Abstract
The neonatal Fc receptor (FcRn) encoded by FCGRT is known to be involved in the pharmacokinetics (PK) of therapeutic monoclonal antibodies (mAbs). Variability in the expression of FCGRT gene and consequently in the FcRn protein level could explain differences in PK observed between patients treated with mAbs. We studied whether the previously described variable number tandem repeat (VNTR) or copy number variation (CNV) of FCGRT are associated with individual variations of PK parameters of cetuximab. VNTR and CNV were assessed on genomic DNA of 198 healthy individuals and of 94 patients treated with the therapeutic mAb. VNTR and CNV were analyzed by allele-specific PCR and duplex real-time PCR with Taqman® technology, respectively. The relationship between FCGRT polymorphisms (VNTR and CNV) and PK parameters of patients treated with cetuximab was studied. VNTR3 homozygote patients had a lower cetuximab distribution clearance than VNTR2/VNTR3 and VNTR3/VNTR4 patients (p = 0.021). We observed no affects of VNTR genotype on elimination clearance. One healthy person (0.5%) and 1 patient (1.1%) had 3 copies of FCGRT. The PK parameters of this patient did not differ from those of patients with 2 copies. The FCGRT promoter VNTR may influence mAbs’ distribution in the body. CNV of FCGRT cannot be used as a relevant pharmacogenetic marker because of its low frequency.
Keywords: FcRn, neonatal Fc receptor, DNA Copy Number Variation, therapeutic monoclonal antibodies, pharmacokinetics, therapeutic drug monitoring, cetuximab, genetic polymorphism
Introduction
Pharmacokinetics (PK) of monoclonal antibodies (mAbs) increasingly appears as a critical determinant of patients’ therapeutic response. There is growing evidence that their concentrations are highly variable and that the degree of exposure to these biopharmaceuticals is associated with the response rate to infliximab,1 adalimumab,2 rituximab,3 cetuximab,4 trastuzumab.5 Based on these observations, empirical attempts to increase doses have led to mixed results. It is therefore of the highest importance to better determine the sources of PK variability in humans to propose new dosing strategies.
Body mass or body surface area,6 sex6,7 and the antigenic burden8,9 are well-known factors that contribute to mAb interindividual PK variability. Another factor responsible for this variability could be FcRn, a widely expressed MHC class I-like receptor that binds IgG and albumin with a higher affinity at acidic pH (< 6.5) than at neutral pH.10-12 FcRn permits the recycling and transcytosis of endogenous IgG, as reviewed by Roopenian et al.13 FcRn is likely to have a crucial role in mAb PK in human, but, to our knowledge, there is no direct proof. We recently reported that serum albumin concentration influences cetuximab clearance,4 suggesting the involvement of FcRn. Another indirect proof of the role of FcRn in humans treated with mAbs resides in the better PK profile and therapeutic activity of chimeric or humanized mAbs in comparison to murine mAbs, which bind poorly to human FcRn.14
An interesting, but not yet documented, approach to understand mAb PK interindividual variability would be to study genetic polymorphisms responsible of variations in FcRn expression. In human, FcRn is encoded by FCGRT, a 14 kb gene located on the long arm of chromosome 19 and comprising 7 exons.15 Few data are available on potential polymorphisms of this gene that are of interest. A VNTR of FCGRT promoter region was described by Sachs et al.16 They observed that homozygous individuals for 3 repetitions of a 37 base pair (bp) VNTR (VNTR3) express 1.66-fold more FCGRT transcripts in their monocytes than heterozygous individuals for 2 and 3 repetitions (VNTR2/VNTR3). Furthermore, they reported a functional effect of this polymorphism, which was that monocytes from VNTR3 homozygous individuals bound IgG at acidic pH more efficiently than heterozygous individuals. In another study, despite the fact that a relationship with the VNTR polymorphism was not observed, common variable immunodeficiency (CVID) patients with low FcRn mRNA level and treated with intravenous immunoglobulins (IVIg) were shown to have a more rapid decline of IgG concentration than patients with higher mRNA level.17 These results were corroborated by our group in recent work showing that IVIg therapy is more efficient in VNTR3/VNTR3 homozygous CVID patients than in VNTR2/VNTR3 patients.18 Nevertheless, to our knowledge the affect of this polymorphism on mAb PK has not yet been studied.
Besides VNTR, copy number variation (CNV) of FCGRT has not been reported yet. CNV has been described as a repetition or deletion of ≥ 1kb genetic region that potentially leads to a variable copy number of a gene.19,20 CNV may affect the protein level and lead to functional consequences. As reviewed by He et al., some CNV are used as pharmacogenetic markers.21 A study using bovine FCGRT transgenic mouse showed that additional copies of FCGRT led to an increase of FcRn mRNA and protein levels and to a lengthening of IgG half-life.22 This suggests that naturally occurring supplemental copies of the gene could have an effect on IgG half-life.
Taken together, these data suggest that the identification of genetic polymorphisms responsible for a variation in FCGRT gene expression is of great interest, particularly in the case of therapeutic mAbs. In this work, we focused on analyzing the influence of VNTR polymorphism and CNV of FCGRT on interindividual PK variability of cetuximab.
Results
VNTR and cetuximab PK
The VNTR polymorphism described by Sachs et al.16 in 2006 was investigated in the present study to assess its affect on mAb PK. We genotyped the 198 healthy individuals and the 94 patients of the FOLFIRICETUX study. Results are summarized in Table 1. Genotype distribution was not different between the 2 cohorts as shown by a chi-square test. We compared volumes of distribution (V1 and V2, respectively) and distribution and elimination clearances (Q and CL, respectively) of cetuximab between homozygous patients (VNTR3/VNTR3 genotype) and heterozygous patients (VNTR2/VNTR3 or VNTR3/VNTR4 genotypes) (Fig. 1). We found a significant difference in distribution clearances (Q) (means of 0.871 L/day and 1.01 L/day for the homozygote group and for the heterozygote group, respectively; p = 0.021). When considering the transfer constants k12 and k21, both are lower in homozygote patients than in heterozygote patients (0.301 d−1 and 0.334 d−1 for k12 and 0.198 d−1 and 0.233 d−1 for k21, respectively). This difference is significant for k21 (p = 0.047), reflecting the fact that cetuximab transfer from peripheral compartment to central compartment is slower in homozygous patients. Consistently, a trend toward a longer distribution half-life (t1/2α) was found (1.335 d vs 1.226 for homozygous and heterozygous patients, respectively; p = 0.058). No difference was found in elimination clearance (means of 0.513 L/day and 0.545 L/day for the homozygote group and heterozygote group, respectively) or distribution volume.
Table 1. Genotypes of FCGRT promoter VNTR in healthy individuals and cetuximab treated patients.
| VNTR genotype | Healthy individuals n (%) |
Cetuximab treated patients n (%) |
|---|---|---|
| VNTR2/VNTR2 | 2 (1.0) | 0 (0) |
| VNTR2/VNTR3 | 32 (16.2) | 13 (13.8) |
| VNTR3/VNTR3 | 160 (80.8) | 78 (83.0) |
| VNTR3/VNTR4 | 3 (1.5) | 3 (3.2) |
| VNTR3/VNTR6 | 1 (0.5) | 0 (0) |
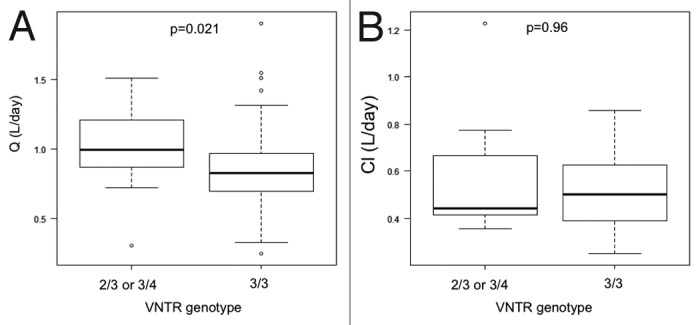
Figure 1. Comparison of cetuximab distribution (Q) and elimination (Cl) clearances according to patients VNTR genotype. Box-plot representation in the two genotypes of distribution clearance (Q) in (A) and elimination clearance (Cl) in (B). Medians, first and third quartiles and 5 and 95 percentiles are represented. Isolated circles are extreme values.
CNV frequency in healthy subjects
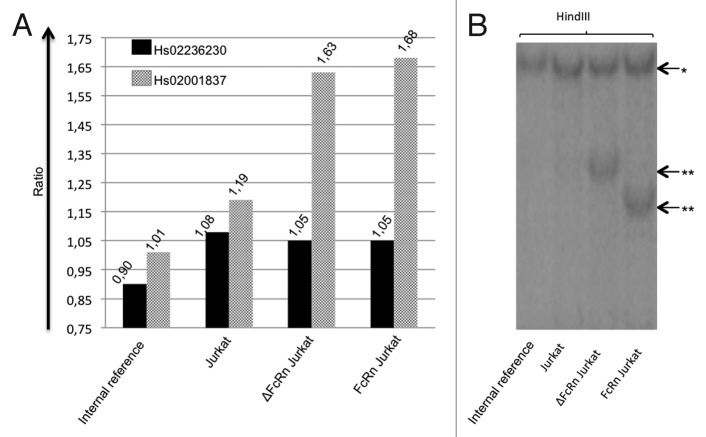
We determined FCGRT copy number by real-time PCR to assess the existence and the frequency of CNV. DNA samples were tested with two different pairs of primers, Hs02236230 and Hs02001837 (Fig. 2) and results are shown in Table 2. The mean ratio obtained was 1.01 ± 0.04 with Hs02236230 primers and 1.02 ± 0.05 with Hs02001837 primers. Theoretical ratios are 0.5; 1; 1.5 and 2 for 1, 2, 3 and 4 copies of FCGRT, respectively. Considering that a ratio between 0.75 and 1.25 corresponds to 2 copies per diploid genome,23 the results obtained in our 198 individuals suggest that FCGRT is rarely affected by CNV. One donor had discordant values between the two primers (1.001 for Hs02236230, 1.262 for Hs02001837). Only one donor (0.5%) apparently had 3 copies of FCGRT because the results obtained with Hs02236230 and Hs02001837 were 1.517 and 1.252 (mean of two experiments), respectively. We confirmed that this patient presented more than two copies of FCGRT with Hs04011945, Hs02317840 and Hs01245147 primers (data not shown). To validate the fact that a third copy of FCGRT leads to a ratio of ~1.5, we conducted a PCR using DNAs extracted from internal control, Jurkat cells, FCGRT coding sequence (CDS)-transfected Jurkat cells and truncated FCGRT CDS-transfected Jurkat cells (Fig. 3A). The ratios obtained by real-time PCR with internal control DNA and DNA from untransfected Jurkat cells were close to 1 (0.9 to 1.19, within the limits described by Nguyen et al.23) using the two sets of primers. DNAs extracted from transfected Jurkat cell lines show data close to 1.5 when considering Hs02001837 primers (1.68 and 1.63 for FCGRT and for ΔCDS FCGRT transfected cell lines, respectively). As expected, they were close to 1 with Hs02236230 primers because this set of primers is partially intronic and, consequently, does not match with transfected CDS of FCGRT. To assert the unique DNA insertion in the transfected cells, we performed a Southern blot analysis after digestion of the various DNA with HindIII and XbaI enzymes. We observed a supplementary band with HindIII digestion (Fig. 3B), in lanes corresponding to DNA of the two transfected Jurkat cell lines compared with the lanes corresponding to internal reference DNA and Jurkat cell DNA. These results are consistent with the insertion of a unique copy of FCGRT in genomic DNA. This was corroborated by the digestion of DNA with the XbaI enzyme (data not shown). Because the presence of one more copy triggers an increase of the ratio substantially equal to 0.5, we can infer that FCGRT is present in 2 copies in our DNA of reference. Taken together, these results show that an additional sequence of the gene results in a ratio close to 1.5 in the real-time PCR assay.
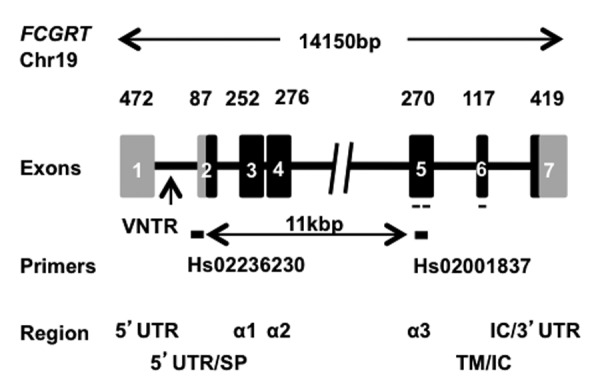
Figure 2. Schematic representation of human FCGRT gene. Chromosome (chr) localization: chr19. Coding and non-coding regions are represented by gray or black boxes respectively. Hs02236230 and Hs02001837 primers used in PCR assays are represented by a black line and probe used in Southern-blot is represented by a dashed line. Exons are identified by box numbers and length in nucleotides. 5′ or 3′UTR: 5′ or 3′ untranslated region, SP: signal peptide, α: α domain 1, 2 and 3; TM: transmembrane region; IC: intracytoplasmic region
Table 2. Determination of FCGRT copy number in 198 healthy blood donors with two different pairs of primers.
| Healthy population | |||
|---|---|---|---|
| Ratio* | Copy Number | Hs02236230 | Hs02001837 |
| < 0.75 0.75 < < 1.25 1.25 < < 1.75 1.25 < < 1.75 > 1.75 |
0–1 2 2 3 > 3 |
0 197 0 1 0 |
0 196 1 1 0 |
*FCGRT fluorescence / RPPH1 fluorescence of the sample reported to FCGRT fluorescence / RPPH1 fluorescence of reference DNA.
Figure 3. Affect of a supplemental coding sequence of FCGRT on PCR ratios. (A) Results of real-time PCR with Hs02236230 (black bars) and Hs02001837 (gray bars). Ratio values are indicated above respective bars. (B) Southern blot analysis following HindIII digestion of corresponding DNA samples. * Bands of identical size corresponding to endogenous FCGRT. ** bands only present in FcRn Jurkat or ΔFcRn Jurkat DNA samples lane corresponding to FCGRT additional insertion.
CNV of FCGRT and cetuximab PK
Because the level of FcRn protein may influence the PK of mAbs, we evaluated the CNV of FCGRT in patients treated with cetuximab for metastatic colorectal cancer during the Phase 2 FOLFIRICETUX study. One of the aims of this study was to analyze the sources and the consequences of interindividual variability in cetuximab PK. The CNV was evaluated by real-time PCR and the results are shown in Table 3. The means of the ratios for the 94 patients were 1.02 ± 0.06 and 1.04 ± 0.08 for Hs02236230 and Hs02001837, respectively. Similarly to the healthy blood donors, we observed that most patients (90 out of 94) presented 2 copies of FCGRT. Three patients had a discrepancy between the results obtained with the two sets of primers and one patient (1.1%) had the two ratios in favor of 3 copies of FCGRT (1.679 and 1.721, respectively; mean of 2 experiments). We confirmed that more than 2 copies of FCGRT were present in the DNA of this patient by using three other primers, Hs01245147, Hs4011945 and Hs02317840 (data not shown). To assess the effect of an additional copy of FCGRT on mAb PK, we then compared the PK profile of cetuximab of this patient to the mean profile of other patients.4 The cetuximab concentrations of this patient did not differ from the mean concentration of the others (Fig. 4).
Table 3. Determination of FCGRT copy number in 94 colorectal cancer patients treated with cetuximab.
| Cetuximab treated patients | ||||
|---|---|---|---|---|
| Ratio* | Copy Number | Hs02236230 | Hs02001837 | |
| < 0.75 0.75 < < 1.25 1.25 < < 1.75 1.25 < < 1.75 > 1.75 |
0–1 2 2 3 > 3 |
0 92 1 1 0 |
0 91 2 1 0 |
|
FCGRT fluorescence / RPPH1 fluorescence of the sample reported to FCGRT fluorescence / RPPH1 fluorescence of reference DNA.
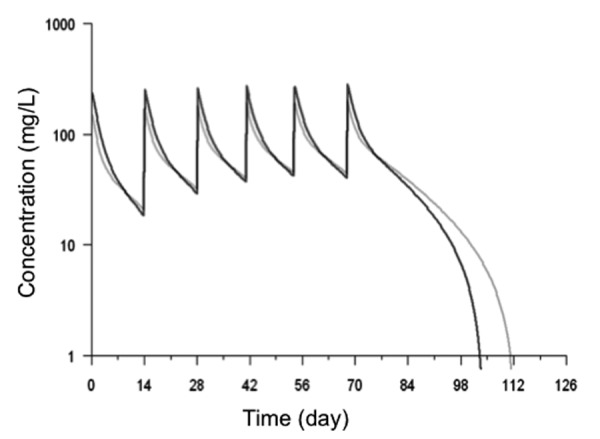
Figure 4. Cetuximab concentrations measured in the patient with 3 copies of FCGRT and mean cetuximab concentration in 93 other patients. Model-predicted concentrations, using the estimated pharmacokinetic parameters, are displayed. Black profile corresponds to the PKs of cetuximab when parameters of patient with 3 copies of FCGRT are used in the model. Grey profile corresponds to the PKs of cetuximab using the mean parameters of 93 patients in the same model.
Discussion
Because of its functions in recycling and transcytosis of IgG, FcRn is a potential source of interindividual variability in PK and its gene, FCGRT, may represent an interesting pharmacogenetic biomarker. Therefore, our study focused on FCGRT polymorphisms potentially affecting FcRn expression because it is likely to affect mAb PK. A VNTR identified within the promoter of FCGRT was associated with variable expression of FcRn in monocytes.16 VNTR3, the most common allele, is associated with an increase in promoter activity and, in turn, an increase of FcRn level compared with VNTR2 allele. Monocytes from VNTR3/VNTR3 patients bind IgG more efficiently. Similarly, we recently found in a cohort of CVID patients treated with IVIg that efficacy of IgG replacement was better in VNTR3/VNTR3 patients than in VNTR2/VNTR3 patients.18 Here, we show that this polymorphism also has a significant effect on cetuximab distribution in the body. Patients with two VNTR3 alleles had a lower distribution clearance; in these patients, the passage of cetuximab from peripheral compartment to central compartment was slower than in heterozygous patients. This may be due to a higher expression of FcRn in homozygous patients, who consequently retain cetuximab more efficiently in peripheral tissues (cells and interstitial fluids).
Other polymorphisms are also likely to result in the modulation of the FcRn expression. Because CNV may affect protein level,24-26 we focused on the not yet precisely studied CNV of FCGRT using real-time PCR assays. We chose primers separated from 11 kb to evaluate the copy number of regions separated from more than 1 kb, the minimal length for a CNV. We validated our real-time PCR results by measuring the affect of an additional copy of the gene on the ratios of fluorescence. In that aim, transfection of Jurkat cells by FCGRT CDS was performed and PCR results were confirmed by a Southern blot experiment. We assessed the presence and frequency of CNV in 198 healthy blood donors and in 94 patients treated with a therapeutic mAb. We found one sample presenting a supplemental FCGRT copy in each cohort. One healthy control and 3 patient samples presented a mismatch for the two probes, suggesting a partial duplication of the gene. No deletion of the gene was apparent. Taken together, our results show that FCGRT is not frequently affected by CNV. Few studies on the FCGRT region are referenced in the Database of Genomic Variants (projects.tcag.ca/variation/). A whole genome study published in Database of Genomic Variants reports deletions in the FCGRT region, but no insertion.27 The authors of the report performed an array comparative genomic hybridization assay on DNA samples from 93 patients, a smaller panel than that of our study. Another array comparative genomic hybridization assay study performed on 30 DNA samples reported deletions of the FCGRT region.28 Our FCGRT specific approach could not reveal such deletion, but has demonstrated that CNV of FCGRT are clearly rare events and cannot be used as a pharmacogenetic marker even if associated with functional consequences on mAb PK. No particular PK profile was observed in the patient with three copies of FCGRT.
Our data show the affect of FcRn on therapeutic mAb distribution in human. Given the functions of FcRn and the present results, an accurate understanding of the regulation of the expression of this receptor is of crucial interest for mAb PK comprehension. The affect of the VNTR polymorphism on the PK of other mAbs should be assessed to confirm this effect.
Material and Methods
Patients
DNA samples were obtained from two cohort studies approved by ethics committees. All human participants gave written informed consent. One hundred and 98 DNA samples of healthy individuals (Etablissement Français du Sang of Tours, cohort number DC-2008–308) and 94 DNA samples from the Phase 2 FOLFIRICETUX study (ClinicalTrials.gov identifier: NCT00559741) were analyzed. Patients of the FOLFIRICETUX study were treated by cetuximab, folinic acid, fluorouracil, and irinotecan for metastatic colorectal cancer. Characteristics of the patients were detailed in our previous paper.4 The study was performed in accordance with the Declaration of Helsinki and was approved by the ethics committee of Angers University Hospital, France.
Cell lines
Jurkat cells were obtained from American Type Culture Collection (ATCC, TIB-152). FcRn or truncated FcRn (ΔFcRn)-expressing Jurkat cells were obtained by stable transfection of Jurkat cells with the full FCGRT CDS containing plasmid or a plasmid containing the FCGRT CDS deleted from 99 nucleotides corresponding to the 33 amino acids in the carboxyl-terminal of the protein, respectively.
DNA extraction
The genomic DNA from all samples, including cell lines, was extracted using the QIAamp® DNA Blood Midi kit according to manufacturer’s instructions (Qiagen, 51185). DNA concentrations were measured with a Nanodrop spectrophotometer (Thermo Fisher Scientific, ND2000) and diluted to 5 ng/µL with nuclease-free water.
FCGRT promoter VNTR polymorphism
PCR assays were performed using primers encompassing the VNTR polymorphism of FCGRT gene described by Sachs et al.16 The 20 µL reaction mixture contained 10 ng of genomic DNA, 250 nM of sense (5′-GGAGCGAGGCTGAAGGGAAC-3′), and antisense (5′-CCCCTGAACTGGATCTCAGTTG-3′) primers, 200 μM of each dNTP (Fermentas, R0181), 1.5 mM MgCl2 and 0.5 U of Taq DNA polymerase in its buffer (Promega, M830B). PCR conditions consisted in 5 min at 94°C followed by 40 cycles, each consisting in 3 steps at 94°C for 1 min, 64.4°C for 1 min and 72°C for 40 sec. PCR complete extension was achieved for 5 min at 72°C. This amplification was performed on the MyCycler thermal cycler (Bio-Rad, 170–9703). PCR products were resolved using 8% acrylamide gel (Invitrogen, EC62152) and visualized after ethidium bromide staining.
CNV determination
To determine the copy number of FCGRT gene, we used the Applied BiosystemsTM CNV TaqMan® Assay kit according to the manufacturer’s instructions. Briefly, a duplex real-time PCR assay was conducted with two pairs of primers and probes in each well. The carboxyfluorescein (FAM) labeled probe matches with FCGRT gene and the VIC labeled probe with RPPH1, the gene encoding the RNase P H1 subunit. The latter is known to present no CNV and serves as the reference gene.29 FAM and VIC fluorescences of the sample were measured and reported to the fluorescence obtained with an internal reference DNA sample identical in each experiment and presenting two copies of FCGRT per diploid genome. Each sample was tested in triplicate with two pairs of FCGRT primers and probes (Hs02236230 and Hs02001837) that were localized at the junction of intron 1 and exon 2, respectively, and within exon 5 (Fig. 2). Some samples were also tested with Hs04011945, Hs02317840 and Hs01245147 that are located in exon 1, exon 4 and exon 7, respectively. Real-time PCR was performed using the LightCycler 2.0 system (Roche Diagnostics, 05015278001). Thermal cycling started with 10 min, 95°C incubation and was followed by 40 cycles of 15 sec denaturation at 95°C and 60 sec elongation at 60°C according to the manufacturer’s protocol. Results were analyzed with LightCycler Software using the ΔΔCt method.
Southern blot
Ten μg of genomic DNA extracted from either Jurkat cells, FcRn expressing Jurkat cells, ΔFcRn expressing Jurkat cells or peripheral blood from healthy individual used as internal control were digested by HindIII and XbaI for 6 h at 37°C. Digestion products were separated on 0.8% agarose gel by electrophoresis with a 1kb DNA ladder (Promega, G5711). Genomic DNA was then transferred to Hybond membrane according to the manufacturer (Amersham, RPN119B). Membranes were hybridized with ULTRAhyb hybridization buffer (Applied Biosystems, AM8670) at 42°C for 16 h with a 32P labeled 348 bp probe corresponding to the bulk of exon 5 and the totality of exon 6 of FcRn cDNA (Fig. 2). It was obtained by PCR amplification of peripheral blood mononuclear cells cDNA with forward primer 5′-CTGGCTTTTCCGTGCTTACC-3′ and reverse primer 5′-CTGGCAGCCCACTCCTCATC-3′ and was labeled using Klenow enzyme (Promega, M2201).
PK analysis
Cetuximab PK analysis was previously described by Azzopardi et al.4 Briefly, cetuximab concentrations were determined by ELISA on blood samples collected before, 2, 3.5 and 5.5 h after the first infusion and 44 h after following infusions.30 Data were analyzed by population PK modeling using MONOLIX 3.1 software. The structural model was bicompartmental combining linear (CL) and nonlinear (k0) elimination of cetuximab. Cetuximab distribution between central volume (V1) and peripheral volume (V2) was described with a clearance Q. Distribution was also quantified in terms of k12 = Q/V1 and k21 = Q/V2 corresponding to transfer of cetuximab from central compartment to peripheral compartment and from peripheral compartment to central compartment respectively.
Statistical analysis
PK parameters estimated in the FOLFIRICETUX study were compared between VNTR groups using the Mann-Whitney test. The results are presented as median and the interquartile range (IQR, the difference between the 75th and the 25th percentiles). VNTR genotype distributions were compared using a chi-square test. Differences were considered significant when p < 0.05.
Acknowledgments
This publication has been funded with support from the French National Research Agency under the program “Investissements d’avenir” Grant Agreement LabEx MAbImprove: ANR-10-LABX-53. Measurement of cetuximab serum concentrations were performed within the CePiBAc platform. CePiBAc is co-financed by the European Union. Europe is committed to the region Centre with the European Regional Development Fund.
Glossary
Abbreviations:
- FcRn
Fc receptor neonatal
- PK
pharmacokinetics
- mAb
monoclonal antibody
- CNV
copy number variation
- VNTR
Variable Number Tandem Repeat
- Q
distribution clearance
- CL
elimination clearance
- CVID
common variable immunodeficiency
- IgG
immunoglobulin G
- MHC
major histocompatibility complex
- CDS
coding sequence
- IVIg
intravenous immunoglobulin
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/24815
References
- 1.St Clair EW, Wagner CL, Fasanmade AA, Wang B, Schaible T, Kavanaugh A, et al. The relationship of serum infliximab concentrations to clinical improvement in rheumatoid arthritis: results from ATTRACT, a multicenter, randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 2002;46:1451–9. doi: 10.1002/art.10302. [DOI] [PubMed] [Google Scholar]
- 2.Bartelds GM, Wijbrandts CA, Nurmohamed MT, Stapel S, Lems WF, Aarden L, et al. Clinical response to adalimumab: relationship to anti-adalimumab antibodies and serum adalimumab concentrations in rheumatoid arthritis. Ann Rheum Dis. 2007;66:921–6. doi: 10.1136/ard.2006.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Igarashi T, Kobayashi Y, Ogura M, Kinoshita T, Ohtsu T, Sasaki Y, et al. IDEC-C2B8 Study Group in Japan Factors affecting toxicity, response and progression-free survival in relapsed patients with indolent B-cell lymphoma and mantle cell lymphoma treated with rituximab: a Japanese phase II study. Ann Oncol. 2002;13:928–43. doi: 10.1093/annonc/mdf155. [DOI] [PubMed] [Google Scholar]
- 4.Azzopardi N, Lecomte T, Ternant D, Boisdron-Celle M, Piller F, Morel A, et al. Cetuximab pharmacokinetics influences progression-free survival of metastatic colorectal cancer patients. Clin Cancer Res. 2011;17:6329–37. doi: 10.1158/1078-0432.CCR-11-1081. [DOI] [PubMed] [Google Scholar]
- 5.Baselga J. Phase I and II clinical trials of trastuzumab. Ann Oncol. 2001;12(Suppl 1):S49–55. doi: 10.1093/annonc/12.suppl_1.S49. [DOI] [PubMed] [Google Scholar]
- 6.Ng CM, Bruno R, Combs D, Davies B. Population pharmacokinetics of rituximab (anti-CD20 monoclonal antibody) in rheumatoid arthritis patients during a phase II clinical trial. J Clin Pharmacol. 2005;45:792–801. doi: 10.1177/0091270005277075. [DOI] [PubMed] [Google Scholar]
- 7.Ternant D, Aubourg A, Magdelaine-Beuzelin C, Degenne D, Watier H, Picon L, et al. Infliximab pharmacokinetics in inflammatory bowel disease patients. Ther Drug Monit. 2008;30:523–9. doi: 10.1097/FTD.0b013e318180e300. [DOI] [PubMed] [Google Scholar]
- 8.Slavin RG, Ferioli C, Tannenbaum SJ, Martin C, Blogg M, Lowe PJ. Asthma symptom re-emergence after omalizumab withdrawal correlates well with increasing IgE and decreasing pharmacokinetic concentrations. J Allergy Clin Immunol. 2009;123:107–13, e3. doi: 10.1016/j.jaci.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 9.Daydé D, Ternant D, Ohresser M, Lerondel S, Pesnel S, Watier H, et al. Tumor burden influences exposure and response to rituximab: pharmacokinetic-pharmacodynamic modeling using a syngeneic bioluminescent murine model expressing human CD20. Blood. 2009;113:3765–72. doi: 10.1182/blood-2008-08-175125. [DOI] [PubMed] [Google Scholar]
- 10.Rodewald R, Kraehenbuhl JP. Receptor-mediated transport of IgG. J Cell Biol. 1984;99:159s–64s. doi: 10.1083/jcb.99.1.159s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simister NE, Rees AR. Isolation and characterization of an Fc receptor from neonatal rat small intestine. Eur J Immunol. 1985;15:733–8. doi: 10.1002/eji.1830150718. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhury C, Mehnaz S, Robinson JM, Hayton WL, Pearl DK, Roopenian DC, et al. The major histocompatibility complex-related Fc receptor for IgG (FcRn) binds albumin and prolongs its lifespan. J Exp Med. 2003;197:315–22. doi: 10.1084/jem.20021829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roopenian DC, Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nat Rev Immunol. 2007;7:715–25. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- 14.Ober RJ, Radu CG, Ghetie V, Ward ES. Differences in promiscuity for antibody-FcRn interactions across species: implications for therapeutic antibodies. Int Immunol. 2001;13:1551–9. doi: 10.1093/intimm/13.12.1551. [DOI] [PubMed] [Google Scholar]
- 15.Mikulska JE, Pablo L, Canel J, Simister NE. Cloning and analysis of the gene encoding the human neonatal Fc receptor. Eur J Immunogenet. 2000;27:231–40. doi: 10.1046/j.1365-2370.2000.00225.x. [DOI] [PubMed] [Google Scholar]
- 16.Sachs UJ, Socher I, Braeunlich CG, Kroll H, Bein G, Santoso S. A variable number of tandem repeats polymorphism influences the transcriptional activity of the neonatal Fc receptor alpha-chain promoter. Immunology. 2006;119:83–9. doi: 10.1111/j.1365-2567.2006.02408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freiberger T, Grodecká L, Ravcuková B, Kurecová B, Postránecká V, Vlcek J, et al. Association of FcRn expression with lung abnormalities and IVIG catabolism in patients with common variable immunodeficiency. Clin Immunol. 2010;136:419–25. doi: 10.1016/j.clim.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Gouilleux-Gruart V, Chapel H, Chevret S, Lucas M, Malphettes M, Fieschi C, et al. DEFI study group Efficiency of immunoglobulin G replacement therapy in common variable immunodeficiency: correlations with clinical phenotype and polymorphism of the neonatal Fc receptor. Clin Exp Immunol. 2013;171:186–94. doi: 10.1111/cei.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–8. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 20.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 21.He Y, Hoskins JM, McLeod HL. Copy number variants in pharmacogenetic genes. Trends Mol Med. 2011;17:244–51. doi: 10.1016/j.molmed.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bender B, Bodrogi L, Mayer B, Schneider Z, Zhao Y, Hammarström L, et al. Position independent and copy-number-related expression of the bovine neonatal Fc receptor alpha-chain in transgenic mice carrying a 102 kb BAC genomic fragment. Transgenic Res. 2007;16:613–27. doi: 10.1007/s11248-007-9108-9. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen DL, Staeker J, Laika B, Steimer W. TaqMan real-time PCR quantification strategy of CYP2D6 gene copy number for the LightCycler 2.0. Clin Chim Acta. 2009;403:207–11. doi: 10.1016/j.cca.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Willcocks LC, Lyons PA, Clatworthy MR, Robinson JI, Yang W, Newland SA, et al. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J Exp Med. 2008;205:1573–82. doi: 10.1084/jem.20072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun YV, Peyser PA, Kardia SL. A common copy number variation on chromosome 6 association with the gene expression level of endothelin 1 in transformed B lymphocytes from three racial groups. Circ Cardiovasc Genet. 2009;2:483–8. doi: 10.1161/CIRCGENETICS.109.848754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breunis WB, van Mirre E, Geissler J, Laddach N, Wolbink G, van der Schoot E, et al. Copy number variation at the FCGR locus includes FCGR3A, FCGR2C and FCGR3B but not FCGR2A and FCGR2B. Hum Mutat. 2009;30:E640–50. doi: 10.1002/humu.20997. [DOI] [PubMed] [Google Scholar]
- 27.Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, et al. A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perry GH, Ben-Dor A, Tsalenko A, Sampas N, Rodriguez-Revenga L, Tran CW, et al. The fine-scale and complex architecture of human copy-number variation. Am J Hum Genet. 2008;82:685–95. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baer M, Nilsen TW, Costigan C, Altman S. Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res. 1990;18:97–103. doi: 10.1093/nar/18.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cézé N, Ternant D, Piller F, Degenne D, Azzopardi N, Dorval E, et al. An enzyme-linked immunosorbent assay for therapeutic drug monitoring of cetuximab. Ther Drug Monit. 2009;31:597–601. doi: 10.1097/FTD.0b013e3181b33da3. [DOI] [PubMed] [Google Scholar]