Abstract
MYC (c-Myc) deregulation has been frequently associated with aggressive lymphomas and adverse clinical outcome in B-cell malignancies. MYC has been implicated in controlling the expression of miRNAs, and MYC-regulated miRNAs affect virtually all aspects of the hallmarks of MYC-driven lymphomas. Increasing evidence has indicated that there is significant cross-talk between MYC and miRNAs, with MYC regulating expression of a number of miRNAs, resulting in widespread repression of miRNA and, at the same time, MYC being subjected to regulation by miRNAs, leading to sustained MYC activity and the corresponding MYC downstream pathways. Thus, these combined effects of MYC overexpression and downregulation of miRNAs play a central regulatory role in the MYC oncogenic pathways and MYC-driven lymphomagenesis. Here, we provide biological insight on the function of MYC-regulated miRNAs, the mechanisms of MYC-induced miRNA repression, and the complicated feedback circuitry underlying lymphoma progression, as well as potential therapeutic targets in aggressive B-cell lymphomas.
Keywords: miRNA, c-MYC, aggressive B-cell lymphomas, epigenetic, methylation
Introduction
MYC is a basic helix–loop–helix leucine zipper transcription factor that coordinates the diverse transcriptional programs necessary for cell growth, proliferation, invasion, expansion, and angiogenesis.1,2MYC’s highly pleiotropic effects are mirrored by thousands of MYC target genes with roles in virtually every aspect of cell biology and oncology.3MYC is one of the most commonly overexpressed oncogene in cancer and one of the most robust and significant prognostic markers for B-cell lymphomas. MYC dysregulation has been implicated in the aggressive transformation of B-cell lymphomas.4 Although MYC has been described as a defining feature and the driving oncogene for Burkitt lymphoma, MYC has also been recognized in other non-Hodgkin B-cell lymphomas. MYC has been detected in 9–14% of diffuse large B-cell lymphomas, associated with an adverse prognosis as a result of chemoresistance and shortened survival.5,6 In mantle cell lymphoma (MCL), increased expression of MYC has been found to be associated with poor prognosis and MCL aggressiveness.7-9 MYC overexpression has been implicated in high-grade large cell transformation in follicular and marginal zone cell lymphomas, supporting the features of MYC in sustaining aggressive transformation of lymphomas.10
However, the underlying mechanisms for MYC action remain elusive in these lymphomas. The direct MYC-induced transcriptional changes that promote cell transformation are still unclear. Important insights into the molecular pathology of MYC-driven B-cell lymphomas could be gained through a better understanding of which targets are responsible for the biological consequences of MYC suppression or induction. It is increasingly clear that the MYC-targeted gene network also includes non-protein coding targets. Among the latter, microRNAs (miRNAs) have attracted the most attention as important regulators of MYC-driven lymphomagenesis. miRNAs are 20- to 22-nucleotide non-coding RNAs found in plants and animals that inhibit gene expression by targeting mRNAs to degradation or inhibiting translation of mRNAs.11 The human genome encodes thousands of miRNAs, which regulate a large fraction of the human transcriptome. MYC has been recently implicated in controlling the expression of a host of miRNAs.12-14 The predominant consequence of MYC activation is widespread repression of miRNA expression.12-14 Here, we will summarize the role of MYC-regulated miRNAs, especially MYC-repressed miRNAs in MYC-mediated oncogenic processes, and discuss the molecular mechanisms of MYC-induced miRNA repression. Finally, we will exploit the MYC-miRNA circuitry as a mechanism to sustain MYC hyperactivity and as a potential therapeutic target for MYC-driven B-cell malignancies.
MYC-Regulated miRNAs Are Associated with the Hallmarks of B-Cell Lymphomas
miRNAs have been shown to be associated with many of the classical hallmarks of cancer, including proliferation, differentiation, angiogenesis, and apoptosis. With their widespread range of influence on biological pathways and implications as either oncogenes or tumor suppressor genes, their dysregulation justifies their significant role in tumorigenesis leading to lymphoma.
The identification and role of MYC-regulated miRNAs were initially established through chromatin immunoprecipitation and miRNA expression array by using MYC-inducible cell lines, human and mouse models of B-cell lymphoma.13,14 Unexpectedly, the predominant consequence of activation of MYC is widespread repression of miRNA expression.14 Moreover, enforced expression of repressed miRNAs diminishes the tumorigenic potential of lymphoma cells in vitro and in vivo, supporting that MYC-repressed miRNAs function as tumor suppressor genes. Indeed, miRNA transcripts repressed by MYC include several with potent tumor suppressor activity, such as miR-15a/16-1, miR-34a, and let-7 family members. Given the ability of a single miRNA to regulate hundreds of targets, it comes as no surprise that MYC can exert pleiotropic effects and cellular functions through reprogramming of miRNA expression. Here, we highlight some recent advances on how miRNA regulation has been integrated into the MYC oncogenic program in the regulation of hallmarks of B-cell malignancies (see Fig. 1).
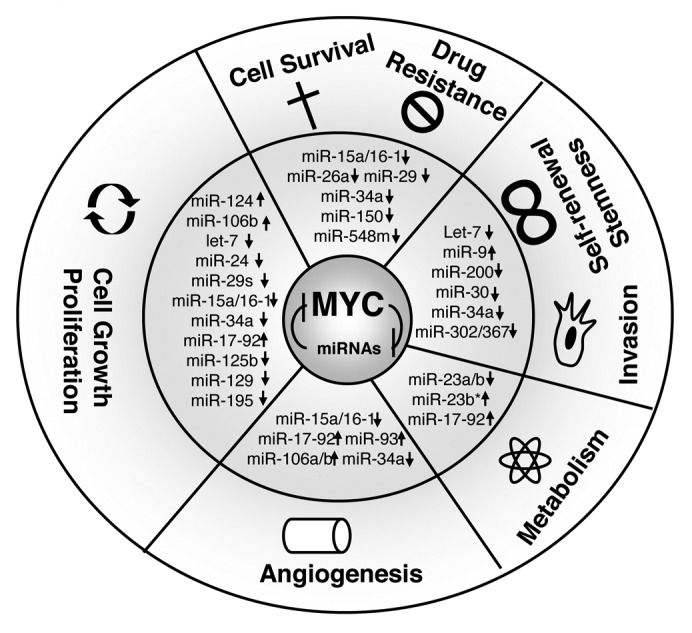
Figure 1. MYC-regulated miRNAs and the hallmarks of B-cell lymphomas. MYC and miRNAs form forward-feedback (double-negative) regulatory loops contributing to sustained MYC activation, miRNA downregulation, and subsequent dramatic deregulation of the hallmarks of lymphoma. “↑”, upregulation; “↓”, repression by MYC. MYC utilizes one or more of the above mechanisms in combination to exert its oncogenic functions.
Cell growth and proliferation
As expected, both induction and repression of specific miRNAs by MYC broadly impact MYC-mediated phenotypes, facilitating cell cycle entry and progression by controlling all levels of the cell cycle-regulatory machinery. The influence of these miRNAs on the cell cycle are mediated through inhibition of cell cycle inhibitors such as INK4 or Cip/Kip families by MYC-induced oncogenic miRNAs or through cell cycle-positive regulators, such as cyclins or cyclin kinases, via MYC-repressed miRNAs. For example, the miR-15a/16-1 cluster directly regulates cell cycle progression and proliferation by controlling the G1 checkpoint proteins. Overexpression of miR-16 leads to induction of G0/G1 arrest in tumor cells by suppressing the identified miR-15a/16-1 targets, including CDK1, CDK2, CDK6, as well as cyclins D and E.15 Thus, loss or repression of miR-15a/16-1, often observed in B-cell lymphomas, resulted in induction of these cell cycle-positive regulators and led to lymphoma cell growth and proliferation. The cell cycle kinase binds cyclins in early G1 phase and participates in the sequential phosphorylation of RB1 by CDK4/6 and CDK2 to repress RB inhibition of E2F, subsequently promoting G1-to-S-phase progression.15-18 CDK4 or CDK6 mRNAs are also regulated by other MYC-regulated miRNAs such as miR-24, miR-29, miR-34a, miR-124, miR-125b, miR-195, and let-7 family members.19,20 In addition, cyclin D levels are downregulated by let-7, miR-15 family, miR-17, miR-19a, miR-20a, and miR-34.21 Moreover, miR-16 and miR-34a downregulate cyclin E to regulate cell growth.18 Thus, when MYC is activated in aggressive B-cell lymphomas, these tumor suppressor miRNAs are inactivated by MYC, resulting in induction of these cell cycle-positive regulators and leading to cell proliferation and growth. On the other hand, MYC-activated miRNAs are also involved in cell proliferation and cell cycle progression. p21Cip1, a p53 target and CDK inhibitor, and pRB (retinoblastoma) are direct targets of miR-17-92 and miR-106b.22,23 By targeting this cell cycle inhibitor, MYC-induced activation of miR-17 and miR-106b promotes cell cycle progression.
Cell survival and drug resistance
A number of MYC-regulated miRNAs are involved in the regulation of lymphoma cell survival and drug resistance. Repression of miRNAs by MYC contributes to cellular survival by activation of anti-apoptotic proteins. For example, miR-15a/16-1, miR-26a, miR-29, miR-34a, and miR-150, which are repressed by MYC, can each activate several survival signaling pathways in B-cell lymphomas. Expression of these miRNAs inhibits cell proliferation, promotes apoptosis of lymphoma cells, and suppresses tumorigenicity both in vitro and in vivo.
The miR-15a/16-1 cluster directly downregulates the anti-apoptotic protein Bcl-2, Mcl-1, CCND1, and WNT3A. Downregulation of these miRNAs has been reported in B-cell malignancies.24 MiR-34a is a direct transcriptional target of p53 and contributes to p53-dependent apoptosis.25,26 Recently, Craig et al. reported that, of the MYC-repressed miRNAs that are downregulated in malignant lymphoma, miR-34a showed the strongest antiproliferative properties when overexpressed, and loss of miR-34a resulted in high proliferation in diffuse large B-cell lymphoma cells. This study further attributes miR-34a’s tumor-suppressive effects to deregulation of its target Foxp1. Our studies revealed that the miR-29 family (miR-29a-c) is inversely correlated with MYC expression and regulates cell growth and survival by targeting CDK6, IGF-1R, TCL-1, PI3K, and MCL1.20,27,28 We and others further demonstrated that miR-26a and miR-548 are repressed by MYC, contributing to lymphoma cell survival through silencing of EZH2 and HDAC6, respectively.20,27 Collectively, these observations highlight the broad impact of MYC-mediated miRNA reprogramming on cellular survival and proliferation pathways.
Angiogenesis and metabolism
In aggressive B-cell lymphomas, as in several other cancers, neo-angiogenesis and production of proangiogenic factors such as vascular endothelial growth factor (VEGF) play a central role in lymphoma progression. MYC may conceivably act as a VEGF transcriptional factor, promoting angiogenesis and vasculogenesis by upregulating the expression of proangiogenic factors.29 Consistent with these activities, MYC deactivation has been shown to contribute to the collapse of tumor.30 The mechanism through which MYC regulates the VEGF axis has not yet been clearly elucidated. Several lines of evidence revealed that MYC controls VEGF by regulating a broad range of miRNAs. VEGF translation is regulated by at least 8 miRNAs, including miR-15a, miR-16, miR-17, miR-20a, miR-34a, miR-93, miR-106a, miR-106b, and all of these miRNAs are under the control of MYC.31 MYC-driven angiogenesis can also be mediated by miRNAs through repression of antiangiogenic factors such as TSP1 (thrombospondin-1) and CTGF (connective tissue growth factor). miR-17-92 family members directly target the transcripts that encode TSP1 and CTGF, respectively, thereby reducing expression of these antiangiogenic proteins, and thus increasing angiogenesis.32
In addition, MYC activates the expression of genes to generate bioenergetic substrates for rapid cell growth and high metabolic activity in aggressive MYC-driven B-cell lymphomas. Both in vitro and in vivo models have provided substantial evidence that MYC induces many genes involved in ribosome biogenesis as well as genes involved in glucose and glutamine metabolism to accommodate to growing lymphoma cells.33 Mitochondrial glutaminase was among the proteins identified that are regulated by MYC, specifically through direct suppression of miR-23a and miR-23b. Suppression of these miRNAs triggers an addiction to glutamine, which is required for bioenergetics, nucleotide biosynthesis, and redox homeostasis in cancer cells. Using MYC-inducible human Burkitt lymphoma model P493-6 cells, it was shown that MYC suppressed proline oxidase expression primarily and regulated proline metabolism through upregulation of miR-23b*.34 Furthermore, the induction of the miR-17-92 cluster by MYC attenuates E2F1 protein expression, such that interruption of this regulatory loop results in DNA replication stress. The miR-17-92 cluster is also involved in glycolysis via potentiating signaling through the PI3K-AKT pathway. It is well established that the PI3K signaling route is engaged in glucose and fatty acid metabolism through multiple mechanisms by increasing glucose transporter surface expression and enhancing glycolytic enzyme.35 AKT also activates ATP citrate lyase to promote glucose-dependent fatty acid synthesis and tumor growth in vivo.36 Taken together, in aggressive B-cell lymphoma, activation of MYC orchestrates the expression of genes and miRNAs to meet the bioenergetic and biosynthetic demands of increased cell growth and proliferation.
Self-renewal, stemness, and invasion
Another biological setting in which MYC and miRNA regulation may converge is stem cell self-renewal and invasion. Genetic studies in mice and in embryonic stem (ES) cells reveled that MYC has been shown to be required for the maintenance of self-renewal.37 Furthermore, MYC family genes, together with OCT4, KLF4, and SOX2, act to reprogram differentiated cells into induced pluripotent stem (iPS) cells with ES cell properties, suggesting that MYC is a driver of pluripotency.38,39 Thus, MYC-targeted miRNAs and targeting-MYC miRNAs are involved in lymphoma self-renewal and invasion. Among the miRNAs, best correlating with a stem cell property (stemness) and self-renewal is miR-34. miR-34 family members downregulate the expression of stemness factors, such as SNAIL1, BMI1, CD44, and CD133. The functional relevance of these miRNAs in cancer cells was further validated by demonstrating that miR-34 is necessary for p53-mediated inhibition of important tumor cell properties, including migration and invasion.40 In lymphomas, loss of miR-34a promoted B-cell accumulation and high-grade transformation, and Foxp1 was a direct target of miR-34a in a 3′-untranslated region (UTR)-dependent fashion.41,42 Another miRNA family with a critical role in tumor self-renewal and invasion is let-7. The let-7 target genes, HMGA2, IMP-1, and LIN28B, as well as Ras and MYC have all been reported to be important in acquisition of cancer cell stemness.43 Indeed, let-7 has been associated with genesis and maintenance of the lymphoma aggressive phenotype in Burkitt lymphoma cells and B-cell differentiation.44,45 Similar to let-7, miR-30 is reduced by MYC, and its target proteins Ubc9 (ubiquitin-conjugating enzyme 9) and ITGB3 (integrin beta3) are markedly upregulated in breast cancer stem cell and associated with tumor progression, metastasis, and post-treatment relapse. Our recent study revealed that miR-30 family members are involved in B-cell differentiation.45
miR-200 members, another MYC-targeted miRNA family, which directly target the self-renewal regulator, polycomb ring finger oncogene BMI1, have been found to be highly downregulated in human breast stem cell as compared with non-tumorigenic cancer cells.46,47 Inhibition of miR-200b upregulates the expression of SUZ12, a subunit of a PRC2 (polycomb repressor complex 2), and is also required for mammosphere growth by repressing E-cadherin expression and increasing cell migration and growth.48 Further, miR-200s promotes the self-renewal and stemness through repressing the expression of ZEB1, ZEB2 (zinc finger E-box binding homeobox 1 and 2). ZEB1 has been described as able to directly suppress the transcription of miR-200 family members via a negative feedback loop, and is thought to regulate epithelial–mesenchymal transition (EMT) and to promote the invasion of cancer cells.49 Similarly, miR-302/367 cluster is regulated by the stem cell transcription factors OCT4 and SOX2, as well as MYC. Overexpression of miR-302 alone is enough to reprogram various somatic cells into induced pluripotent stem cells.50 MYC also induces expression of miR-9.51 Through targeting E-cadherin, miR-9 promotes tumor cell migration and invasion, B-lymphocyte differentiation, and MYC-driven lymphomagenesis.45,52 In conclusion, miRNAs are emerging as important markers and key regulators of cancer stem cell (CSC) by suppressing CSC-specific genes and activities, including self-renewal, invasion, and lymphoma aggressive transformation.
Mechanisms of miRNA Repression by MYC
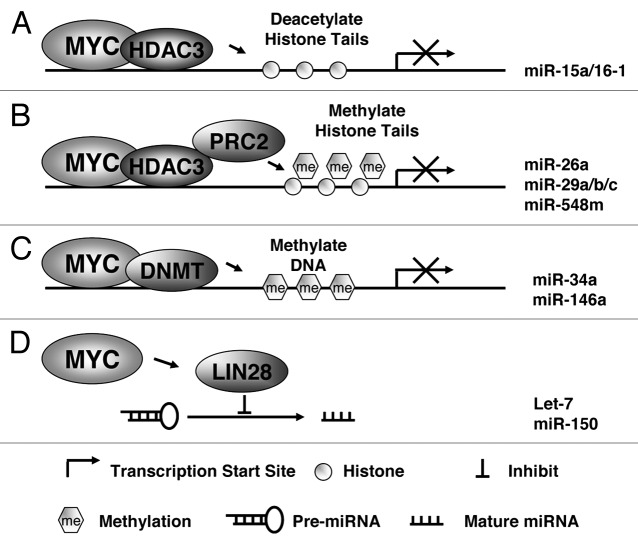
Although the mechanisms by which MYC activates gene transcription are relatively well known, less is known about how MYC represses transcription of target genes, including miRNAs. While genetic alteration of miRNA loci can be one cause of miRNA downregulation, it is likely that MYC hyperactivity also contributes to widespread downregulation of miRNA expression through transcriptional and post-transcritional regulation. The mechanisms through which MYC represses miRNAs are therefore of particular significance, since reversing these effects could have important therapeutic implications. Our study revealed that MYC acts as a repressor of miR-15a/16-1 by recruiting HDAC3.53 To investigate the role of histone acetylation and HDAC in MYC-induced miRNA repression, we performed miRNA expression profiling in lymphoma cells after pan-HDAC inhibitor, suberoylanilide hydroxamic acid (SAHA) treatment. We observed that SAHA indeed induced expression of a set of MYC-regulated miRNAs, including miR-29c, miR-26a, miR-30, and miR-15a/16-1. These findings suggest that histone deacetylation is involved in MYC-mediated transcriptional repression. Moreover, we recently demonstrated that MYC, HDAC3, and PRC2 form a repressive complex tethered to miR-29 and miR-26a promoter elements to epigenetically repress transcription of these miRNAs in MYC-expressing lymphoma cells.20 These results indicated that EZH2-mediated histone methylation is involved in MYC regulation of miRNA expression. This is further supported by our miRNA array study showing that EZH2 inhibition induced expression of a number of MYC-regulated miRNA such as miR-101, miR-200, miR-494, and miR-548.20,54 Collectively, these findings reveal that epigenetic histone acetylation and/or methylation are novel mechanisms for MYC-mediated miRNA transcriptional repression (Fig. 2).
Figure 2. Potential mechanisms of miRNA repression by MYC. (A) MYC recruits HDAC complex to miRNA promoter (of miR-15a/16-1) to induce histone tail deactylation, compact chromatin, and lead to miRNA transcription suppression. (B) PRC2 is tethered to miRNA promoter (of miR-26a, miR-29 and miR-548 min) with HDAC3 by MYC to methylate and deacetylate histone tail and subsequently inhibits miRNA transcription. (C) DNMT3 is recruited to miRNA promoter (of miR-34a and miR-146a) by MYC to methylate DNA and lead to repression of miRNA transcription. (D) MYC induces Lin-28B expression through direct association with the Lin-28 promoter, and Lin-28 proteins act as negative regulators of miRNAs (let-7 and miRNA-150) maturation and biogenesis.
One of the most common causes of the loss of tumor-suppressor miRNAs in B-cell lymphomas is the silencing of their primary transcripts by CpG island promoter hypermethylation. One line of evidence suggests that MYC recruits the DNA methyltransferase DNMT3a to the promoters of its negatively regulated target genes, an example of which is p21. DNA methylation-based regulation of miRNAs has also been recently described.55 To investigate whether miR-34a promoter methylation contributes to its repression in aggressive gastric lymphoma, Craig et al. performed methylation-specific PCR analyses of a CpG island that surrounds the transcriptional start site of the miR-34a.42 Primary miRNA-34a exhibited promoter hypermethylation in high-grade large B-cell lymphoma, indicating that epigenetic silencing through DNA methylation may contribute to miR-34a repression and constitute another mechanism for MYC-induced miRNA repression.42 Promoter methylation of miR-146a, another MYC-downregulated miRNA has been described in primary aggressive NK/T-cell lymphomas and is associated with low expression level and poor prognosis.56 Therefore, methylation may occur in promoter-associated CG islands of multiple miRNAs regulating MYC signaling pathways and functions (Fig. 2). Another layer of complexity for MYC miRNA regulation is miRNA processing (Fig. 2). Once transcribed, miRNAs are processed and exported from the nucleus to the cytoplasm. Alterations in the processing machinery can also lead to deregulation of functional miRNAs. Chang et al. revealed that MYC-mediated transactivation of the RNA-binding protein Lin-28B is necessary for MYC’s ability to post-transcriptionally repress let-7 family members.57 The study showed that MYC induces Lin-28B expression through direct association with the Lin-28 promoter, and that Lin-28 proteins act as negative regulators of let-7 maturation and biogenesis. Lin-28 regulates the expression of let-7 by binding to the precursors and blocking their maturation. More recently, Jiang et al. showed that the maturation of another MYC-repressed miRNA, miR-150, is also regulated by MYC-LIN28 axis, and mixed lineage leukemia (MLL) fusion proteins negatively regulate production of miR-150 by blocking miR-150 precursors from being processed to mature miRNAs through MYC/LIN28 functional axis.58 In summary, these data uncover an orchestration of transcriptional and posttranscriptional mechanisms in MYC-mediated reprogramming of miRNA expression (Fig. 2). Epigenetic silencing through DNA methylation or histone acetylation and/or methylation and disruption of miRNA biogenesis contribute to MYC-induced miRNA repression, and MYC utilizes one or more of the above mechanisms in combination to exert its oncogenic functions.
In addition, MYC also upregulates expression of a set of oncogenic miRNAs. Perhaps the most well-studied oncogenic miRNA induced by MYC is the miR-17-92 cluster, also known as Oncomir-1. This miRNA cluster is located within the noncoding gene C13orf25 at 13q31.3, which is frequently amplified in B-cell lymphomas and overexpressed in a variety of other tumors.59 Unlike most protein coding genes, miR-17-92 is a polycistronic miRNA cluster that contains multiple miRNA components, each of which has a function to regulate hundreds of target mRNAs. Transgenic expression of this cluster in mice leads to a lymphoproliferative disorder,60 while its genetic ablation impairs normal B-cell development.61 Six mature miRNAs, miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1, are encoded in this cluster. When overexpressed in the lymphoid compartment, miR-17-92 cluster-derived miRNAs cooperate with MYC in inducing lymphomas in the Eµ-myc mouse lymphoma model. Recently, miR-19 was identified as the key oncogenic component of the cluster in this model. In addition, miR-18 was also shown to have some oncogenic potential. The tumor suppressor PTEN and the proapoptotic protein Bim have emerged as important targets repressed by oncomir-1-derived miRNAs in hematopoietic system.59 Hence, the miRNAs in the miR-17-92 cluster regulate multiple functions involved in lymphomagenesis. Ectopic expression of miR-17-92 cooperates with the MYC oncogene in a mouse model of B-cell lymphomas.62 The functional interplay between miR-17-92 and MYC is further supported by the finding that MYC itself is a potent and direct transcriptional activator of miR-17-92,13 thus supporting that miR-17-92 may contribute to the oncogenic properties of MYC. The exact molecular basis underlying miRNA-mediated gene induction is not entirely clear. Our CHIP analysis revealed, in contrast to MYC-repressed miRNAs, much stronger MYC binding and weaker or no HDAC3 or/and EZH2 bindings to the E-box regions of miR-17-92 cluster gene promoter. The different binding complex of MYC and epigenetic modifiers (HDAC and EZH2) is likely attributed to E-box context of different miRNAs and dictates the transcriptional activation or repression function of MYC on miRNAs expression.
MYC Regulation by miRNAs and MYC–miRNA Circuitry as a Mechanism of Sustaining MYC Activity
The interaction between MYC and miRNAs is mutual, as MYC itself is targeted by miRNAs. When interactions between downregulated miRNAs and overexpressed target protein-coding genes were mapped in murine and human lymphomas, MYC was identified as the upregulated gene with the highest interaction with downregulated miRNAs.19 The functionally best-characterized MYC-targeting miRNA is miR-34a, which is located at chromosome 1p36, a region frequently deleted in MYC-associated lymphomas.14 Overexpression of miR-34a in lymphoma cell lines decreased MYC levels, inhibited proliferation, and induced apoptosis.14,63 The let-7 tumor suppressor miRNA, another MYC-repressed miRNA, is also able to downregulate MYC, reverting MYC-induced growth in Burkitt lymphoma cells.44 In addition to miR-34a and let-7, we revealed that miR-135a, miR-186, miR-494, miR-200c, miR-374a/b, miR-101, and miR-548 target MYC. To further validate that these miRNAs target the MYC 3′-UTR directly, we cloned the full-length of MYC 3′-UTR and constructed a luciferase reporter plasmid (p-miR-MYC-3′-UTR-WT). Overexpression with each of the aforementioned pre-miRNAs reduced the luciferase activity and diminished MYC levels, which reduced proliferation and clonogenic growth.20,54 On the other hand, miRNAs regulate MYC indirectly through targeting other MYC-regulatory proteins or miRNAs. The miRNA target proteins regulate MYC expression at transcriptional and posttranscriptional levels. For example, the miR-26a-regulated PRC2 protein, EZH2, modulates MYC expression through EZH2-mediated miR-494 expressions.20,27 We and others have also shown that a combination of MYC-targeting miRNAs has a stronger effect in MYC downregulation than individual miRNAs. Given widespread miRNA repression, including a panel of miRNAs that target MYC in aggressive lymphomas, it is predicted that downregulation of these miRNAs coordinately contribute to MYC activation and induction of MYC downstream oncogenic pathways, leading to lymphoma aggressive progression.
On another note, MYC is present at very low levels in normal cells, both the short-lived MYC protein and its equally short-lived mRNA, indicating that MYC levels are tightly regulated by transcriptional and posttranscriptional mechanisms. However, constitutive MYC activation has been reported in aggressive lymphomas.4 A regulatory mechanism that enhances the strength and prolongs the duration of MYC activity is required for these lymphomas. Indeed, accumulating evidence has shown that many of these MYC-targeting (direct or indirect) miRNAs are silenced by MYC through binding to E-boxes in the promoter regions of these target genes, thus generating forward-feedback (double-negative) regulatory loops between MYC and miRNAs in aggressive B-cell lymphomas (Fig. 1). In contrast to low MYC activation and high expression of MYC-repressed miRNAs in normal and reactive B lymphocytes, in aggressive MYC-associated lymphomas, lymphoma cells acquire diverse genetic or epigenetic alterations that result in MYC overexpresssion. Amplified and overexpressed MYC leads to low levels of these miRNAs, with the negative feedback loops for regulating MYC being abrogated. Thus, MYC-miRNA circuitry through autocrine/paracrine loops contributes to sustained MYC activation and subsequent dramatic deregulation of the cell cycle, protein translation, and metabolism among other cellular processes for lymphomas aggressive progression.
Summary and Perspectives
MYC is a potent oncogene initially identified as the hallmark and driving force in Burkitt lymphoma. Increasing evidence has supported that MYC gene alterations, which have been identified in other mature B-cell neoplasms, are usually associated with aggressive clinical behavior. The advent of functional and structural genomics with advances in immunology analyses and new animal models has greatly accelerated our understanding of oncogenic mechanisms in these MYC-associated lymphomas. The current findings of widespread downregulation of the miRNA transcriptome in aggressive lymphomas clearly indicate a central role of miRNA deregulation in lymphoma initiation and progression. The identification of the MYC-repressed tumor-suppressor miRNAs underlies the molecular mechanism of MYC-induced lymphomagenesis.
MYC has the ability to activate genes that increase the malignancy of the tumor and at the same time has the ability to repress genes such as tumor suppressors. Although the transcription activator mechanisms of MYC are relatively well known, few studies have been conducted to explain how MYC can exert its transcription repression function. One interesting aspect of that emerges from our own studies, showing that MYC can directly interact with many chromatin components, particularly HDAC3 and PRC2 proteins (EZH2, SUZ12), as a novel, genuine, and critical mechanism of MYC-mediated transcriptional repression.20,53 Thus, our findings provide a rational to redirect therapeutic effort by reactivating these MYC-repressed tumor suppressor miRNAs through inhibition of HDAC and/or EZH2. Indeed, we demonstrated that the combination of HDAC and EZH2 inhibitors (vorinostat and DZNep) induced miR-29 gene expression, resulting in the synergistic reduction of oncogenic protein levels of CDK6 and IGF-1R and subsequent inhibition of cell survival and lymphoma formation in vitro and in vivo. Further, the identification of the reciprocal MYC-miRNA feedback circuits added another layer of complexity to the molecular basis of sustaining MYC oncogenic signal and driving lymphoma aggressive progression.20,54 Thus, disruption of the MYC-miRNA loop to suppress aggressive B-cell lymphoma survival and growth can be a novel promising therapeutic approach. Indeed, recently, using a novel small-molecule BET bromodomain inhibitor, JQ1, and the EZH2 inhibitor, DZNep, we demonstrated that combined treatment of JQ1 and DZNep cooperatively disrupted MYC-miRNA, resulting in a greater MYC reduction, restoration of tumor suppressor miRNAs such as miR-26a, and a synergistic suppression of lymphoma growth and clonogenicity in aggressive lymphoma cells.27 Taken together, it is now becoming increasingly evident that interplay between MYC and miRNAs plays a crucial role in aggressive lymphomas. The control of MYC-miRNA interaction is therefore a therapeutic target for control of MYC and MYC downstream pathways and lymphoma therapy. From this perspective, a deep understanding of the nature of the genetic and epigenetic MYC regulation mechanisms and continued efforts to unravel the specific contribution of miRNA deregulation will be needed for improved therapy of aggressive lymphomas.
Disclosure of Potential Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by grants from the National Cancer Institutes (R01 CA137123, to JT), Maher Fund (to JT), Lymphoma Research Foundation (to JT), National Functional Genomics Center Programmatic Research grand and American Hematology Society Bridge Grant (to JT).
Glossary
Abbreviations:
- MCL
mantle cell lymphoma
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/27646
References
- 1.Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–30. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 2.Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–66. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ott G, Rosenwald A, Campo E. Understanding MYC-driven aggressive B-cell lymphomas: pathogenesis and classification. Blood. 2013;122:3884–91. doi: 10.1182/blood-2013-05-498329. [DOI] [PubMed] [Google Scholar]
- 5.Savage KJ, Johnson NA, Ben-Neriah S, Connors JM, Sehn LH, Farinha P, Horsman DE, Gascoyne RD. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood. 2009;114:3533–7. doi: 10.1182/blood-2009-05-220095. [DOI] [PubMed] [Google Scholar]
- 6.Valera A, López-Guillermo A, Cardesa-Salzmann T, Climent F, González-Barca E, Mercadal S, Espinosa I, Novelli S, Briones J, Mate JL, et al. Grup per l’Estudi dels Limfomes de Catalunya i Balears (GELCAB) MYC protein expression and genetic alterations have prognostic impact in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Haematologica. 2013;98:1554–62. doi: 10.3324/haematol.2013.086173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernández L, Hernández S, Beà S, Pinyol M, Ferrer A, Bosch F, Nadal A, Fernández PL, Palacín A, Montserrat E, et al. c-myc mRNA expression and genomic alterations in mantle cell lymphomas and other nodal non-Hodgkin’s lymphomas. Leukemia. 1999;13:2087–93. doi: 10.1038/sj.leu.2401599. [DOI] [PubMed] [Google Scholar]
- 8.Nagy B, Lundán T, Larramendy ML, Aalto Y, Zhu Y, Niini T, Edgren H, Ferrer A, Vilpo J, Elonen E, et al. Abnormal expression of apoptosis-related genes in haematological malignancies: overexpression of MYC is poor prognostic sign in mantle cell lymphoma. Br J Haematol. 2003;120:434–41. doi: 10.1046/j.1365-2141.2003.04121.x. [DOI] [PubMed] [Google Scholar]
- 9.Hartmann E, Fernàndez V, Moreno V, Valls J, Hernández L, Bosch F, Abrisqueta P, Klapper W, Dreyling M, Hoster E, et al. Five-gene model to predict survival in mantle-cell lymphoma using frozen or formalin-fixed, paraffin-embedded tissue. J Clin Oncol. 2008;26:4966–72. doi: 10.1200/JCO.2007.12.0410. [DOI] [PubMed] [Google Scholar]
- 10.Slack GW, Gascoyne RD. MYC and aggressive B-cell lymphomas. Adv Anat Pathol. 2011;18:219–28. doi: 10.1097/PAP.0b013e3182169948. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 12.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–5. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 14.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–52. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeshita F, Patrawala L, Osaki M, Takahashi RU, Yamamoto Y, Kosaka N, Kawamata M, Kelnar K, Bader AG, Brown D, et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol Ther. 2010;18:181–7. doi: 10.1038/mt.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, Fu XD, Zhou Y, Zhang Y. Down-regulation of the cyclin E1 oncogene expression by microRNA-16-1 induces cell cycle arrest in human cancer cells. BMB Rep. 2009;42:725–30. doi: 10.5483/BMBRep.2009.42.11.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 2011;1812:592–601. doi: 10.1016/j.bbadis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Zhao X, Fiskus W, Lin J, Lwin T, Rao R, Zhang Y, Chan JC, Fu K, Marquez VE, et al. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-Cell lymphomas. Cancer Cell. 2012;22:506–23. doi: 10.1016/j.ccr.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Bui TV, Mendell JT. Myc: Maestro of MicroRNAs. Genes Cancer. 2010;1:568–75. doi: 10.1177/1947601910377491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–74. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK, et al. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–81. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17:215–20. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 25.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–43. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 27.Zhao X, Lwin T, Zhang X, Huang A, Wang J, Marquez VE, Chen-Kiang S, Dalton WS, Sotomayor E, Tao J. Disruption of the MYC-miRNA-EZH2 loop to suppress aggressive B-cell lymphoma survival and clonogenicity. Leukemia. 2013;27:2341–50. doi: 10.1038/leu.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao JJ, Lin J, Lwin T, Yang H, Guo J, Kong W, Dessureault S, Moscinski LC, Rezania D, Dalton WS, et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115:2630–9. doi: 10.1182/blood-2009-09-243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knies-Bamforth UE, Fox SB, Poulsom R, Evan GI, Harris AL. c-Myc interacts with hypoxia to induce angiogenesis in vivo by a vascular endothelial growth factor-dependent mechanism. Cancer Res. 2004;64:6563–70. doi: 10.1158/0008-5472.CAN-03-3176. [DOI] [PubMed] [Google Scholar]
- 30.Sodir NM, Evan GI. Finding cancer’s weakest link. Oncotarget. 2011;2:1307–13. doi: 10.18632/oncotarget.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El Baroudi M, Corà D, Bosia C, Osella M, Caselle M. A curated database of miRNA mediated feed-forward loops involving MYC as master regulator. PLoS One. 2011;6:e14742. doi: 10.1371/journal.pone.0014742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–5. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479–83. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, Phang JM. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci U S A. 2012;109:8983–8. doi: 10.1073/pnas.1203244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–21. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009;28:3157–70. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 39.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–24. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 40.Liu C, Tang DG. MicroRNA regulation of cancer stem cells. Cancer Res. 2011;71:5950–4. doi: 10.1158/0008-5472.CAN-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao DS, O’Connell RM, Chaudhuri AA, Garcia-Flores Y, Geiger TL, Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33:48–59. doi: 10.1016/j.immuni.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Craig VJ, Cogliatti SB, Imig J, Renner C, Neuenschwander S, Rehrauer H, Schlapbach R, Dirnhofer S, Tzankov A, Müller A. Myc-mediated repression of microRNA-34a promotes high-grade transformation of B-cell lymphoma by dysregulation of FoxP1. Blood. 2011;117:6227–36. doi: 10.1182/blood-2010-10-312231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–52. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–70. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 45.Lin J, Lwin T, Zhao JJ, Tam W, Choi YS, Moscinski LC, Dalton WS, Sotomayor EM, Wright KL, Tao J. Follicular dendritic cell-induced microRNA-mediated upregulation of PRDM1 and downregulation of BCL-6 in non-Hodgkin’s B-cell lymphomas. Leukemia. 2011;25:145–52. doi: 10.1038/leu.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bai JX, Yan B, Zhao ZN, Xiao X, Qin WW, Zhang R, Jia LT, Meng YL, Jin BQ, Fan DM, et al. Tamoxifen represses miR-200 microRNAs and promotes epithelial-to-mesenchymal transition by up-regulating c-Myc in endometrial carcinoma cell lines. Endocrinology. 2013;154:635–45. doi: 10.1210/en.2012-1607. [DOI] [PubMed] [Google Scholar]
- 48.Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39:761–72. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–9. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, Ying SY. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–24. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–56. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Onnis A, De Falco G, Antonicelli G, Onorati M, Bellan C, Sherman O, Sayed S, Leoncini L. Alteration of microRNAs regulated by c-Myc in Burkitt lymphoma. PLoS One. 2010;5:5. doi: 10.1371/journal.pone.0012960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X, Chen X, Lin J, Lwin T, Wright G, Moscinski LC, Dalton WS, Seto E, Wright K, Sotomayor E, et al. Myc represses miR-15a/miR-16-1 expression through recruitment of HDAC3 in mantle cell and other non-Hodgkin B-cell lymphomas. Oncogene. 2012;31:3002–8. doi: 10.1038/onc.2011.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lwin T, Zhao X, Cheng F, Zhang X, Huang A, Shah B, Zhang Y, Moscinski LC, Choi YS, Kozikowski AP, et al. A microenvironment-mediated c-Myc/miR-548m/HDAC6 amplification loop in non-Hodgkin B cell lymphomas. J Clin Invest. 2013;123:4612–26. doi: 10.1172/JCI64210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–43. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 56.Paik JH, Jang JY, Jeon YK, Kim WY, Kim TM, Heo DS, Kim CW. MicroRNA-146a downregulates NFκB activity via targeting TRAF6 and functions as a tumor suppressor having strong prognostic implications in NK/T cell lymphoma. Clin Cancer Res. 2011;17:4761–71. doi: 10.1158/1078-0432.CCR-11-0494. [DOI] [PubMed] [Google Scholar]
- 57.Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A. 2009;106:3384–9. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang X, Huang H, Li Z, Li Y, Wang X, Gurbuxani S, Chen P, He C, You D, Zhang S, et al. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22:524–35. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–14. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–14. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cannell IG, Bushell M. Regulation of Myc by miR-34c: A mechanism to prevent genomic instability? Cell Cycle. 2010;9:2726–30. doi: 10.4161/cc.9.14.12182. [DOI] [PubMed] [Google Scholar]