44−PDGFRα+ cells from CL-treated mice (Fig. 2F). Together, data suggest that CD44 expression can be used to distinguish activated pre-adipocytes from quiescent progenitors, and to identify progenitors at the earliest stages of adipogenesis in vivo.
Figure 2. CD44 expression defines differentiating adipocyte progenitors. (A–E) FACS analysis of CD44 expression and lipid accumulation in PDGFRα+ cells in gonadal WAT (gWAT) from control mice and mice treated with CL for 3 d. Representative flow profiles (A) and quantification (B) of lipid+CD44+ and lipid+CD44- cells (mean ± SEM; n = 3–4; *P < 0.05, **P < 0.01). (C) Mean fluorescence intensity (MFI) analyses of PDGFRα+ cells demonstrate a positive correlation between CD44 expression levels and cellular lipid content. (D) Images of PDGFRα+ cells collected using ImageStream flow cytometry demonstrate that high CD44 expression predicted intracellular lipid droplet formation as indicated by LipidTox staining and bright field morphology. Intensity values are displayed below each image. Nuclei were counterstained with 7-AAD. (E) Flow profiles of PDGFRα+ cells showing a positive correlation between lipid staining and Bright Detail Intensity (cellular texture feature) or CD44 expression. Linear regression analyses were performed with log-transformed data (intensity values). (F) Quantitative PCR analysis of adipogenic marker expression in FACS-isolated cells from gWAT of control mice or mice treated with CL for 3 d (n = 3–4, mean ± SEM; *P < 0.05, **P < 0.01).
We note that PDGFRα+ CD44+cells were also detected in control conditions, albeit at much lower frequency. Importantly, these cells also had higher lipid content (Fig. 2A and B), indicating that CD44 expression likely identifies progenitors undergoing adipogenesis during homeostatic turnover as well.
Adipose tissue macrophages proliferate in situ, and clear lipid remains during ADRB3 agonist-induced WAT remodeling
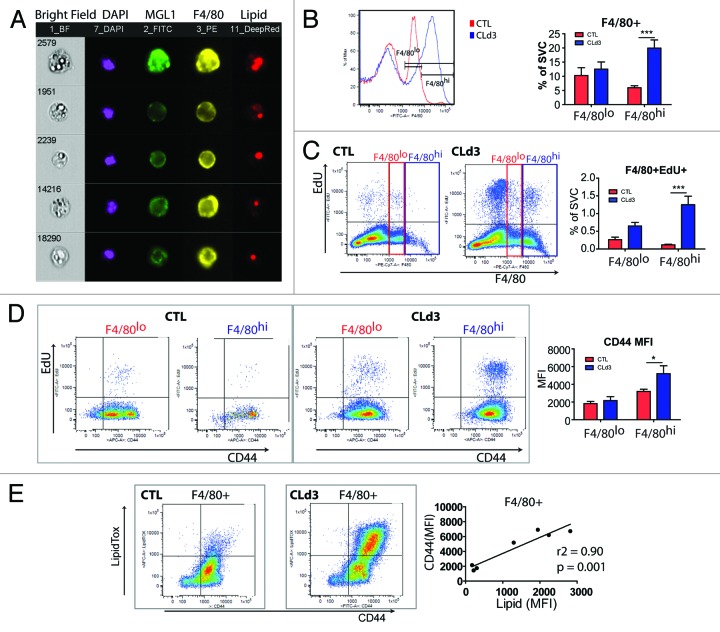
ADRB3 stimulation-induced adipocyte death appears to be a major cellular event that triggers WAT remodeling in abdominal WAT. Because adipocytes are largely occupied by lipids, adipose tissue remodeling requires efficient removal of the lipid remnants from dead adipocytes. Indeed, histological and flow cytometric analyses identified lipidhi macrophages.19 Imaging flow cytometry confirmed cytoplasmic lipid droplets in numerous MGL1+ M2 macrophages (Fig. 3A). We previously reported that CL treatment increases macrophage number in abdominal WAT, although the mechanisms of macrophage recruitment remained unclear.19 Evidence suggests that macrophage recruitment involves 2 distinct mechanisms: (1) local proliferation of tissue macrophages; and (2) infiltration from bone marrow-derived monocytes and subsequent differentiation into macrophages.13,24 For example, tissue-resident macrophages in several anatomic locations, including brain, liver, and lung, maintain steady-state pool size by replacing lost cells without influx of circulating monocytes derived from bone marrow hematopoiesis.25 In contrast, upon bacterial infection, pro-inflammatory macrophages can be recruited from circulating Ly6C+ monocytes.13,26 Although multiple cell surface markers are required to define tissue macrophages at various anatomic location,27 tissue-resident macrophages are often characterized by high expression of F4/80.24 Interestingly, we observed significant expansion of F4/80hi macrophages during ADRB3 stimulation (Fig. 3B). After flash labeling with EdU (i.e., 2 h of EdU exposure), 5.8% of F4/80+ macrophages incorporated EdU and F4/80hi macrophages showed much greater mitogenic responses compared with F4/80lo macrophages (Fig. 3C). These data indicate that macrophage recruitment during CL treatment involves local proliferation of resident macrophages in adipose tissue.
Figure 3. Characterization of macrophage recruitment during ADRB3 stimulation. (A) Single-cell images of MGL1+F4/80+ M2 macrophages obtained from ImageStream. Cytoplasmic lipid droplets are visualized in bright field images and LipidTox staining. Nuclei were counterstained with DAPI. (B–E) Flow cytometric characterization of F4/80+ macrophages in gWAT from control mice and mice treated with CL for 3 d. Mice were injected with EdU 2 h before sacrifice (n = 4–6 per condition, mean ± S.E.M). (B) Representative histograms and quantification of F4/80hi and F4/80lo macrophages in SVC from gWAT. Contents of F4/80hi macrophages significantly increased after 3 d of CL treatment (***P < 0.001). (C) Analysis of EdU incorporation in F4/80hi and F4/80lo macrophages, showing a significant increase in the mitotic index of F4/80hi macrophages (***P < 0.001). (D) Analysis of CD44 expression in F4/80hi and F4/80lo macrophages, showing upregulation of CD44 expression in F4/80hi macrophages after 3 d of CL treatment (*P < 0.05). (E) Analysis of lipid content and CD44 expression in F4/80+ macrophages. MFI of lipid staining and CD44 expression showed a strong positive correlation.
CD44 has been reported to play a crucial role in immune responses, including resolution of inflammation after injury and removal of apoptotic cells.28,29 We also observed upregulation of CD44 expression in F480hi macrophages (Fig. 3D), suggesting that CD44 expression may define recruited macrophages that actively clear residual lipid. Further flow cytometric characterization demonstrated that CD44 expression correlated strongly with macrophage lipid content (Fig. 3E). Together, these data indicate that ADRB3-induced adipose tissue remodeling involves proliferation of tissue-resident macrophages that are specialized to catabolize the lipid remains of dead adipocytes.
Discussion
Adipogenesis involves the commitment of mesenchymal stem cells to the adipogenic lineages and their subsequent differentiation into pre-adipocytes and mature adipocytes.1,30 In vitro studies using pre-adipocyte cell lines (e.g 3T3-L1) have provided a great deal of information on transcriptional controls of adipogenesis. However, in vivo events occurring in adipocyte progenitors are poorly characterized. In this report, we identified CD44 as a marker that can discriminate differentiation-primed progenitors from quiescent progenitors. Thus, characterization of CD44+PDGFRα+ cells can be utilized as a powerful tool to examine the early steps of adipogenesis in vivo. Furthermore, in conjunction with lipid content and PDGFRα expression levels, this may allow elucidation of a cellular hierarchy in adipogenesis and identification of stage-specific differentiation markers.
PDGFRα+ progenitors are distributed evenly throughout adipose tissue under basal conditions. This cell type has distinct morphology, with long cytoplasmic processes that appear to contact numerous cells in the local environment, including the vascular cells, adipocytes, and immune cells. These morphological characteristics may allow progenitors to sense and respond to changes in microenvironment. Most PDGFRα+ cells are quiescent under basal conditions, and it is possible that quiescence is maintained by anti-adipogenic signals, such as WNT and hedgehog modulators from stromal cells and adipocytes.31 Beside signals from niche cells, intercellular contacts or contact with the extracellular matrix may be required for maintenance of quiescent progenitors.32
We have shown that a common progenitor can give rise to brown or white adipocytes in abdominal adipose tissue.8 Interestingly, the fate of PDGFRα+ cells is determined by the nature of inductive signals. Under ADRB3 stimulation, PDGFRα+ cells become brown adipocytes, whereas HFD feeding recruits white adipocytes from PDGFRα+ cells.8 Our expression profiling studies indicate that ADRB3 is expressed in CD44+PDGFRα+ cells (unpublished data), implying that adrenergic signaling may direct the induction of the brown adipocyte phenotype during this early stage of differentiation. PKA signaling during proliferation of progenitors may have a critical role in determining metabolic characteristics of the resulting new fat cells, possibly by inducing epigenetic reprogramming.
Macrophages are a constituent of the mononuclear phagocyte system and function in the innate immune response and homeostatic tissue maintenance.33 Macrophages have been categorized into functionally distinct subsets based on cell surface markers and patterns of cytokine expression.34,35 Beyond the heterogeneity between tissues, macrophages are often classified according to their activation status. In general, M1 macrophages are activated by bacterial infections and secrete proinflammatory cytokines, such as TNFα and CCL2.13,36 In contrast, M2 macrophages are associated with tissue repair, parasite infection, and remodeling.13,36
Our results indicated that M2-polarized macrophages perform important functions during adipose tissue remodeling, including adipocyte efferocytosis and the recruitment and differentiation of adipocyte progenitors. As mentioned above, effective clearance of dead adipocytes by M2 macrophages is a remarkable feature of catabolic remodeling of WAT.19 The M2 CLS are widespread during the first 3 d of remodeling, yet are completely resolved by 7 d. In contrast, CLS found in adipose from obese individuals seem to be slower in eliminating remains of dying adipocytes and display a chronic inflammatory reaction, indicated by the sporadic appearance of giant multinucleated macrophages.37 It is likely that differences may exist in lipid metabolizing capacity of macrophage subsets. For example, it has been reported that M2 macrophages favor oxidative phosphorylation as their metabolic energy source, whereas M1 macrophages exhibit a propensity for aerobic glycolysis.38 Furthermore, M2 polarization promotes FFA uptake and oxidation, which may stimulate efficient lipid clearance during adipose tissue remodeling.
Although a phenotypic shift into the proinflammatory M1 polarization has been proposed to be a pathological feature of obesity,18,39 recent work indicates that the obese state recruits macrophages with a complex mixture of M1 and M2 phenotypes.19,40,41 It is presently unclear whether this heterogeneity represents differences in anatomical distribution, for example cells in CLS vs. those scattered throughout the tissue.22,42 This is an important issue, since obesity involves adipocyte turnover and recruitment, yet is also associated with generalized proinflammatory signaling that contributes to insulin resistance.43,44 We have observed PDGFRα+ progenitors within CLS of obese mice, suggesting that macrophages in CLS are important in recruiting new adipocytes from progenitors.8,19 Alternatively, adipose tissue macrophages may possess specialized molecular characteristics, where the M1–M2 classification system is not relevant. For example, recent work identified populations of macrophages from obese animals that have a mixed M1/M2 profile and display elevated lysosomal lipid metabolism.41 These cells appear to suppress adipocyte lipolysis while promoting lysosome-mediated fat clearance. It is presently unclear whether activation of lysosome biogenesis is specific to hypertrophied adipose tissue, or if it is a general mechanism of lipid catabolism in adipose tissue macrophages. It would be informative to characterize the catabolic phenotype of recruited macrophages under various adipogenic and lipolytic conditions (e.g., fasting, calorie restriction, adrenergic remodeling).15,19 A cell-isolation strategy based on CD44 expression and lipid content may allow more precise isolation of the relevant subpopulations.
In addition to their lipid scavenging role, macrophages release OPN as a chemotactic signal to recruit progenitors. Macrophages may produce other cytokines and growth factors that induce progenitor proliferation and differentiation. Macrophages play a critical role in the organogenesis of several tissues,45 and recent work indicates that loss of tissue-resident M2 macrophages results in lipodystrophy.46 Importantly, lipid metabolites from macrophages, such as peroxisome proliferator activated receptor (PPAR) ligands and prostaglandins, may function as adipogenic factors. Analysis of the transcriptional signatures and lipid metabolism in the subpopulation of CD44hiLipidhi macrophages may provide mechanistic insights into healthy adipose tissue remodeling.
In summary, our results indicate that progenitor recruitment by macrophages is a universal feature of adipose tissue remodeling and restoration. Future studies on activated progenitor populations will assist identification of regulatory networks that govern in vivo adipogenesis, while characterization of the origin and metabolic phenotypes of adipose tissue macrophages may facilitate development of new strategies to improve adipose tissue function.
Materials and Methods
Mice
C57BL/6J mice (Jackson Laboratory, Stock # 000664) were used for all experiments. For continuous ADRB3 stimulation, mice were infused with CL316.243 (0.75 nmol/h) by osmotic pumps (ALZET, 1003D) for 3 d and injected with 5-ethynyl-2’-deoxyuridine (EdU, Invitrogen, 10 nmol/mouse, i.p.) 2 h before sacrifice. All animal protocols were approved by the Institutional Animal Care and Use Committee at Wayne State University.
WAT stromovascular cell fractionation and flow cytometry
SVC fractions from mouse gonadal WAT (gWAT) were isolated and analyzed by flow cytometry, as previously described.19 Antibodies used for FACS analysis were anti-PDGFRα-PE (Biolegend, rat, 1:200), F4/80-PE, FITC, or PE/Cy7 (Biolegend, rat, 1:200), MGL1-FITC (Abserotec, rat, 1:100), CD44-FITC, APC, or PE/Cy5 (Biolegend, rat, 1:100). Species-matched IgG isotypes were used as nonspecific controls. HCS LipidTOX™ Deep Red Neutral Lipid Stain (Invitrogen, H34477) was used for lipid staining. Click-it EdU Pacific Blue or Alexa Flour 488 detection kit (Invitrogen, C-10418, C-10425) were used for EdU detection. Flow cytometry was performed at the Microscopy, Imaging, and Cytometry Resources Core of Wayne State University. Cell sorting and analytic cytometry were performed using FACS Vantage SE SORP and BD LSR II (BD Biosciences) flow cytometers, respectively. Raw data were processed using FlowJo software (Tree Star).
Imaging flow cytometry was performed using ImageStream (EMD Millipore-Amnis) with 40× magnification, and the acquired images were analyzed with IDEAS software (EMD Millipore-Amnis). A gradient RMS (root mean square) and a scatter plot of aspect ratio/area were used to gate for cells in focus and single cells, respectively. Events positive for PDGFRα-PE were identified by an intensity feature that quantifies the relative brightness of a given fluorochrome on a cell and are calculated by taking the sum of the pixel intensities within a given channel mask normalized for camera dark-current. A cellular texture feature was quantified using Bright Detail Intensity R3, which computes the intensity of localized bright spots that are 3 pixels in radius or less within the masked area in the single cell image.
Statistical analysis
Statistical analyses were performed with GraphPad Prism 5. Data are presented as mean ± SEM. Statistical significance between two groups was determined by unpaired t test or Mann–Whitney test, as appropriate. Comparison among groups was performed using 1-way ANOVA or 2-way ANOVA, with Bonferroni posttests to determine the relevant P values.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Drs T Leff, J Wang, M Sanders, and members of CIMER for discussions. This study was supported by NIH grants (RO1DK62292 and RO1DK76629) to JGG. The Microscopy, Imaging, and Cytometry Resources Core was supported, in part, by NIH Center grant P30CA022453 to the Karmanos Cancer Institute, Wayne State University, and the Perinatology Research Branch of the National Institutes of Child Health and Development, Wayne State University.
Glossary
Abbreviations:
- CL
CL-316,243
- CLS
crown-like structure
- FFA
free fatty acids
- gWAT
gonadal white adipose tissue
- HFD
high-fat diet
- SVC
stromovascular cells
- WAT
white adipose tissue
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/27647
References
- 1.Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–56. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135:240–9. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 3.Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, Graff JM. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–6. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci U S A. 2011;108:143–8. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billon N, Iannarelli P, Monteiro MC, Glavieux-Pardanaud C, Richardson WD, Kessaris N, Dani C, Dupin E. The generation of adipocytes by the neural crest. Development. 2007;134:2283–92. doi: 10.1242/dev.002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez-Gurmaches J, Hung C-M, Sparks CA, Tang Y, Li H, Guertin DA. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab. 2012;16:348–62. doi: 10.1016/j.cmet.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scimè A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–7. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y-H, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab. 2012;15:480–91. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voog J, Jones DL. Stem cells and the niche: a dynamic duo. Cell Stem Cell. 2010;6:103–15. doi: 10.1016/j.stem.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones DL, Wagers AJ. No place like home: anatomy and function of the stem cell niche. Nat Rev Mol Cell Biol. 2008;9:11–21. doi: 10.1038/nrm2319. [DOI] [PubMed] [Google Scholar]
- 12.Cinti S. The adipose organ. Prostaglandins Leukot Essent Fatty Acids. 2005;73:9–15. doi: 10.1016/j.plefa.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 13.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–55. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen KD, Qiu Y, Cui X, Goh YPS, Mwangi J, David T, Mukundan L, Brombacher F, Locksley RM, Chawla A. Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature. 2011;480:104–8. doi: 10.1038/nature10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosteli A, Sugaru E, Haemmerle G, Martin JF, Lei J, Zechner R, Ferrante AW., Jr. Weight loss and lipolysis promote a dynamic immune response in murine adipose tissue. J Clin Invest. 2010;120:3466–79. doi: 10.1172/JCI42845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–20. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han MS, Jung DY, Morel C, Lakhani SA, Kim JK, Flavell RA, Davis RJ. JNK expression by macrophages promotes obesity-induced insulin resistance and inflammation. Science. 2013;339:218–22. doi: 10.1126/science.1227568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y-H, Petkova AP, Granneman JG. Identification of an adipogenic niche for adipose tissue remodeling and restoration. Cell Metab. 2013;18:355–67. doi: 10.1016/j.cmet.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–30. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortega Martinez de Victoria E, Xu X, Koska J, Francisco AM, Scalise M, Ferrante AW, Jr., Krakoff J. Macrophage content in subcutaneous adipose tissue: associations with adiposity, age, inflammatory markers, and whole-body insulin action in healthy Pima Indians. Diabetes. 2009;58:385–93. doi: 10.2337/db08-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cinti S, Mitchell G, Barbatelli G, Murano I, Ceresi E, Faloia E, Wang S, Fortier M, Greenberg AS, Obin MS. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res. 2005;46:2347–55. doi: 10.1194/jlr.M500294-JLR200. [DOI] [PubMed] [Google Scholar]
- 23.Granneman JG, Li P, Zhu Z, Lu Y. Metabolic and cellular plasticity in white adipose tissue I: effects of β3-adrenergic receptor activation. Am J Physiol Endocrinol Metab. 2005;289:E608–16. doi: 10.1152/ajpendo.00009.2005. [DOI] [PubMed] [Google Scholar]
- 24.Sieweke MH, Allen JE. Beyond stem cells: self-renewal of differentiated macrophages. Science. 2013;342:1242974. doi: 10.1126/science.1242974. [DOI] [PubMed] [Google Scholar]
- 25.Yona S, Kim K-W, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–61. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat Immunol. 2013;14:986–95. doi: 10.1038/ni.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teder P, Vandivier RW, Jiang D, Liang J, Cohn L, Puré E, Henson PM, Noble PW. Resolution of lung inflammation by CD44. Science. 2002;296:155–8. doi: 10.1126/science.1069659. [DOI] [PubMed] [Google Scholar]
- 29.Kawana H, Karaki H, Higashi M, Miyazaki M, Hilberg F, Kitagawa M, Harigaya K. CD44 suppresses TLR-mediated inflammation. J Immunol. 2008;180:4235–45. doi: 10.4049/jimmunol.180.6.4235. [DOI] [PubMed] [Google Scholar]
- 30.Tang QQ, Lane MD. Adipogenesis: from stem cell to adipocyte. Annu Rev Biochem. 2012;81:715–36. doi: 10.1146/annurev-biochem-052110-115718. [DOI] [PubMed] [Google Scholar]
- 31.Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol. 2011;12:722–34. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watt FM, Huck WTS. Role of the extracellular matrix in regulating stem cell fate. Nat Rev Mol Cell Biol. 2013;14:467–73. doi: 10.1038/nrm3620. [DOI] [PubMed] [Google Scholar]
- 33.Galli SJ, Borregaard N, Wynn TA. Phenotypic and functional plasticity of cells of innate immunity: macrophages, mast cells and neutrophils. Nat Immunol. 2011;12:1035–44. doi: 10.1038/ni.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–64. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 35.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Immunological Genome Consortium Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–28. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–37. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murano I, Barbatelli G, Parisani V, Latini C, Muzzonigro G, Castellucci M, Cinti S. Dead adipocytes, detected as crown-like structures, are prevalent in visceral fat depots of genetically obese mice. J Lipid Res. 2008;49:1562–8. doi: 10.1194/jlr.M800019-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.O’Neill LAJ, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–55. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- 39.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117:175–84. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaul ME, Bennett G, Strissel KJ, Greenberg AS, Obin MS. Dynamic, M2-like remodeling phenotypes of CD11c+ adipose tissue macrophages during high-fat diet--induced obesity in mice. Diabetes. 2010;59:1171–81. doi: 10.2337/db09-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu X, Grijalva A, Skowronski A, van Eijk M, Serlie MJ, Ferrante AW., Jr. Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18:816–30. doi: 10.1016/j.cmet.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishimura S, Manabe I, Nagasaki M, Hosoya Y, Yamashita H, Fujita H, Ohsugi M, Tobe K, Kadowaki T, Nagai R, et al. Adipogenesis in obesity requires close interplay between differentiating adipocytes, stromal cells, and blood vessels. Diabetes. 2007;56:1517–26. doi: 10.2337/db06-1749. [DOI] [PubMed] [Google Scholar]
- 43.Glass CK, Olefsky JM. Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab. 2012;15:635–45. doi: 10.1016/j.cmet.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Odegaard JI, Chawla A. Pleiotropic actions of insulin resistance and inflammation in metabolic homeostasis. Science. 2013;339:172–7. doi: 10.1126/science.1230721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pollard JW. Trophic macrophages in development and disease. Nat Rev Immunol. 2009;9:259–70. doi: 10.1038/nri2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Satoh T, Kidoya H, Naito H, Yamamoto M, Takemura N, Nakagawa K, Yoshioka Y, Morii E, Takakura N, Takeuchi O, et al. Critical role of Trib1 in differentiation of tissue-resident M2-like macrophages. Nature. 2013;495:524–8. doi: 10.1038/nature11930. [DOI] [PubMed] [Google Scholar]