Abstract
Zhangfei/CREBZF, a basic region-leucine zipper (bLZip) transcription factor, is a potent suppressor of growth and the unfolded protein response (UPR) in some cancer cell lines, including the canine osteosarcoma cell line, D-17. However, the effects of Zhangfei are not universal, and it has no obvious effects on untransformed cells and some cancer cell lines, suggesting that Zhangfei may act through an intermediary that is either not induced or is defective in cells that it does not affect. Here we identify the tumor suppressor protein p53 as this intermediary. We show the following: in cells ectopically expressing Zhangfei, the protein stabilizes p53 and co-localizes with it in cellular nuclei; the bLZip domain of Zhangfei is required for its profound effects on cell growth and interaction with p53. Suppression of p53 by siRNA at least partially inhibits the effects of Zhangfei on the UPR and cell growth. The effects of Zhangfei on D-17 cells is mirrored by its effects on the p53-expressing human osteosarcoma cell line U2OS, while Zhangfei has no effect on the p53-null osteosarcoma cell line MG63. In U2OS cells, Zhangfei displaces the E3 ubiquitin ligase mouse double minute homolog 2 (Mdm2) from its association with p53, suggesting a mechanism for the effects of Zhangfei on p53.
Keywords: cell cycle, protein domains, p53, osteosarcoma, protein translocation, Zhangfei/CREBZF, unfolded protein response, Mdm2, basic-leucine zipper domain
Introduction
Zhangfei/CREBZF/SMILE was first discovered as a binding partner for host cell factor (HCF), a co-activator of the herpes simplex virion transcription factor VP16.1 The primary structure of the protein contains a leucine zipper, a basic region that lacks an asparagine residue conserved in most bLZip proteins, 3 potential nuclear factor binding domains (LLXXLL, where L is a leucine residue and X is any amino acid), and a domain for binding HCF. Zhangfei interacts with several proteins, possibly through its nuclear receptor and HCF binding domains as well as its leucine zipper. While Zhangfei can activate gene expression through factors such as p532 and ATF4,3 it suppresses the activity of a number of transcription factors, which includes nuclear receptors,4-6 bLZip containing proteins such as CREBH,7 Luman/CREB3,8 Xbp1, and SMAD 1,5,8,9 and the HCF-binding VP16.10 Zhangfei also has a profoundly suppressive effect on the unfolded protein response (UPR), at least partly because it targets Xbp1s, an important UPR mediator, for proteasomal degradation.11
We have detected Zhangfei protein in differentiated neurons, but not in developing neurons or cells of neuronal tumors,10 nor in osteosarcoma cell lines. The ectopic expression of Zhangfei in several tumor cells lines derived from medulloblastomas (ONS-76, UW22812) and osteosarcomas (D-1713) causes the cells to stop growing and display markers of apoptosis. However, the suppressive effect of Zhangfei on the growth of cells is not universal. While the protein has a profound effect on some cells, it has no effect on others, such as MRC5 fibroblasts.12 This suggests that Zhangfei may act through an intermediary that is either not induced or is defective in cells that it does not affect. The objective of this study was to identify such an intermediary and to determine how Zhangfei exerted its effect. Herein, we identified that Zhangfei suppressed cell growth and the UPR in osteosarcoma cells through direct interaction with tumor suppressor protein p53. We demonstrated that Zhangfei stabilized p53 and promoted its nuclear retention by displacing the E3 ubiquitin ligase, Mdm2. Overall, our findings reveal a novel mechanism by which Zhangfei may inhibit tumor growth and metastasis. This may provide an alternative modality for the therapy of certain types of osteosarcoma, and perhaps other tumors with functional p53.
Results
Leucine zipper is required for the suppressive effects of Zhangfei on both cell growth and UPR
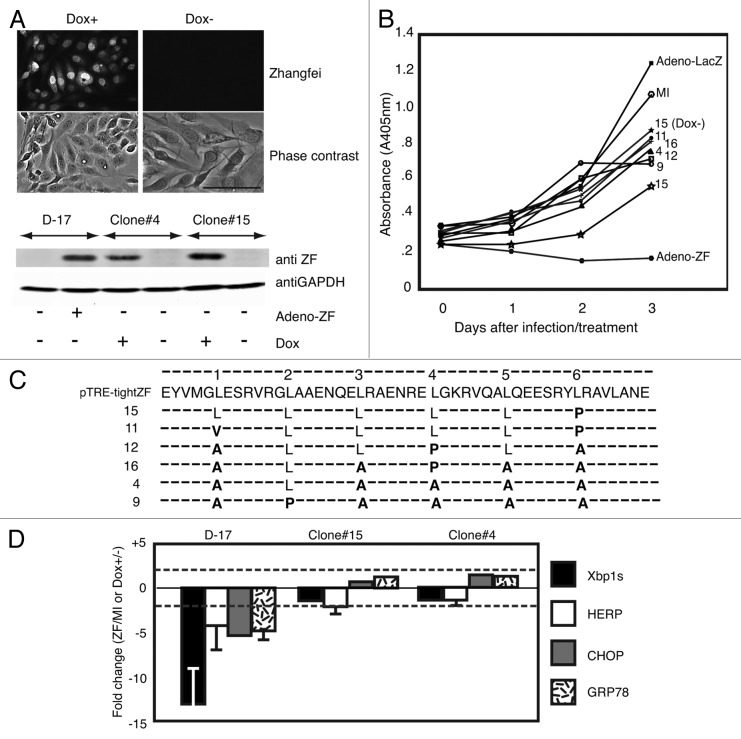
We have previously shown that Zhangfei has a profound effect on the growth and the induction of the UPR in the canine osteosarcoma cell line, D-17.13 We have also shown that Zhangfei requires its leucine zipper to suppress the UPR in some cells.11 To better characterize the molecular mechanism by which Zhangfei suppresses the growth of these cells, we attempted to generate clones of D-17 cells in which Zhangfei could be induced. Six clones that produced stable and detectable Zhangfei only in the presence of the inducer, doxycycline (Fig. 1A shows evidence from representative clones) were analyzed further. While the original D-17 cells responded to the ectopic expression of Zhangfei by complete cessation of growth (Fig. 1B, compare cells infected with adenovirus vectors expressing either Zhangfei [Adeno-ZF] or the control protein β-galactosidase [Adeno-LacZ] and mock-infected cells [MI]) the induction of Zhangfei had little or no effect on most of the clones (Fig. 1B, clones 4, 9, 11, 12, and 16). The induction of Zhangfei had a partial effect on growth on clone 15. We next amplified the complete coding sequences for Zhangfei by PCR from RNA purified from the induced cell clones as well as from pTRE-tightZF (the plasmid used to develop the doxycycline-inducible clones) and determined the nucleotide sequence of the products. While the derived amino acid sequence of the leucine zipper (LZip) region from pTRE-TightZF was identical to the published sequence, one or more leucines from the LZip regions of all clones had been replaced with other amino acids (Fig. 1C, bold). No other changes were observed in the entire Zhangfei coding sequences, suggesting that the leucine mutations were not random changes. To determine if the Zhangfei mutants retained the ability to suppress the UPR, we induced the UPR in the mutant cell clones with thapsigargin, with and without doxycycline, and compared stable levels of transcripts for 4 UPR-related genes: Xbp1s, HERP, CHOP, and GRP78. As controls, we compared the original D-17 cells, mock-infected or infected with Adeno-ZF. While Zhangfei suppressed the UPR transcripts in infected cells, doxycycline-induction of the protein in the cell clones had no effect on them Fig. 1D). These results suggest that Zhangfei requires a functional leucine zipper for suppressing cell growth as well as inhibiting the UPR.
Figure 1. Spontaneous mutation of leucine residues in the bLZip domain of Zhangfei in D-17 cells stably expressing the protein in the presence of tetracycline. (A) Doxycycline induces the expression of Zhangfei protein in D-17 cell clones. Twenty-four hours after induction with 1 µg/ml doxycycline, Zhangfei protein in D-17 clones was detected by immunofluorescence and immunoblotting. (B) Growth of D-17 cells after induction of Zhangfei. Six D-17 clones were treated with 1µg/ml doxycycline to induce Zhangfei expression. Cell growth characteristics were monitored at different time points using WST-1 assay. Growth characteristics of the original D-17 cells, mock-infected (MI) or infected with adenovirus vectors expressing either ZF (Adeno-ZF) or β-galactosidase (Adeno-LacZ) are shown for comparison. (C) Mutations in the leucine zipper of Zhangfei-expressing D-17 clones. Amino acid sequence of the LZip region for Zhangfei recovered by PCR from the vector used to develop the clones and the doxycycline-inducible Zhangfei expressing clones. Dashes (−) indicate no changes. Leucine residues (L) comprising the zipper are shown and mutations are in bold. (D) Induction of mutant Zhangfei has no effect on the UPR. Clones expressing ZF with mutations in the leucine zipper did not inhibit the UPR. Original D-17 cells and clones 4 and 15 were treated with 1 µg/ml doxycycline for 24 h, followed by 4 h of treatment of thapsigargin. Then cells were harvested and UPR transcripts were estimated by qRT-PCR. The original D-17 cells, mock-infected or infected with Adeno-ZF were also analyzed for comparison. The values represent fold changes in levels of transcripts between Adeno-ZF infected cells with mock-infected cells (in original D-17 cells) or between doxycycline treated with untreated cells (in Tet-on clones). Error bars indicated standard deviations from means of 3 individual experiments. Dashed lines indicate fold changes of less or more than 2-fold and changes exceeding these limits were arbitrarily regarded as significant. Bar = 100 µm.
Zhangfei regulates p53 at a post-translational level and promotes p53 nuclear retention
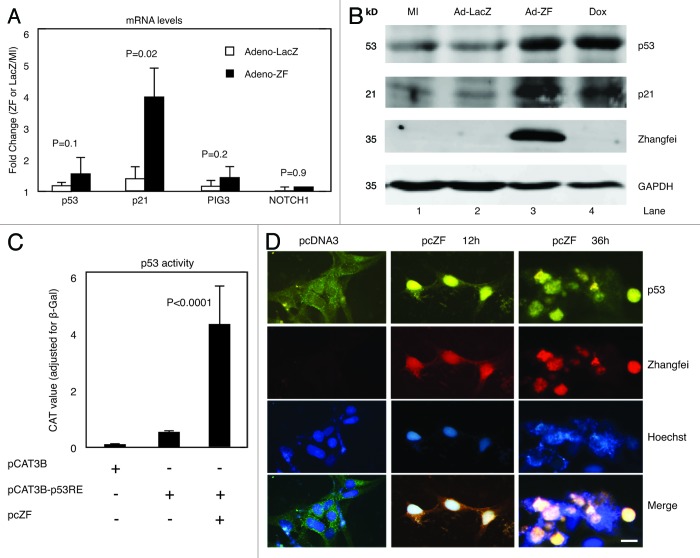
Zhangfei stabilizes p53 in some cancer cells,2 although the mechanisms by which Zhangfei mediates its effects on p53 have not been characterized. To investigate if Zhangfei regulates the expression of endogenous p53 and its target genes, we infected D-17 cells with Adeno-ZF or Adeno-LacZ. We observed that Zhangfei increased transcripts for p21, a well-characterized p53-target gene, but had no significant effects on p53 transcripts as well as its other target genes, PIG3 and NOTCH1 (Fig. 2A). However, Zhangfei did increase the protein levels of both endogenous p53 and p21 comparable to that induced by the drug doxorubicin, a known inducer of p5314 (Fig. 2B, compare lanes 3 and 4 with 1 and 2), suggesting that Zhangfei increased p53 protein by post-translational mechanisms. We also assessed the ability of Zhangfei to indirectly induce transcription from a promoter containing 2 copies of p53-responsive elements (pCAT3B-p53RE). Zhangfei induced the expression of a reporter gene, CAT, linked to the promoter by about 20-fold (Fig. 2C).
Figure 2. Zhangfei regulates p53 in a post-translational level and induces p53 nuclear localization. (A) Zhangfei enhances the expression of p21, a p53-dependant gene. D-17 cells were mock-infected or infected with either Adeno-ZF or Adeno-LacZ, 24 h after infection, transcripts for p53, p21, PIG3, and NOTCH1 were determined by qRT-PCR. The values represented fold changes of transcripts between Adeno-LacZ (white bar) or Adeno-ZF (black bar) infected cells with mock-infected cells. Error bars indicated standard deviations from means. (B) Zhangfei stabilizes p53 and p21 proteins. p53, p21, Zhangfei, and GAPDH were detected by immunoblotting in D-17 cells either mock-infected (MI) or infected with either Adeno-LacZ or Adeno-ZF. As a positive control, cells were treated with 0.5 µM doxorubicin for 6 h. (C) Zhangfei activates p53-dependent transactivation. D-17 cells were transfected with a reporter plasmid containing the coding sequence for CAT linked to a promoter with 2 copies of p53-responsive element (pCAT3B-p53RE, 0.5 µg) in the presence or absence of a plasmid expressing Zhangfei (pcZF, 1 µg). The promoter-less parental reporter plasmid, pCAT3B was included as a control to show basal CAT activity. All samples also contained, as a control, a plasmid expressing β-galactosidase. 24 h after transfection, the CAT activity was determined. Values represented the relative CAT activity (normalized to the internal control, β-galactosidase) of different treatments. Standard deviations from means of three individual experiments are and the significant P values from ANOVA tests were noted above the bars. (D) Zhangfei alters the subcellular localization of p53. D-17 cells were transfected with 1 µg of pcZF or a control (pcDNA3), and 12 h and 36 h after transfection, endogenous p53 as well as Zhangfei were visualized by immunofluorescence with anti-p53 and anti-ZF antibody. The nucleus was detected by Hoechst staining (bar = 10 µm). The means and standard deviations of representative experiments (n = 3) were shown. P < 0.05 were considered to be significant.
The protein p53 possesses nuclear localization and nuclear export signals, enabling it to shuttle between the nucleus and the cytoplasm.15,16 To investigate the impact of Zhangfei on p53 nucleo-cytoplasmic shuttling, we monitored the p53 localization in ZF-expressing D-17 cells. We observed that, compared with the negative control (pcDNA3), the nuclear staining of endogenous p53 increased by 12 h after transfection of the cells with a plasmid expressing Zhangfei (pcZF) with a concomitant decrease in cytoplasmic staining (Fig. 2D). By 36 h following transfection, endogenous p53 was predominantly in the nucleus and cells displayed features of apoptosis (diffuse DNA staining by Hoechst and membrane blebbing).
Basic-region leucine zipper domain (bLZip) of Zhangfei is required for the regulation of p53
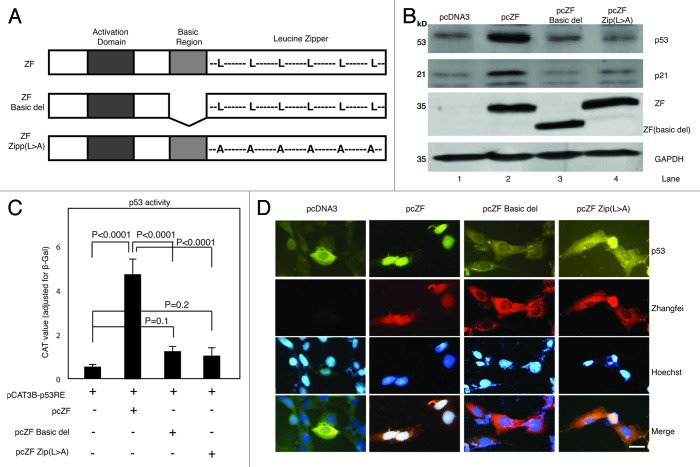
Given that the bLZip domain plays an important role in the inhibitory ability of Zhangfei on cell growth and the UPR, as described above, we next sought to examine whether this domain was also required for the regulation of p53. We found that transfection of plasmids expressing Zhangfei with a deleted basic region (pcZF Basic del) or a mutated leucine zipper (pcZF Zip[L > A]) (Fig. 3A) did not increase the protein levels of either p53 or p21 (Fig. 3B, compare lane 2 with lanes 3 and 4). The increase of p53 transcriptional activity induced by wild-type Zhangfei was also significantly reduced in cells expressing the mutated proteins (Fig. 3C). In addition, the mutant Zhangfei proteins were unable to increase nuclear localization of p53 (Fig. 3D). These results indicate that bLZip domain is an important functional region of Zhangfei, required for its regulatory effects on cell growth, the UPR, as well for its interaction with p53.
Figure 3. The basic-region leucine zipper domain (bLZip) of Zhangfei is required for its effect on p53. (A) Schematic representation of the structures of Zhangfei (ZF) and Zhangfei mutants: ZF Basic del, basic region was deleted; ZF Zip (L > A), all leucines in the leucine-zipper domain were replaced with alanines. (B) The bLZip domain of Zhangfei is required for stabilization of p53 and p21 proteins. D-17 cells were transfected with 1 µg of plasmid expressing Zhangfei (pcZF) or mutants (pcZF Zip(L > A) or pcZF Basic del). Twenty-four h after transfection endogenous p53 and p21 proteins were detected by immunoblotting. (C) The bLZip domain of Zhangfei is required for p53-dependent transactivation. D-17 cells were transfected with 0.5 µg of p53 response element containing reporter plasmid pCAT3B-p53RE and 1 µg of pcZF or mutants (pcZF Zip [L > A] or pcZF Basic del). Twenty four hours after transfection, the CAT activity was determined. The means and standard deviations of experiments (n = 3) were shown. P < 0.05 were considered to be significant. (D) The bLZip domain of Zhangfei is required for p53 nuclear retention. D-17 cells were treated as described in (A), and endogenous p53 as well as Zhangfei were visualized by immunofluorescence. The nucleus was detected by Hoechst staining (bar = 10 µm).
p53 is the key molecule responsible for mediating suppressive regulation of Zhangfei on D-17 cell growth and the UPR
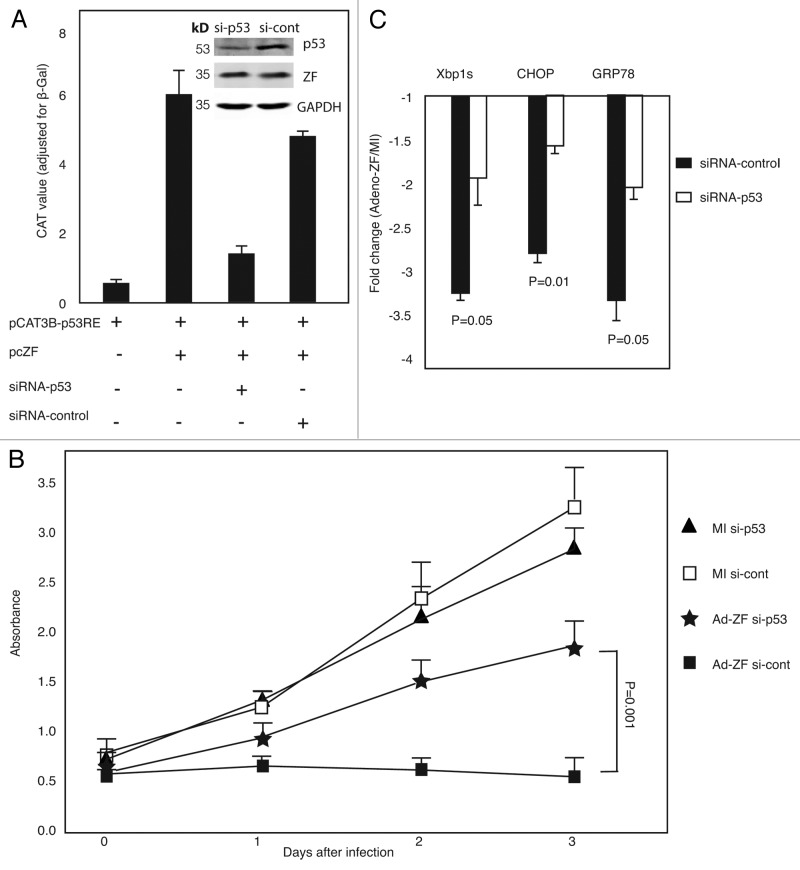
The tumor suppressor p53 limits cellular proliferation by inducing cell cycle arrest and apoptosis in response to cellular stresses, such as DNA damage, hypoxia, nutrient deprivation, and oncogene activation (review refs. 17 and 18), and these stresses also activate the UPR. The results shown above demonstrated that Zhangfei downregulated cell growth and UPR, but upregulated p53 through its bLZip domain. To explore whether Zhangfei expression influence cell proliferation and the UPR through p53, we used p53-sepcific siRNA to knockdown p53 transcripts and protein. While Zhangfei enhanced transcription from a promoter with p53 response elements, siRNA against p53 suppressed its effects (Fig. 4A). Suppression of p53 also partially restored the growth of D-17 cells inhibited by Zhangfei (Fig. 4B) and partially restored the suppression of UPR transcripts (Fig. 4C). These results indicate that the inhibitory influences of Zhangfei on D-17 cells that we have described previously, at least in part, are mediated by p53.
Figure 4. Zhangfei regulates p53-mediated cell growth and UPR. (A) Zhangfei-induced transcription from a promoter with p53 response elements is mediated by p53. D-17 cells were transfected with 0.5 µg of p53 response element containing promoter (pCAT3B-p53), 1 µg of plasmid expressing Zhangfei (pcZF) and 100 pM of si-RNA against p53 (si-p53) or non-targeting siRNA control (si-cont), 48 h after transfection, cells were analyzed for CAT activity or p53, Zhangfei and GAPDH proteins (inset). (B) Zhangfei-mediated suppression of cell growth is mediated through p53. D-17 cells were transfected with 100 pM of si-RNA against p53 (siRNA-p53) or non-targeting siRNA control (siRNA-cont). Eight hours after transfection, cells were either mock-infected or infected with Adeno-ZF, and growth characteristics were measured using WST-1 assay at 0 h, 24 h, 48 h, and 72 h after infection. (C) Zhangfei-mediated suppression of UPR genes is mediated by p53. D-17 cells were transfected 100 pM of siRNA-p53 or non-targeting siRNA-control. Forty-eight hours after transfection, cells were treated with thapsigargin for 4 h and harvested. Differences in levels of transcripts (Xbp1s, CHOP, and GRP78) were determined by qRT-PCR. The results were expressed as fold changes relative to mock-infected cells. The means and standard deviations of experiments (n = 3) are shown. P < 0.05 were considered to be significant.
Zhangfei suppresses the growth and UPR in p53-expressing, but not in p53-null human osteosarcoma cells
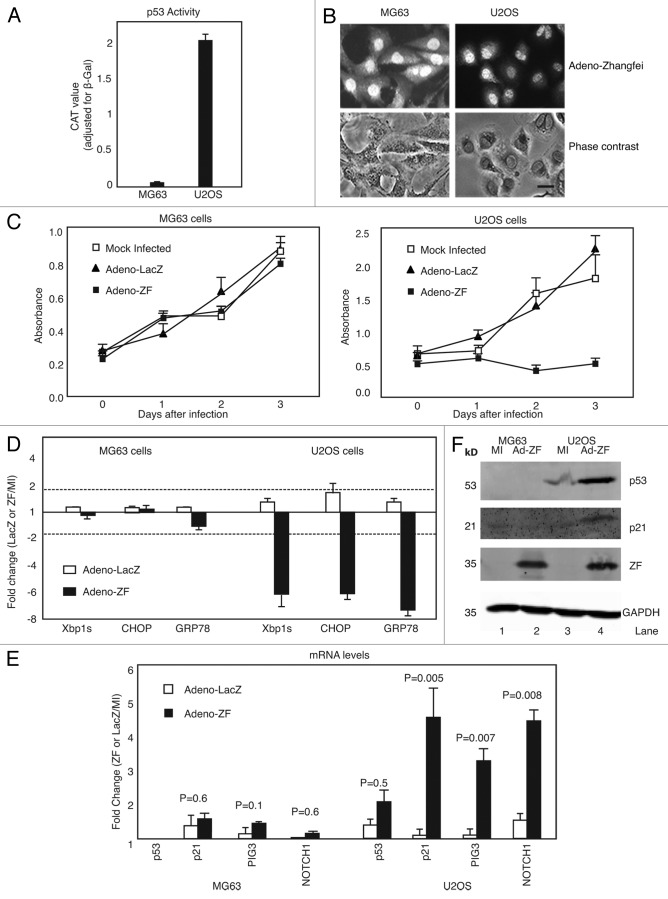
To determine if our results linking p53 as the mediator of the effects of Zhangfei on canine osteosarcoma cells applied to human osteosarcoma cells as well, we examined 2 human cells lines: U2OS cells possess and express a functional p53 protein, whereas MG63 cells do not.19 We confirmed this by detecting and comparing p53 transcriptional activities of both cell lines (Fig. 5A). Both osteosarcoma cell lines have been used extensively to assess the role of p53 in several phenomena.
Figure 5. Zhangfei suppresses cell growth and UPR in wild-type p53-expressing U2OS cells, but not in p53-null MG63 cells. (A) p53-dependent transactivation in U2OS and MG63 cells. Cells were transfected with 0.5 µg of p53 reporter (pCAT3B-p53RE) for 24 h, then p53 transcriptional activity was determined by CAT ELISA assay. (B) U2OS and MG63 cells express Zhangfei when infected with adenovirus vector expressing the protein. Cells were infected with Adeno-ZF, and 24 h later Zhangfei protein was visualized by immunofluorescence (bar = 10 µm). (C) Ectopic expression of ZF suppresses cell growth in p53-wild type U2OS cells, but has no effect in p53-null MG63 cells. U2OS and MG63 human osteosarcoma cells were mock-infected or infected with Adeno-ZF or Adeno-LacZ and their growth rates were assessed at different time points after infection as absorbance at 405 nm after treatment with WST-1. (D) Zhangfei negatively regulates the UPR in U2OS cells but not in MG63 cells. U2OS and MG63 human cells were either mock-infected or infected with Adeno-ZF or Adeno-LacZ. Twenty-four hours after infection, cells were treated with thapsigargin for 4 h, and then harvested. The differences in levels of transcripts for UPR genes (Xbp1s, CHOP, and GRP78) were determined by qRT-PCR. (E) Zhangfei enhances p53-dependent transcripts in U2OS cells but not in MG63 cells. Transcripts for p53, p21, PIG3, and NOTCH1 in U2OS and MG63 cells either mock-infected or infected with Adeno-LacZ or Adeno-ZF were measured by pRT-PCR. (F) Zhangfei increases p53 and p21 proteins in U2OS cells but not in MG63 cells. p53 and p21 proteins were detected by immunoblotting in U2OS and MG63 cells either mock-infected or infected with Adeno-ZF. The values in (D and E) represented fold changes of transcripts between Adeno-LacZ (white bar) or Adeno-ZF (black bar) infected cells with mock-infected cells. The means and standard deviations of experiments (n = 3) were shown. P < 0.05 were considered to be significant. Error bars indicate standard deviations from means. Horizontal dotted line indicated a 2-fold change. A change of more than 2-fold was arbitrarily regarded as significant.
As with D-17 cells, Zhangfei suppressed the growth of U2OS cells but had little effect on MG63 cells (Fig. 5C). Immunofluorescent detection of Zhangfei in these cells showed that both cells expressed Zhangfei when infected with adenovirus vector expressing the protein (Fig. 5B). Also, as in D-17 cells, ectopic expression of Zhangfei suppressed UPR-related transcripts while increasing levels of p53-related transcripts in U2OS cells but not in MG63 cells (Fig. 5D and E). Figure 5F further supports these observations by showing that, as in D-17 cells, in U2OS cells p53 and p21 proteins increased, but, as expected, they did not in MG63 cells.
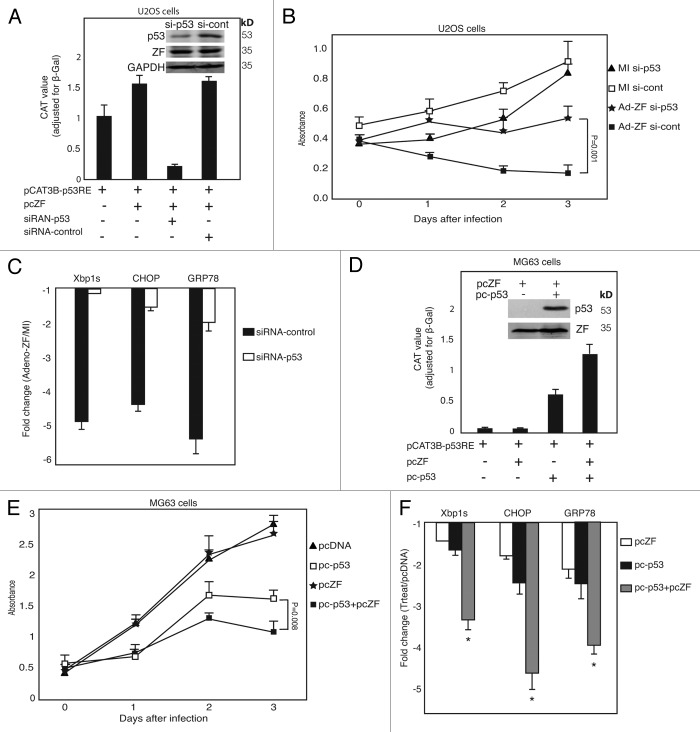
To confirm that the suppressive effects of Zhangfei on growth and UPR of U2OS cells was indeed mediated through p53, we suppressed p53 in these cells with siRNA. Figure 6A shows that treatment with siRNA directed against p53 suppressed activation of transcription from a promoter with p53 response elements and reduced levels of p53 protein. The specific suppression of p53 had a significant effect on the ability of Zhangfei to inhibit the growth of U2OS cells (Fig. 6B) and UPR-related transcripts (Fig. 6C). To further evaluate the role of p53 in cell growth arrest and UPR suppression induced by Zhangfei, studies were performed to determine the effects of introducing p53 using a plasmid expressing wild-type p53 (from U2OS) into p53-null MG63 human cells in absence or presence of Zhangfei. The overexpression of exogenous p53 (confirmed by immunoblotting and CAT reporter assay in Fig. 6D) had a strong inhibitory effect on cell growth and led to cell death within 24–36 h (Fig. 6E, open squares), which is consistent with a previous report that the infection of adenovirus vector expressing wild-type p53 in p53-null Saos-2 osteosarcoma cells induced these cells to commit apoptosis,20 and Zhangfei further increased this suppression (Fig. 6E, solid squares). In contrast, Zhangfei alone had no effect on cell growth in these cells (Fig. 6E, stars). For the influence on UPR, p53 expression in MG63 cells led to decreases (about 1.5–2.5-fold) in the levels of UPR transcripts induced by thapsigargin (Fig. 6F, black bars). And ectopic expression of Zhangfei suppressed UPR-related transcripts more potently (about 3.5–5-fold decrease) in p53-expressing MG63 cells than that in p53-null MG63 cells (Fig. 6F), suggesting p53 acts as an intermediary in the suppressive events induced by Zhangfei.
Figure 6. p53 mediates the suppressive effects of Zhangfei on cell growth and UPR in human osteosarcoma cells. (A) Zhangfei-mediated transcription from a p53 response element-containing promoter is mediated by p53 in U2OS cells. U2OS cells were transfected with 0.5 µg of p53 reporter plasmid (pCAT3B-p53RE) and 1 µg of plasmids expressing Zhangfei (pcZF) in the presence of 100 pM of siRNA-p53 or non-targeting siRNA-control. Forty eight hours after transfection CAT reporter activity and p53 protein were detected by ELISA and immunoblotting (inset). (B) Suppression of p53 by siRNA in U2OS cells restored Zhangfei-induced suppression of cell growth. U2OS were transfected with 100 pM of siRNA-p53 or non-targeting siRNA-control. Eight hours after transfection, cells were either mock-infected or infected with Adeno-ZF and measured growth rates as absorbance at 405 nm after treatment with WST-1. (C) Suppression of p53 prevents Zhangfei-induced inhibition of UPR genes. U2OS cells were transfected with 100 pM of siRNA-p53 or non-targeting siRNA-control. Forty eight hours after transfection, cells were treated with thapsigargin for 4h and harvested. Differences in levels of transcripts for UPR genes (Xbp1s, CHOP, and GRP78) were determined by qRT-PCR. The values represented fold changes of transcripts between Adeno-LacZ (white bar) or Adeno-ZF (black bar) infected cells with mock infected cells. (D) Transactivation activity and expression of p53 protein in p53-null MG63 cells. p53-null MG63 cells were transfected with 0.5 µg of p53 reporter plasmid and 1 µg of plasmids expressing Zhangfei (pcZF) in the presence of 1µg of plasmids expressing wild-type p53 (pc-p53, from U2OS cells) for 24 h, and then CAT reporter activity and p53 protein were detected by ELISA and immunoblotting. (E) Overexpression of p53 in p53-null MG63 cells suppressed cell growth and activated growth arrest induced by Zhangfei. MG63 cells were transfected with 3 µg of pcZF in the absence or presence of 3 µg of pc-p53 and measured growth rates as absorbance at 405 nm after treatment with WST-1. (F) Overexpression of p53 in p53-null MG63 cells activated the UPR suppression induced by Zhangfei. MG63 cells were transfected with 3 µg of pcZF in the absence or presence of 3 µg of pc-p53. Twenty-four hours after transfection, cells were treated with thapsigargin for 4 h and harvested. Differences in levels of transcripts for UPR genes (Xbp1s, CHOP, and GRP78) were determined by qRT-PCR. The values represented fold changes of transcripts between pcZF (white bar), pc-p53 (black bar), or pcZF plus pc-p53 (gray bar) transfected cells with pcDNA3 transfected cells. The total amount of DNA in each transfection above was made up to 5 μg with pcDNA3. The means and standard deviations of experiments (n = 3) were shown. *P < 0.05 were considered to be significant.
Zhangfei interacts with p53
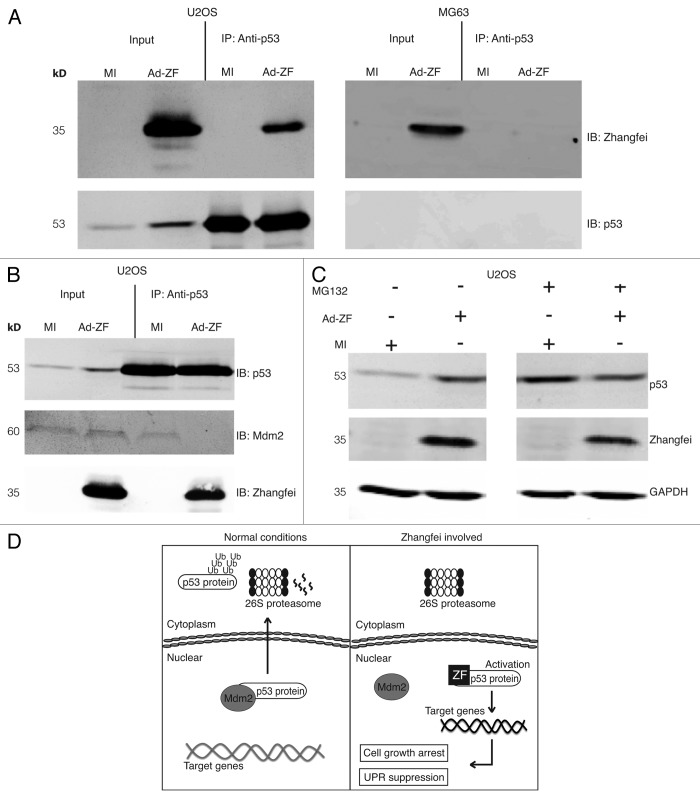
Next, we confirmed that p53 and Zhangfei interacted in U2OS cells by co-immunoprecipitating the proteins in cells expressing Zhangfei. Immunoprecipitation of p53 from U2OS cells also precipitated Zhangfei (Fig. 7A). As expected, in MG63 cells, which lack p53, no p53 was detected and, consequently, no Zhangfei was precipitated.
Figure 7. In vitro interaction of Zhangfei and p53. (A) Zhangfei associates with p53 in U2OS cells. Cell lysate from U2OS and MG63 human cells previously infected with adenovirus vector expressing Zhangfei (Ad-ZF) or mock infected (MI) were incubated with 2 µg of mouse anti-p53 antibody for 12 h at 4 °C, followed by an additional 4 h incubation with 100 µl of Protein A/G agarose beads (IP). The associated proteins were detected by immunoblotting (IB) using rabbit anti-Zhangfei antibody and mouse anti-p53 antibody. Input represented 1/10 of lysate used for immunoprecipitation. (B) Zhangfei displaces Mdm2 from p53 in U2OS cells. Cell lysates as in (A) were immunoprecipitated with anti-p53 antibody followed by immunoblotting with either anti-Mdm2 or anti-Zhangfei antibody. (C) Inhibition of proteasomal degradation reverses degradation of p53 in the absence of Zhangfei. Cells infected as in (A) were either treated with the proteasomal inhibitor MG132 (5 µM) or left untreated. Proteins in the cell lysates were separated by SDS-PAGE, and p53, Zhangfei and GAPDH (loading control) were detected by immunoblotting. (D) Schematic diagram of a model for the proposed mechanism by which Zhangfei inhibits cell growth and UPR. Under normal conditions, Mdm2 translocates the p53 protein out of the nucleus for degradation via the ubiquitin-dependent pathway. In Zhangfei-expressing cells, Zhangfei interacts with p53 and prevents it from binding to Mdm2. This results in p53 stabilization and nuclear accumulation, which, in turn, activates its target genes and suppresses tumor cell growth and the UPR. These results illustrate how Zhangfei controls the activity of p53 toward the cell growth and the UPR, and offer an explanation to why p53 is upregulated by Zhangfei in tumor cells.
Zhangfei displaces Mdm2 from p53, protecting it from proteolysis
In unstressed cells p53 is associated with the E3 ubiquitin ligase, mouse double minute homolog 2 (Mdm2), that facilitates its nuclear export 21,22and proteasomal degradation.23 To determine if Zhangfei inhibited Mdm2–p53 interactions, we immunoprecipitated p53 from mock-infected and Adeno-ZF-infected U2OS cells and probed the precipitates for Mdm2 and Zhangfei (Fig. 7B). p53 was precipitated from both mock-infected and Zhangfei-expressing cells, but it was only associated with Mdm2 in the absence of Zhangfei. In addition, while the proteasome inhibitor MG132 (5 µM) prevented the degradation of p53 in mock-infected cells, it had little effect on p53 when Zhangfei was present (Fig. 7C). These results indicate that Zhangfei displaces Mdm2 from p53, thereby preventing Mdm2-mediated nuclear export and subsequent proteasomal degradation of p53 (Fig. 7D).
Discussion
The protein p53 is a key tumor suppressor protein. Various cellular stresses, such as UV radiation, DNA damage, hypoxia, and oncogene activation, activate p53, which functions as a transcription factor, regulating the expression of a large and disparate group of target genes to initiate apoptosis, cell cycle arrest, DNA repair, cellular senescence, as well as differentiation. To overcome this suppression, tumor development is often accompanied by mutation or loss of p53. The transcription factor Zhangfei activates the mitogen-activated protein kinase (MAPK) pathway that directs medulloblastoma cells to commit apoptosis.24 In addition to this pathway, here we demonstrate another tumor repressor role for Zhangfei through its ability to directly interact with tumor suppressor p53 and to promote p53 protein stabilization and its nuclear retention in canine and human osteosarcoma cells. Of note, the knockdown of endogenous p53 partly, but significantly, counteracted Zhangfei-induced arrest of cell growth in p53 wild-type osteosarcoma cells (D-17 and U2OS), while exogenous expression of p53 enhanced this process in p53-null osteosarcoma (MG63).
In normal cells, p53 is a short-lived protein and functions to control excessive cell proliferation. Under unstressed conditions, low intranuclear concentrations of p53 protein are maintained by its binding to E3 ubiquitin ligases such as Mdm2, COP1 and pirh2, which keep p53 in check by ubiqitination, nuclear export, and proteasomal degradation (review ref. 25). In our studies, we found that the suppressive effect of Zhangfei on the growth of cells was not universal. In some normal cells, such as MRC5 fibroblasts,12 or p53-null tumor cells, such as MG63 (Fig. 5C), Zhangfei had no effect on cell proliferation. This selective effect of Zhangfei could be due either to the tight regulation of p53 by means of post-translational modifications, cofactor binding, and subcellular localization, or due to a lack of functional p53. As shown here, the growth of U2OS osteosarcoma cells were dramatically inhibited by Zhangfei (Figs. 5C and 6B), likely because the cells express wild-type p53. However, our results are not in agreement with those of Lopez-Mateo and others who in a recent study2 showed that tetracycline induction of Zhangfei/CREBH in inducible clones derived from U2OS cells resulted in no effect on the proliferation of the cells, nor did the cells display obvious morphological changes. We speculate that there could be 2 reasons for the differences in our results: (1) leaky expression of tetracycline-inducible Zhangfei in U2OS cells2 may have resulted in the selection of clones with structural and functional mutations in Zhangfei. This would be consistent with our observations (Fig. 1) that suggest that the growth suppressive effects of Zhangfei in cells expressing functional p53 exert strong selective pressure and lead to the selection of cells expressing non-functional Zhangfei; (2) the cell line studied by Lopez and coworkers may have spontaneously lost its functional p53. We confirmed that the U2OS cell line we used in this study does indeed have a functional p53 (Fig. 5A).
The protein p53 has been reported to antagonize the UPR and inhibit ground glass hepatocyte development during ER stress.26 In the present study, we determined that p53, at least in part, mediates Zhangfei-induced suppression of the UPR (Figs. 4C and 6C and F). Induction of UPR is a protective mechanism utilized by cells to adapt to ER stress. In normal cells, the UPR is related to tissue preservation or organ protection against ER stress. In neoplastic cells, however, the adaptive function of the UPR is implicated in immune resistance, cancer progression, and drug resistance (review refs. 27 and 28). Therefore, the inhibition of UPR, mediated by p53, represents a potential strategy by which Zhangfei could be used to inhibit cancer. We show that Zhangfei inhibits the UPR and induces apoptosis by promoting p53 stabilization and nuclear retention. Interestingly, in keeping with our results, ER stress and the resulting activation of the UPR induces p53 cytoplasmic localization and prevents p53-dependent apoptosis,29 suggesting the existence of an ER stess-p53-UPR regulatory loop (see model in Fig. 8). Although the precise relationship between p53 stabilization and UPR remains unclear, glycogen synthase kinase-3 is shown to be a mediator of ER stress-induced p53 cytoplasmic localization,29 and the ubiquitination and importin-α3 binding of p53 are also associated with stress-mediated p53 translocalization.30,31 In the case of Zhangfei, improved understanding of how exactly p53 suppresses the UPR still should be further investigated.
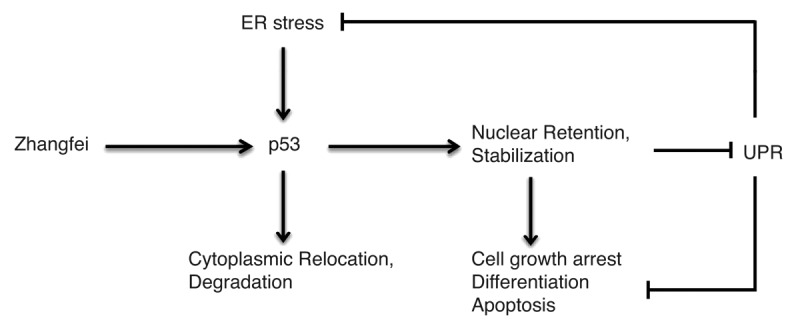
Figure 8. Zhangfei and ER stress have opposing effects on p53. In Zhangfei-expressing cells, p53 is targeted for nuclear localization and stabilization, and further to prevent cell growth, induce cell apoptosis, as well as inhibit the UPR in response to ER stress. In contrast, ER stress promotes the cytoplasmic localization and degradation of p53.
We had previously shown that ectopic expression of Zhangfei in medulloblastoma12 and osteosarcoma cells13 suppressed the cell growth and the UPR. In addition, the effects of this protein were also observed in 4 other canine osteosarcoma cell lines (not published). To study the mechanisms involved, we tried to develop cell lines in which the expression of Zhangfei could be induced by doxycycline. We were successful in developing several clones that expressed Zhangfei protein, as detected by immunofluorescence and immunoblots, only in the presence of the inducer (Fig. 1A). However, none of the clones behaved as did cells in which the ectopic expression of Zhangfei was induced by infection with adenovirus vectors (Fig. 1B and D). On further examination we found that in the clones, 1 to as many as all 6 leucines had been replaced with other amino acids (Fig. 1C). The clone with only one substitution had growth properties most similar to the cells expressing adenovirus-Zhangfei. No other mutations in the entire Zhangfei coding sequences were observed in any of the clones. We interpret this to mean that even small amounts of intact Zhangfei, which might be expressed in the absence of the inducer, was sufficient to bock cell division, and only cells in which the zipper had been disabled survived.
While our results clearly identify the leucine zipper of Zhangfei as important for its interaction with p53 and subsequent p53-mediated suppression of the UPR, we have also shown that Zhangfei interacts with the bLZip UPR transcription factor Xbp1s, targeting it for proteosomal degradation,11 and that the leucine zipper of Zhangfei is also important for this interaction. Hence, modulation of UPR by Zhangfei may require a combination of several coordinated events, including direct interaction with UPR mediators and indirect mediation through p53.
Like Zhangfei, several other bLZip proteins, such as ATF3,32,33 ING2,34 K-bZIP,35 and CREB,36 are known to interact with p53 through their leucine zippers. The nature of these interactions as well as their consequences, however, vary. Interactions between p53 and Zhangfei, ATF3, and CREB lead to enhanced transcriptional activity and stabilization of p53. Our results show that Zhangfei displaced Mdm2 from p53 (Fig. 7B) and prevented its degradation to a similar extent as the proteasomal inhibitor MG132 (Fig. 7C). The effects of MG132 and Zhangfei were not additive. It is likely that the combined toxic effects of MG132 and suppressive effects of Zhangfei cause a global reduction of new protein synthesis in cells. These effects would be more noticeable for a protein like p53 that is routinely turned over rapidly than for the more stable GAPDH, the internal control. In contrast to the effects of Zhangfei on p53, ATF3 neither displaces Mdm2, nor does it suppress its activity. ATF3 binds directly to the C terminus of p53 and prevents ubiquitination of key lysine residues in the region.33 CREB binds to the N terminus of p53 and acts as a bridge directing CREB binding protein to p53-responsive promoters37 and suppresses transcription of Mdm2.38 Unlike the bLZip–p53 interactions mentioned above, which enhance p53 activity, the association of the Kaposi sarcoma herpesvirus protein K-bZIP with p53 suppresses its activity and may contribute to tumorigenesis by the virus.35
Materials and Methods
Cells and tissue culture
The canine osteosarcoma D-17 cells were obtained from the American Type Tissue Culture Collection and were grown in MEM-Alpha containing 10% fetal bovine serum. Human MG63 and U2OS osteosarcoma cells were obtained from Dr Douglas H Thamm (Associate Professor of Oncology, Animal Cancer Center, Colorado State University) and grown in Dulbecco minimal essential medium containing penicillin and streptomycin and 10% newborn calf serum. All media, serum, and antibiotics were purchased from Invitrogen. D-17 cells were treated with doxorubicin (0.5 µM, WCVM’s Accredited Veterinary Pharmacy, University of Saskachewan) for p53 induction. Stock solutions of MG132/Z-L-L-L-A (10 mM, Sigma, C2211) were prepared in dimethyl sulfoxide (DMSO) and used at a concentration of 5 µM. Equivalent volumes of DMSO were added to untreated cultures.
Plasmids
The construction of pcZF,1 a plasmid that expresses Zhangfei in mammalian cells, has been described. The plasmids expressing Zhangfei mutant pcZF Zip (L > A), in which all leucines in the leucine-zipper domain were replaced with alanines, was constructed by subcloning a 265 bp synthetic DNA fragment (IDT) bracketed by NotI and SgrA1sites into the corresponding coding sequences of Zhangfei between unique Not1 and SgrA1 sites in pcZF. The plasmids expressing Zhangfei mutant pcZF Basic del, in which basic region was deleted, was constructed by site-directed loop-out mutagenesis using 36 base complementary oligonucleotides that bracketed the 25 codons for the basic region of Zhangfei. The CAT reporter plasmid pCAT3B-p53RE was constructed by transferring 2 copies of p53-responsive element, GGTCAAGTTG GGACACGTCC AAGAGCTAAG TCCTGACATG TCT, to pCAT3Basic (Promega). Oligonucleotides representing the p53-responsive elements with overhanging 5′ terminal KpnI and 3′ terminal BglII sites were purchased from IDT. The oligonucleotides were annealed and ligated pCAT3Basic cut with the same enzymes. The plasmids expressing the full length of wild-type p53 were constructed by subcloning the HindIII-XhoI cDNA fragment containing the entire coding sequence of p53 from U2OS cells between the HindIII and XhoI sites in the multiple restriction site cloning regions of the mammalian expression vector pcDNA3 (Invitrogen, V79020). The specific primers for p53 amplification are: forward: 5′-GACACGCTTC CCTGGATTGG C-3′, reverse: 5′- TCAGTCTGAG TCAGGCCCTT-3′. The veracity of all cloned wild-type and mutants coding sequences were confirmed by sequencing
Transfection and CAT assays
Cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen, 11668-019) as described in manufacturer’s instructions. Five μg of DNA was used for 5 × 105 cells for chloramphenicol acetyl transferase (CAT) assays. For CAT assays, 250 ng of pCMVBGal, a plasmid specifying β-galactosidase, were added to each transfection. Lysates were assayed for β-galactosidase39 and for CAT using an enzyme-linked immunosorbent assay kit (Roche Applied Science, 11-363-727-001). CAT values were adjusted for transfection efficiency using β-galactosidase values. In figures for CAT assays, each datum point is the average of duplicate transfections with the bar representing the range. The data are representative of several (at least 2) independent experiments that gave the same results.
RNA interference
p53-specific siRNA (Invitrogen, TP53VHS40366) or Stealth RNAi™ siRNA negative control (Invitrogen, 12935-300) (1 pmol/5000 cells) were transfected into cells with Lipofectamine 2000 (Invitrogen, 11668-019) as described in manufacturer’s instructions. These p53-specific siRNAs are homologous to human as well as canine p53, which was confirmed by comparing the sequence of human or canine p53 with siRNA sequence.
Adenovirus vectors expressing Zhangfei and β-galactosidase (LacZ)
These vectors were constructed, grown, and purified using the Adeno-X Expression System (Clontech, K1650-1) as described earlier.8 Cells were infected with adenovirus vectors expressing either Zhangfei (Adeno-ZF) or a control protein, E. coli β-galactosidase, (Adeno-LacZ) or were mock-infected. A multiplicity of infection (MOI) of 100 plaque-forming units (pfu) per cell was used.
Antibodies, immunoblotting, and immunofluorescence
The antibodies used were rabbit anti-Zhangfei serum, mouse anti-p53 (Santa Cruz Biotechnology, DO-1 sc-126), rabbit anti-p21 (Santa Cruz Biotechnology, C-19, sc-397), rabbit anti-Mdm2 (Santa Cruz Biotechnology, C-18, sc-812), and mouse anti-GAPDH (Sigma, G8795-200UL). Secondary antibodies were goat anti-mouse Alexa488 (Invitrogen, A-11001), goat anti-rabbit Alexa546 (Invitrogen, A-11035) and goat anti-rabbit Cy5 (Invitrogen, A-10523). Cells were processed for immunoblotting and immunofluorescence as described previously.1,13 Images were captured using a digital camera attached to a Zeiss Axioskop microscope (Axiovert 135) and Northern Eclipse software (EMPIX Imaging). Captured images were processed using Adobe Photoshop and Illustrator CS6 software.
Quantitative real-time PCR
Total RNA was extracted using RNeasy Plus Mini Kit (Qiagen, 74136). Gene expression was analyzed by RT-PCR using Brilliant II SYBR Green QPCR Master Mix Kit (Agilent Technologies, 600828). The primers used were: primers for Xbp1, HERP, CHOP, GRP78, and GAPDH has been described13; p53-forward: CTCTCCTCAA CAAGTTGTTT TG, p53-reverse: CTACAGTCAG AGCAGCGTTC ATGG, p21-forward: GCAGACCAGC ATGACAGATT T, p21-reverse: GGATTAGGGC TTCCTCTTRG A, PIG3-forward: AMTGTCAGAG ACAAGGCCRR TA, PIG3-reverse: TCCCCRATCT TCCAGTGYCC, NOTCH1-forward: GAACTGCCCA TGACCACTAC CCAGTTC, NOTCH1-reverse: GGGTGTTGTC CACAGGTGA, All qRT-PCR reactions satisfied MIQE guidelines.40 Disassociation profiles in reactions that yielded products contained single homogeneous peaks. In all reactions, GAPDH was used as a normalizer. In previous experiments24 qRT-PCR arrays comparing Zhangfei expressing and non-expressing cells five housekeeping genes were analyzed. The levels of GAPDH were not affected by Zhangfei expression.
Co-immunoprecipitation
U2OS and MG63 cells in 6-well dishes were infected with Adeno-ZF or mock infected. Twenty-four h after infection, cells were washed with cold PBS and lysed in 250 µl/well-cold lysis buffer (50 mM Tris, pH7.5, 150 mM NaCl, 1 mM EDTA, and 0.1% TritonX-100) containing protease inhibitor cocktail (Sigma, P8340). After centrifugation at 13 000 xg at 4 °C, 20 µl of cell lysate supernatant were frozen as pre-immunoprecipitated sample, and mouse anti-p53 antibody (2 µg, Santa Cruz Biotechnology, DO-1 sc-126) was added to the remaining supernatant (230 µl) and the sample incubated for 12 h with constant gentle agitation. Protein A/G agarose beads (100 µl, Pierce, Fisher Scientific, 20421) was added, and the samples were incubated for an additional 4 h at 4 °C. Agarose beads were collected by centrifugation at 13 000 × g at 4 °C and washed 4 times in lysis buffer before boiling in SDS-PAGE sample buffer. Proteins in samples of the unfractionated cell lysate or immunoprecipitates were separated by SDS-PAGE, transferred to membranes, and probed with rabbit anti-Zhangfei antisera, mouse anti-p53, or rabbit anti-Mdm2 (Santa Cruz Biotechnology, C-18 sc-821). Antibodies were visualized after incubation with Alexa488-labeled anti-rabbit or anti-mouse antibody.
Statistical analysis
Statistical analysis was performed by t test or ANOVA test using IBM SPSS statistics version 21.0.0 software. ANOVA tests with LSDpost hoc comparison was used to analyze the differences between multi-group means and their associated procedures by adding individuals as a treatment variable, and a paired t test was used to evaluate the effects of one treatment compared with no treatment/control. A P value of less than 0.05 was considered to be statistically significant for ANOVA tests and t tests.
Acknowledgments
The authors thank Noreen Rapin for technical assistance. This work was supported by a Discovery grant to VM from the Natural Sciences and Engineering Research Council (NSERC) of Canada; and a research grant from the Western College of Veterinary Medicine, Companion Animal Health Fund. RZ was supported by scholarships from the Government of China (China Scholarship Council, RZ-2010635007) and the University of Saskatchewan College of Graduate Studies and Research.
Glossary
Abbreviations:
- ANOVA
analysis of variance
- bLZip
basic region, leucine zipper domain
- CAT
chloramphenicol-acetyl-transferase
- CHOP
CAAT enhancer-binding protein homology protein
- CREBZF
cyclic AMP response element binding protein-Zhangfei
- EDTA
ethylenediaminetetraacetic acid
- ER
endoplasmic reticulum
- GAPDH
glyceraldehyde 6 phosphate dehydrogenase
- GRP78
glucose regulated protein 78 000 MW
- HCF
host cell factor
- HERP
homocysteine-induced endoplasmic reticulum protein
- ING2
inhibitor of growth protein 2
- K-bZIP
Kaposi sarcoma herpes virus bLZip protein
- L
leucine
- MAPK
mitogen-activated protein kinase
- Mdm2
mouse double minute homolog 2
- MI
mock-infected
- NOTCH1
Drosophila Notch family protein 1
- P53
protein 53 000 molecular weight
- PIG3
p53-inducible protein 3
- SMAD
small body size- mothers against decapentaplegic
- UPR
unfolded protein response
- VP16
virion protein #16
- Xbp1
X-factor binding protein 1
- Xbp1s
Xbp1 derived from cytoplasmic spliced mRNA
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/27053
References
- 1.Lu R, Misra V. Zhangfei: a second cellular protein interacts with herpes simplex virus accessory factor HCF in a manner similar to Luman and VP16. Nucleic Acids Res. 2000;28:2446–54. doi: 10.1093/nar/28.12.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.López-Mateo I, Villaronga MA, Llanos S, Belandia B. The transcription factor CREBZF is a novel positive regulator of p53. Cell Cycle. 2012;11:3887–95. doi: 10.4161/cc.22133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogan MR, Cockram GP, Lu R. Cooperative interaction of Zhangfei and ATF4 in transactivation of the cyclic AMP response element. FEBS Lett. 2006;580:58–62. doi: 10.1016/j.febslet.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 4.Xie YB, Lee OH, Nedumaran B, Seong HA, Lee KM, Ha H, Lee IK, Yun Y, Choi HS. SMILE, a new orphan nuclear receptor SHP-interacting protein, regulates SHP-repressed estrogen receptor transactivation. Biochem J. 2008;416:463–73. doi: 10.1042/BJ20080782. [DOI] [PubMed] [Google Scholar]
- 5.Xie YB, Nedumaran B, Choi HS. Molecular characterization of SMILE as a novel corepressor of nuclear receptors. Nucleic Acids Res. 2009;37:4100–15. doi: 10.1093/nar/gkp333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie YB, Park JH, Kim DK, Hwang JH, Oh S, Park SB, Shong M, Lee IK, Choi HS. Transcriptional corepressor SMILE recruits SIRT1 to inhibit nuclear receptor estrogen receptor-related receptor gamma transactivation. J Biol Chem. 2009;284:28762–74. doi: 10.1074/jbc.M109.034165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misra J, Chanda D, Kim DK, Li T, Koo SH, Back SH, Chiang JY, Choi HS. Curcumin differentially regulates endoplasmic reticulum stress through transcriptional corepressor SMILE (small heterodimer partner-interacting leucine zipper protein)-mediated inhibition of CREBH (cAMP responsive element-binding protein H) J Biol Chem. 2011;286:41972–84. doi: 10.1074/jbc.M111.274514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misra V, Rapin N, Akhova O, Bainbridge M, Korchinski P. Zhangfei is a potent and specific inhibitor of the host cell factor-binding transcription factor Luman. J Biol Chem. 2005;280:15257–66. doi: 10.1074/jbc.M500728200. [DOI] [PubMed] [Google Scholar]
- 9.Lee JH, Lee GT, Kwon SJ, Jeong J, Ha YS, Kim WJ, Kim IY. CREBZF, a novel Smad8-binding protein. Mol Cell Biochem. 2012;368:147–53. doi: 10.1007/s11010-012-1353-4. [DOI] [PubMed] [Google Scholar]
- 10.Akhova O, Bainbridge M, Misra V. The neuronal host cell factor-binding protein Zhangfei inhibits herpes simplex virus replication. J Virol. 2005;79:14708–18. doi: 10.1128/JVI.79.23.14708-14718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang R, Rapin N, Ying Z, Shklanka E, Bodnarchuk TW, Verge VMK, Misra V. Zhangfei/CREB-ZF - A Potential Regulator of the Unfolded Protein Response. PLoS One. 2013;8:e77256. doi: 10.1371/journal.pone.0077256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valderrama X, Rapin N, Verge VM, Misra V. Zhangfei induces the expression of the nerve growth factor receptor, trkA, in medulloblastoma cells and causes their differentiation or apoptosis. J Neurooncol. 2009;91:7–17. doi: 10.1007/s11060-008-9682-6. [DOI] [PubMed] [Google Scholar]
- 13.Bergeron T, Zhang R, Elliot K, Rapin N, Macdonald V, Linn K, Simko E, Misra V. The effect of Zhangfei on the unfolded protein response and growth of cells derived from canine and human osteosarcomas. Vet Comp Oncol. 2013;11:140–50. doi: 10.1111/j.1476-5829.2011.00310.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, Konorev EA, Kotamraju S, Joseph J, Kalivendi S, Kalyanaraman B. Doxorubicin induces apoptosis in normal and tumor cells via distinctly different mechanisms. intermediacy of H(2)O(2)- and p53-dependent pathways. J Biol Chem. 2004;279:25535–43. doi: 10.1074/jbc.M400944200. [DOI] [PubMed] [Google Scholar]
- 15.Shaulsky G, Goldfinger N, Ben-Ze’ev A, Rotter V. Nuclear accumulation of p53 protein is mediated by several nuclear localization signals and plays a role in tumorigenesis. Mol Cell Biol. 1990;10:6565–77. doi: 10.1128/mcb.10.12.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Xiong Y. A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science. 2001;292:1910–5. doi: 10.1126/science.1058637. [DOI] [PubMed] [Google Scholar]
- 17.Sharpless NE, DePinho RA. p53: good cop/bad cop. Cell. 2002;110:9–12. doi: 10.1016/S0092-8674(02)00818-8. [DOI] [PubMed] [Google Scholar]
- 18.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 19.Park YB, Park MJ, Kimura K, Shimizu K, Lee SH, Yokota J. Alterations in the INK4a/ARF locus and their effects on the growth of human osteosarcoma cell lines. Cancer Genet Cytogenet. 2002;133:105–11. doi: 10.1016/S0165-4608(01)00575-1. [DOI] [PubMed] [Google Scholar]
- 20.Marcellus RC, Teodoro JG, Charbonneau R, Shore GC, Branton PE. Expression of p53 in Saos-2 osteosarcoma cells induces apoptosis which can be inhibited by Bcl-2 or the adenovirus E1B-55 kDa protein. Cell Growth Differ. 1996;7:1643–50. [PubMed] [Google Scholar]
- 21.Roth J, Dobbelstein M, Freedman DA, Shenk T, Levine AJ. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–64. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carter S, Bischof O, Dejean A, Vousden KH. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol. 2007;9:428–35. doi: 10.1038/ncb1562. [DOI] [PubMed] [Google Scholar]
- 23.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–7. doi: 10.1016/S0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 24.Bodnarchuk TW, Napper S, Rapin N, Misra V. Mechanism for the induction of cell death in ONS-76 medulloblastoma cells by Zhangfei/CREB-ZF. J Neurooncol. 2012;109:485–501. doi: 10.1007/s11060-012-0927-z. [DOI] [PubMed] [Google Scholar]
- 25.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–83. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 26.Dioufa N, Chatzistamou I, Farmaki E, Papavassiliou AG, Kiaris H. p53 antagonizes the unfolded protein response and inhibits ground glass hepatocyte development during endoplasmic reticulum stress. Exp Biol Med (Maywood) 2012;237:1173–80. doi: 10.1258/ebm.2012.012140. [DOI] [PubMed] [Google Scholar]
- 27.Moenner M, Pluquet O, Bouchecareilh M, Chevet E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007;67:10631–4. doi: 10.1158/0008-5472.CAN-07-1705. [DOI] [PubMed] [Google Scholar]
- 28.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 29.Qu L, Huang S, Baltzis D, Rivas-Estilla AM, Pluquet O, Hatzoglou M, Koumenis C, Taya Y, Yoshimura A, Koromilas AE. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3beta. Genes Dev. 2004;18:261–77. doi: 10.1101/gad.1165804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becker K, Marchenko ND, Maurice M, Moll UM. Hyperubiquitylation of wild-type p53 contributes to cytoplasmic sequestration in neuroblastoma. Cell Death Differ. 2007;14:1350–60. doi: 10.1038/sj.cdd.4402126. [DOI] [PubMed] [Google Scholar]
- 31.Marchenko ND, Hanel W, Li D, Becker K, Reich N, Moll UM. Stress-mediated nuclear stabilization of p53 is regulated by ubiquitination and importin-alpha3 binding. Cell Death Differ. 2010;17:255–67. doi: 10.1038/cdd.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Mo P, Ren S, Yan C. Activating transcription factor 3 activates p53 by preventing E6-associated protein from binding to E6. J Biol Chem. 2010;285:13201–10. doi: 10.1074/jbc.M109.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan C, Lu D, Hai T, Boyd DD. Activating transcription factor 3, a stress sensor, activates p53 by blocking its ubiquitination. EMBO J. 2005;24:2425–35. doi: 10.1038/sj.emboj.7600712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Wang J, Li G. Leucine zipper-like domain is required for tumor suppressor ING2-mediated nucleotide excision repair and apoptosis. FEBS Lett. 2006;580:3787–93. doi: 10.1016/j.febslet.2006.05.065. [DOI] [PubMed] [Google Scholar]
- 35.Park J, Seo T, Hwang S, Lee D, Gwack Y, Choe J. The K-bZIP protein from Kaposi’s sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J Virol. 2000;74:11977–82. doi: 10.1128/JVI.74.24.11977-11982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okoshi R, Kubo N, Nakashima K, Shimozato O, Nakagawara A, Ozaki T. CREB represses p53-dependent transactivation of MDM2 through the complex formation with p53 and contributes to p53-mediated apoptosis in response to glucose deprivation. Biochem Biophys Res Commun. 2011;406:79–84. doi: 10.1016/j.bbrc.2011.01.114. [DOI] [PubMed] [Google Scholar]
- 37.Giebler HA, Lemasson I, Nyborg JK. p53 recruitment of CREB binding protein mediated through phosphorylated CREB: a novel pathway of tumor suppressor regulation. Mol Cell Biol. 2000;20:4849–58. doi: 10.1128/MCB.20.13.4849-4858.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okoshi R, Ozaki T, Yamamoto H, Ando K, Koida N, Ono S, Koda T, Kamijo T, Nakagawara A, Kizaki H. Activation of AMP-activated protein kinase induces p53-dependent apoptotic cell death in response to energetic stress. J Biol Chem. 2008;283:3979–87. doi: 10.1074/jbc.M705232200. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Russell DW. Molecular Cloning, a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2001. [Google Scholar]
- 40.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]