Abstract
Cytokine signaling is mediated by the combinatorial usage of seven STAT proteins that form homo- or heterodimers involved in the regulation of specific transcriptional programs. Among STATs, STAT2 is classically known to dimerize with STAT1 and together with IRF9 forms the ISGF3 transcription factor complex that has long been considered a hallmark of activation by type I and type III interferons. However, accumulating evidence reveal distinct facets of STAT2 and IRF9 activity mediated by the segregation in alternative STAT1-independent complexes/pathways that are thought to trigger different transcriptional programs. The goal of this review is to summarize our current knowledge of the stimuli, regulatory mechanisms, and function of these alternative pathways.
Keywords: IRF9, ISGF3, ISGs, STAT, TNFα, gene expression, interferon, signaling, synergism
Introduction
Initially discovered half a century ago, cytokines now count over 50 members that orchestrate cellular communication in an autocrine, juxtacrine, and paracrine fashion through binding to distinct families of receptors. Intriguingly, the fate of the response triggered by binding of cytokines to their cognate receptor is mainly determined by the specific combinatorial usage of only seven different signal transducer and activator of transcription (STAT) proteins. STAT proteins activated through phosphorylation by the receptor-bound Janus kinases (JAK) act as homo- or heterodimers that translocate to the nucleus to regulate the transcription of numerous target genes. Over the years, in vitro and in vivo studies mostly performed using single cytokine stimulation have provided a picture of STAT(s) activation by specific cytokines. Our current knowledge of these cytokine/STAT combinations was recently extensively reviewed and therefore will not be reiterated here.1 In this review we will rather focus on the specific activation and function of STAT2, which has long been considered a hallmark of activation by type I interferons (IFNα/β) and type III IFNs (IFNλ1)/interleukin (IL) 29, IFNλ2/IL28A and IFNλ3/IL28B). Response of immune and non-immune cells to type I and III IFNs is classically known to trigger the formation of heterotrimers containing STAT2, STAT1, and the interferon regulatory factor (IRF) 9, a transcription factor complex known as IFN-stimulated gene factor 3 (ISGF3). While activation of STAT2/IRF9 to form the ISGF3 complex has been widely considered a hallmark of type I and III IFN activation, accumulating evidence highlights a far more complex activation and function beyond ISGF3. The characterization of alternative signaling encompassing activation of STAT2/IRF9 suggests their segregation in distinct complexes (Fig. 1), which might be a crucial determinant in the specific activation of different transcriptional programs.
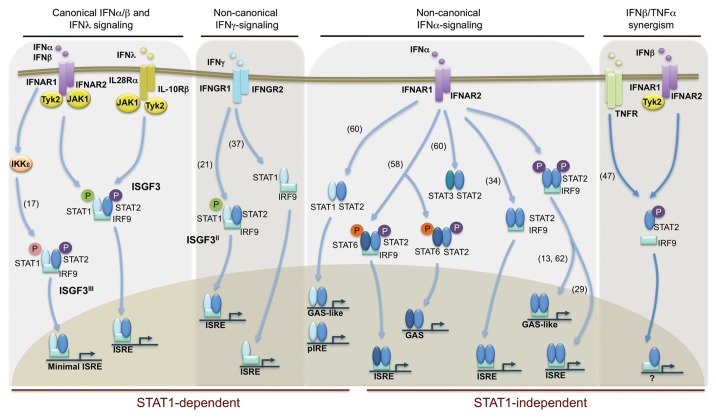
Figure 1. STAT2- and/or IRF9-containing complexes. The canonical type I and type III IFN signaling leads to the formation of the classical ISGF3 complex composed of STAT1, STAT2, and IRF9. In this complex, STAT1 is phosphorylated on Tyr701 (green) and STAT2 is phosphorylated on Tyr690 (violet) as a result of JAK1 and Tyk2 activation. ISGF3 regulates the expression of hundreds of ISGs containing an interferon stimulated response element (ISRE) in their promoter region. In the recent years, alternative complexes containing STAT2 and/or IRF9 have been described in various contexts either in combination with STAT1 (STAT1-dependent) or without STAT1 (STAT1-independent). Some STAT1-dependent complexes are distinct from the ISGF3 complex by their phosphorylation. For instance, in the ISGF3II complex STAT2 is not phosphorylated on Tyr690. In the ISGF3III complex, STAT1 is phosphorylated on Ser708 (pink). Additionally, alternative complexes are formed by association with different STAT partners, including STAT6 and STAT3. In some STAT6-containing complexes, STAT6 is also phosphorylated on a yet unknown residue (orange). These different complexes regulate gene expression though diverse consensus sequences, including ISRE, IFNγ-activated sequence (GAS), GAS-like, and palindromic IFN response element (pIRE). References are indicated in brackets.
The ISGF3 Complex: the Classical Role of STAT2 and IRF9 Engaged by Type I and III IFNs
IFNβ is a pleiotropic cytokine with potent antiproliferative, antiviral and host innate and adaptive immune regulatory functions.2 Based on extensive studies of IFNβ single cytokine stimulation, the binding of IFNβ to its receptor (IFNAR) is well characterized to engage a cascade of signaling events initiated by the rapid activation of the Tyk2 and JAK1 members of JAK kinases, which phosphorylate the intracellular domain of IFNAR on tyrosine (Tyr), thereby providing a docking site for the SH2 domains of the latent STAT1 and STAT2. Subsequent phosphorylation of STAT1 and STAT2 induces their heterodimerization and association with IRF9 to form the ISGF3 complex that translocates to the nucleus.3 Signaling of type III IFNs, which bind to a distinct heterodimeric receptor (IL28Rα/IL-10Rβ) through the ISGF3 complex, has been unveiled more recently with the discovery and growing interest of type III IFNs in the antiviral response.4,5
The formation of the heterotrimeric ISGF3 complex is mechanistically unique among STAT-dependent pathways, in part because among the STAT factors, STAT2 is distinct in that it does not recognize a DNA target site as a homodimer. Rather, STAT2 provides the transcriptional activation domain (TAD) essential for the induction of target gene transcription.6 The DNA-binding component of ISGF3 provides the specificity for binding to the consensus IFN-stimulated response element (ISRE), 5′-AGTTTCNNTT TCNC/T-3′,7 and not to the IFNγ-activated site (GAS) like other STAT dimers,8 present in the promoter of hundreds of IFN-stimulated genes (ISGs) encoding proteins with antiviral activities.9 The interaction with the core of the ISRE consensus is mediated by IRF9, but DNA contacts of STAT1 and STAT2 are required to provide stability of the interaction.10-13 Although a large majority of type I IFN-induced gene expression is attributable to the activation of the canonical ISGF3, it has become clear that its sole activation cannot completely explain the pleiotropic biological effects of type I IFNs. Likely part of the equation explaining these multiple functions is the increasing body of evidence showing that alternative STAT complexes composed of homo- or heterodimers of other members of the STAT proteins, also form upon type I IFN stimulation in a cell-specific fashion1,3 (Fig. 1).
Advances in our understanding of the regulation of ISGF3 have started to uncover the existence of heterogenous ISGF3 complexes, which are likely to participate in the regulation of specific functions. The paradigm has long been that the ISGF3 complex is composed of STAT1 phosphorylated on Tyr701 and of STAT2 on Tyr689. Additional phosphorylation of STAT1 on Serine (Ser) 727 provides full transcriptional activity of the ISGF3 complex.14,15 In contrary, Ser phosphorylation of STAT2 on Ser287 was shown to negatively regulate the activity of the ISGF3 complex.16 However, interesting reports suggest the existence of distinct pools of ISGF3 complexes involving heterogeneous phosphorylation of STAT1 or STAT2. Additional phosphorylation of STAT1 on Ser708 by the IκB kinase ε (IKKε) was shown to be required for the expression of a subset of ISGF3-regulated genes driven by minimal ISRE sites in mice.17 IKKε-mediated STAT1 Ser708 phosphorylation at late time points in the IFN response was found to be crucial for IFN-induced protein with tetratricopeptide repeats 2 (IFIT2) gene expression to control West Nile virus infection in mouse models. In this same study the author showed STAT1 Tyr701 phosphorylation temporally precedes Ser708 phosphorylation and that these two phosphorylations are mutually exclusive suggesting that the order of STAT1 phosphorylation during the course of IFN stimulation could be an important contributor to the kinetics of ISG expression.18 We propose to name the ISGF3 complex containing Ser708-phosphorylated STAT1 ISGF3III to distinguish it from the classical ISGF3 and the ISGF3II complexes (Fig. 1). ISGF3II complex was surprisingly identified in response to IFNγ. Few studies have reported that IFNγ is capable of inducing STAT2 Tyr phosphorylation.19,20 More recently, stimulation of A549 human lung epithelial cells with IFNγ was found to trigger an early and delayed peak of STAT1 Tyr701 phosphorylation. Unexpectedly, the delayed peak corresponds to the formation of an ISGF3 complex, named ISGF3II, containing phosphorylated STAT1, but unphosphorylated STAT2 on Tyr689, which is associated with double-stranded RNA-dependent protein kinase (PKR) antiviral gene expression21 (Fig. 1). This novel complex might reflect the observation that mice lacking IRF9 are impaired not only in their type I IFN response, but also in their IFNγ-induced ISRE-dependent gene expression.22 Whether the ISGF3II complex exists in response to type I and type III IFNs and/or exhibit cell type specificity remains to be determined.
STAT1-Independent Functions of STAT2 and IRF9
As described above, STAT2- and IRF9-dependent transcriptional activation of target genes has long been considered to reflect the involvement of the ISGF3 complex. More importantly, the initial observation that STAT1 knockout mice are defective in type I and type II IFN responses,23,24 and are now known to be defective in type III IFN responses as well, are highly susceptible to virus and bacterial infections,24-26 strongly supported that STAT1 is essential for antiviral gene expression. STAT2 knockout mice also exhibit severe susceptibility to viruses and this phenotype has largely been considered to reflect impaired ISGF3 activation.27 Thus, the function of STAT2 and IRF9 beyond ISGF3 has long been underestimated and it is only recently that it has started to be documented. The observation that STAT1 knockout mice are resistant to Dengue virus infection suggested the existence of STAT1-independent antiviral mechanisms,28 which were recently shown using STAT1 and STAT1/STAT2 knockout mice to be mediated by STAT2.29 Importantly, a fundamental difference in the role of STAT1 vs. STAT2 and IRF9 was further strengthened by the report that lymphocytic choriomeningitis virus (LCMV) infection in wild-type mice or in mice lacking either STAT2 or IRF9 is nonlethal, while in STAT1-deficient mice it is lethal.26 An interesting twist in the existence of these STAT1-independent functions was revealed in in vivo mouse models of measles virus and LCMV infections. Both viruses were found to evade the known antiviral effect of type I IFN by suppressing dendritic cell development, a process that is initiated through type I IFN-mediated STAT2 activation, which is STAT1-independent.30
Additional evidence of the existence of STAT2/IRF9 functions beyond ISGF3 is also accumulating through the thorough analysis of ISGs expression. Comparison of the expression of IRFs transcription factors in the brain of uninfected and LCMV-infected mice showed that IRF7 gene expression is induced by LCMV infection in a STAT2-dependent, but STAT1-independent, mechanism involving the engagement of type I IFN receptor.31 Using murine embryonic fibroblasts deficient in various JAKs or STATs, IFNα-mediated induction of adenosine deaminase acting on RNA 1 (ADAR1) A-to-I editing enzyme encoding gene was shown to be JAK1- and STAT2-dependent, but independent of STAT1.32 Analysis of apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) gene expression has recently highlighted cell type and stimulus specificity in the involvement of STAT1 in the regulation of target genes. In contrary to typical ISGs, such as PKR, IFNα induced APOBEC3G expression in primary hepatocytes and hepatocellular carcinoma cell lines and in primary macrophages, but not in primary CD4+ T cells.33 Mechanistically, while IFNγ-induced APOBEC3G expression in hepatocellular carcinoma cell lines was STAT1-dependent, IFNα-mediated induction was found to be dependent on STAT2 and IRF9, but despite a functional activation of STAT1, STAT1 itself was dispensable. Interestingly, analysis of the expression of typical ISGs, including PKR and MX1, also revealed that they were induced in a STAT1-independent, but STAT2- and IRF9-dependent manner.33 Similar experiments performed in the human embryonic kidney 293T cell line showed that PKR and MX1 expression were dependent on STAT1, highlighting the cell-type specificity of the involvement of STAT1 in gene regulation.33 Additional important proof for the biological significance of STAT2/IRF9-dependent, but STAT1-independent, regulation of gene expression was provided by the finding that STAT2 and IRF9 can effectively drive the transcription of the retinoic acid-induced gene G (RIG-G) gene in response to all-trans retinoic acid (ATRA) through a STAT1-independent mechanism.34
Interestingly, IRF9 was shown to bind to the IFNβ promoter independently of the binding of the larger ISGF3 complex.35,36 However, it remains to be determined whether the alternative IRF9/STAT2 pathway is involved in this regulation. The existence of alternative IRF9-dependent pathways independent on STAT2 has also been suggested. For instance, IRF9 in a complex with STAT1 that does not contain STAT2 regulates the transcription of the CXCL10 chemokine encoding gene in 2fTGH human fibrosarcoma cells.37 Additionally, overexpression of IRF9 observed in a significant number of breast and uterine tumors provides resistance to antimicrotubule agents through transcriptional activation of ISGs independently of STAT1 and STAT2.38
IFNβ and TNFα: Two Cytokines Synergizing to Specifically Trigger an IRF9/STAT2-Dependent pathway
Over the past decades, most studies aimed at characterizing the discovery of ligands, receptors, downstream signaling mechanisms and biological functions of type I and III IFNs have been performed in relation to single cytokine stimulation. However, this is a very unlikely physiological situation, as a cell rather simultaneously responds to a “cocktail of cytokines”. The events occurring downstream of concomitant cytokine stimulation have been barely studied, but one can expect that the fate of the gene expression response requires that cytokine-induced signaling pathways “work” together or at least exhibit significant cross-talk. The transcriptional program resulting from stimulation with a combination of cytokines might result from their synergistic action. Three different scenarios are currently proposed to contribute to the synergism of two cytokines mostly based on large-scale analysis of gene expression following stimulation with IFNγ and IFNβ,39 but also specific gene expression analysis following costimulation with IFNγ and TNFα or IFNβ and TNFα.40-43 The most intuitive mechanism of synergy, referred to as “independent action”, consists in the induction of two independent signaling cascades that ultimately lead to the induction of distinct sets of genes whose combined activities lead to a synergistic outcome.39 A second mechanism, referred to as “cooperative action”, results in the enhanced expression of a shared gene that is expressed, although to a lower extent, by single cytokine stimulation. This is mainly due to the cooperative action of different transcription factors activated independently by each cytokine.39 An unexpected third mechanism, named “cooperative induction”, has been recently unveiled by the group of G McFadden and is characterized by the synergistic expression of genes that are not induced by either cytokine itself or only induced to a lower level by either one of the cytokine.44
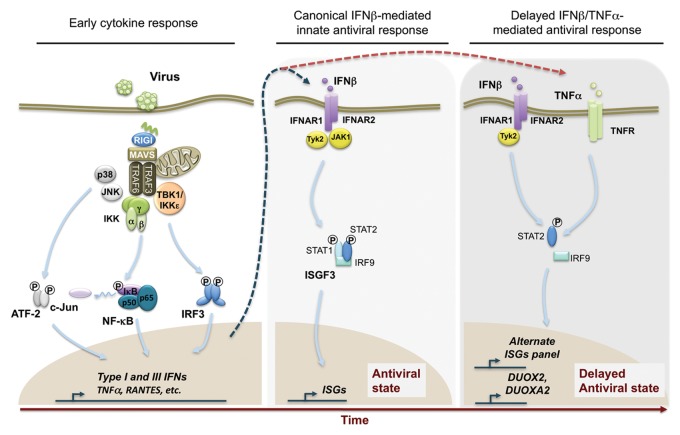
The “cooperative induction” model was identified in the context of costimulation with IFNβ and TNFα, two cytokines simultaneously produced early following innate recognition of invading viruses to trigger a specific transcriptional program dictating the subsequent immune response. The synergistic antiviral response triggered by IFNβ and TNFα was originally described as early as 1988.40 Subsequent reports have confirmed the synergistic action of IFNβ and TNFα in the regulation of ISGs.45 However, it was only in 2009, through the use of microarray gene expression profiling, that a first hint of a specific mechanism underlying the synergistic action of IFNβ and TNFα was revealed. Costimulation of human primary fibroblasts by IFNβ and TNFα induced the expression of a distinct panel of delayed antiviral genes that were either not responsive to IFNβ nor TNFα or that were only responsive to either one of the cytokine when used separately.46 This specific synergy-dependent antiviral response was necessary and sufficient to completely abrogate the productive replication and spreading of Myxoma virus in primary human fibroblasts. In contrast, the replication of Vaccinia and Tanapox poxviruses was only partially inhibited, whereas their spreading to neighboring cells was efficiently blocked.46 The importance of this delayed antiviral response triggered by the synergistic action of IFNβ and TNFα was further strengthened by the observation that it also takes place in the context of infection by the paramyxoviruses, Sendai virus, and respiratory syncytial virus, in lung epithelial cells.47 In this context, the resulting delayed antiviral response includes the induction of the expression of the DUOX2 NADPH oxidase that ultimately produces extracellular H2O2, an event that controls the levels of type I and type III IFNs at late time points of infection by a yet to be characterized mechanism47 (Fig. 2).
Figure 2. Canonical and non-canonical functions of STAT2 and IRF9 in the antiviral response. Cells respond to virus infection through the activation of multiple signaling pathways, leading to the activation of AP-1 (ATF-2/c-jun), NFκB and interferon regulatory factor 3 (IRF3). These transcription factors regulate the expression of various proinflammatory and antiviral cytokines and chemokines, including IFNβ and TNFα. Binding of IFNβ to its cognate receptor activates the “classical” antiviral pathway through activation of Tyk2 and JAK1 kinases ultimately leading to the formation of ISGF3. ISGF3 regulates the expression of multiple interferon stimulated genes (ISGs). Additionally, the synergism between IFNβ and TNFα induces late gene expression through a non-canonical antiviral STAT2- and IRF9-dependent, but STAT1-independent, pathway. This pathway is dependent on Tyk2 kinase activity and requires phosphorylation of STAT2 on Tyr690. This pathway triggers the delayed expression of an alternative ISG panel and, at least in airway epithelial cells, of the DUOX2/DUOXA2 genes encoding for a functional NADPH oxidase enzyme.
Early reports had offered a possible explanation of how the “cooperative induction” synergy between IFNβ and TNFα might be achieved. Several reports have shown that the synergy between IFNβ and TNFα is mechanistically grounded in the TNFα-mediated autocrine induction of IFNβ, a mechanism that can occur in an IRF1-dependent manner.45,48,49 However, in the context of infection with poxviruses and paramyxoviruses, a role of this autocrine pathway was excluded. Rather, the specificity of the crosstalk between IFNβ and TNFα is mirrored at the level of intracellular signal transduction. Silencing of the components of the ISGF3 complex using siRNA revealed that IFNβ/TNFα-induced expression of the DUOX2 gene in A549 cells was dependent on STAT2 and IRF9, but was STAT1-independent47 (Fig. 2). This suggests that the synergistic action of IFNβ and TNFα engages a non-canonical STAT2 and IRF9-dependent, but STAT1-independent, signaling pathway. Further analysis of this non-canonical pathway revealed that in the presence of TNFα, IFNβ induces enhanced Tyk2-mediated phosphorylation of STAT2 at Tyr690 over an extended period of time compared with IFNβ stimulation. Based on the use of AG490 JAK inhibitor and Bayer18 Tyk2 inhibitor, it was concluded that Tyk2 kinase activity was essential to mediate STAT2 phosphorylation in this non-canonical pathway (Fig. 2). Additionally, stimulation with TNFα led to IRF9 induction, suggesting that this could also contribute to the synergism with IFNβ. Thus, IRF9 induction and enhanced/extended STAT2 phosphorylation likely contribute to the specific activation of the non-canonical STAT2/IRF9 pathway during costimulation by IFNβ/TNFα.47 Of note, previous reports have demonstrated activation of JAK kinases, notably Tyk2, following stimulation with TNFα. JAK kinases were found to be recruited to type 1 TNF receptor (TNFR1) via a box 1-like membrane proximal proline rich motif thereby inducing activation of JAKs by autophosphorylation and ultimately promoting phosphorylation of STATs.50-52 Thus, it would be interesting to determine whether a TNFα-JAK axis contributes to the enhanced/extended STAT2 phosphorylation observed in the context of the synergistic action of IFNβ and TNFα. The specific recruitment of JAKs to TNFR1 suggests the existence of distinct pools of JAKs, which could have distinct functions in the “cooperative induction” synergy induced by IFNβ and TNFα.
The NFκB pathway is widely known to be engaged downstream of TNFR. Synergistic activation of NFκB was reported in the context of IFNγ and TNFα treatment53 and it would therefore seem intuitive that NFκB could play a role in the synergistic action of IFNβ and TNFα. An IFNβ-induced signal was found to synergize with NFκB through an uncharacterized mechanism to trigger the expression of genes, such as the CXCL10 and CXCL9, in a context where IFNβ and TNFα synergy is mediated by the autocrine induction of IFNβ in response to TNFα.45 However, overexpression of the super repressor (IκBα2NΔ4) of NFκB that inhibits the classical pathway of NFκB failed to block IFNβ/TNFα-induced DUOX2 expression, suggesting that NFκB is not involved in the “cooperative induction” synergism.47
Whether the non-canonical STAT2/IRF9 signaling pathway is responsible for the regulation of all other genes previously identified to be specifically responsive to the combination of IFNβ and TNFα 46 remains to be determined. Importantly, how exactly induction of the non-canonical STAT2- and IRF9-dependent pathway engaged downstream of IFNβ/TNFα costimulation diverge from the classical IFNβ-induced ISGF3 complex containing STAT1 remains unknown. However, an interesting report hints a possible mechanism. In HeLa cells, STAT1 was associated with TNFR1 and the signaling factors TRADD and FADD following TNFα stimulation. This mechanism was proposed to balance the pro- and anti-apoptotic signals induced by TNFα, as STAT1 recruitment to TNFR1 led to a decreased activation of NFκB, thereby promoting TNFα-induced apoptosis.54 Thus, it is a relevant hypothesis that in the context of IFNβ/TNFα costimulation, TNFα could induce the sequestration of STAT1 by interaction with TNFR1, thereby creating an environment where the STAT2- and IRF9-dependent, but STAT1-independent, non-canonical pathway could be specifically activated.
The understanding of how the synergism between IFNβ and TNFα ultimately lead to the activation of a STAT2/IRF9 pathway different from ISGF3, is even more elusive if one keeps in mind that several reports mentioned above have reported that type I IFN alone is capable of engaging STAT2- and IRF9-dependent, but STAT1-independent gene expression. Considering the synergism observed in the context of the costimulation it is tempting to hypothesize that the non-canonical STAT2/IRF9 pathway acting downstream of the IFNβ/TNFα costimulation differs from the STAT1-independent pathway observed downstream of type I IFN signaling in various contexts. This would imply the existence of heterogeneous STAT2/IRF9 pathways that could result from association with different protein partners or from different regulatory phosphorylation events targeting STAT2. Alternatively, these pathways could be identical, but potentiated by the presence of TNFα. Interestingly, the synergism of IFNγ and TNFα induces CXCL10 and CXCL9 by a “cooperative action” type of synergism. Mechanistic analysis revealed that this synergy is mediated via the enhanced recruitment of the transcriptional coactivator CBP and of the RNA polymerase II to the promoters.55,56 Whether this type of mechanism takes place in the “cooperative induction” synergy between TNFα and IFNβ remains unknown. Further biochemical and molecular studies are required to fine-tune our understanding of the regulation and function of STAT2 and IRF9.
Evidence of the Existence of STAT2/IRF9 Complex(es) Independent on STAT1
In many of the studies cited above, the dependency on STAT2 and IRF9 was not accompanied by the demonstration that both factors were present in a complex. However, several other studies have reported the existence of STAT2 and IRF9 containing complexes independently of STAT1 (Fig. 1). The initial observations of the existence of distinct STAT2 and IRF9 complexes were made concomitantly by the groups of D Levy and N Reich in 1997. Phosphorylated STAT2 was found to be capable of forming homodimers in response to IFNα. These STAT2 homodimers were shown to interact with IRF9 to form an ISGF3-like complex.13,57 Alternative complexes containing STAT6 as a partner of STAT2 and IRF9 have also been described. Formation of ISGF3-like complexes containing phophorylated STAT2, phosphorylated STAT6 and IRF9 were observed in IFNα-stimulated B cells.58 Formation of a STAT2/STAT6 heterodimer was also shown in type I IFN-treated hepatoma cells, but the association with IRF9 in this context has not been evaluated.59 Alternatively, STAT2/STAT3 complexes were shown to form in response to type I IFN in myeloma cells.60 Additional studies have illustrated the capacity of STAT2 and IRF9 to form a complex in the absence of STAT1 without full characterization of the complex composition. In all-trans retinoic acid (ATRA)-treated cells, the autocrine/paracrine action of secreted IFNα mediates the regulation of RIG-G by a STAT2/IRF9-containing complex independently of the STAT2 phosphorylation status.34 The observation that a hybrid protein consisting of IRF9 fused to the C-terminal TAD of STAT2 was targeted to endogenous ISGF3 target loci and was sufficient to recapitulate the type I IFN biological response, inducing an antiviral transcriptional program61 suggest that an IRF9/STAT2 complex is sufficient to mediate specific gene transcription, which is consistent with the existence of the STAT2 homodimer/IRF9 complex.
The first analysis of the capacity of STAT2/IRF9 to bind DNA independently of STAT1 revealed only limited DNA-binding affinity for the typical ISRE sequence targeted by the ISGF3 complex.13 To date, no specific DNA-binding consensus sequence for the non-canonical STAT2/IRF9 complex(es) has been reported. By comparing the IFN-regulated gene expression profile of cells expressing intact STAT2 with that of cells expressing a mutated STAT2 lacking the DNA-binding domain, a subset of genes was found to be selectively regulated by ISGF3-independent, STAT2-containing complexes. Promoter analysis revealed that these genes contain GAS elements, but not ISRE, suggesting that a GAS element could be the target of the STAT2-containing complexes.62 However, direct binding to the GAS element has not been demonstrated. In contrary, chromatin immunoprecipitation analysis using primers specific to ISRE sites confirmed the association of STAT2 with the promoter of antiviral genes induced in response to Dengue virus in STAT1-deficient mice.29 These studies were performed on a restricted number of genes and more comprehensive studies will be required to further characterize promoter elements involved in STAT2-dependent, STAT1-independent gene regulation. In this line, attempts to identify enrichment in specific transcription factor binding sites in the promoter of genes specifically regulated by the IFNβ/TNFα synergism through bioinformatic analysis failed.46 However, as discussed above, it is not yet clear that the non-canonical STAT2/IRF9 pathway is responsible for all IFNβ/TNFα-dependent gene expression. Nevertheless, the possibility that STAT2/IRF9 uses a yet uncharacterized consensus responsive element cannot be excluded.
Concluding Remarks
From the aforementioned studies, it is obvious that the role of STAT2 and IRF9 in the regulation of specific transcriptional programs is not restricted to their involvement in the classical ISGF3 complex. Whether it is in the context of alternative ISGF3-like complexes formed from distinct pools of phosphorylated STAT1 or STAT2, or whether it is in the context of alternative complexes that do not contain STAT1, the specific mechanisms allowing the segregation of STAT2 and IRF9 in distinct complexes are awaiting discoveries. In particular, it will be of high interest to depict the upstream mechanisms allowing IFNβ and TNFα to synergize. Further biochemical analyses of the components of the various complexes will also be required to pave the way to the characterization of their regulatory mechanisms and their transcriptional specificity in conjunction with a particular DNA target sites. Cell-specific mechanisms might also need to be considered in this quest and might constitute the biggest challenge. For instance, it will be easier to isolate STAT2/IRF9 complexes from cell with a STAT1-deficient background. While several cell types might be available, although to a limited extent, from knockout mice, it will be more difficult to address these same questions in human cells. Cells derived from 2fTGH fibrosarcoma cells and lacking the different STATs have proven to be very useful to study these mechanisms, but they do not allow identifying complexes that are cell-type specific. The use of transcription activator-like effector nucleases (TALEN), zinc finger nucleases (ZFNs), or clustered regulatory interspaced short palindromic repeat (CRISPR)/Cas approaches for targeted gene disruption in cultured human cells63 might represent powerful strategy to redefine the boundaries of the experimental design required to achieve cell-specific studies.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Dr N Arbour for critical reading of the manuscript. The work in N Grandvaux laboratory is funded by the Canadian Institutes of Health Research (CIHR) and by the Natural Sciences and Engineering Research Council of Canada (NSERC) to N Grandvaux. K Fink was recipient of studentships from CIHR and N Grandvaux was a recipient of Tier II Canada Research Chairs.
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/27521
References
- 1.Delgoffe GM, Vignali DA. STAT heterodimers in immunity: A mixed message or a unique signal? JAKSTAT. 2013;2:e23060. doi: 10.4161/jkst.23060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207:2053–63. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 4.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 5.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–8. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 6.Wesoly J, Szweykowska-Kulinska Z, Bluyssen HA. STAT activation and differential complex formation dictate selectivity of interferon responses. Acta Biochim Pol. 2007;54:27–38. [PubMed] [Google Scholar]
- 7.Au-Yeung N, Mandhana R, Horvath CM. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT. 2013;2:e23931. doi: 10.4161/jkst.23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darnell JE, Jr., Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–21. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 9.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–25. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy DE, Kessler DS, Pine R, Darnell JE., Jr. Cytoplasmic activation of ISGF3, the positive regulator of interferon-alpha-stimulated transcription, reconstituted in vitro. Genes Dev. 1989;3:1362–71. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- 11.Veals SA, Santa Maria T, Levy DE. Two domains of ISGF3 gamma that mediate protein-DNA and protein-protein interactions during transcription factor assembly contribute to DNA-binding specificity. Mol Cell Biol. 1993;13:196–206. doi: 10.1128/mcb.13.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qureshi SA, Salditt-Georgieff M, Darnell JE., Jr. Tyrosine-phosphorylated Stat1 and Stat2 plus a 48-kDa protein all contact DNA in forming interferon-stimulated-gene factor 3. Proc Natl Acad Sci U S A. 1995;92:3829–33. doi: 10.1073/pnas.92.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bluyssen HA, Levy DE. Stat2 is a transcriptional activator that requires sequence-specific contacts provided by stat1 and p48 for stable interaction with DNA. J Biol Chem. 1997;272:4600–5. doi: 10.1074/jbc.272.7.4600. [DOI] [PubMed] [Google Scholar]
- 14.Wen Z, Zhong Z, Darnell JE., Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–50. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 15.Pilz A, Ramsauer K, Heidari H, Leitges M, Kovarik P, Decker T. Phosphorylation of the Stat1 transactivating domain is required for the response to type I interferons. EMBO Rep. 2003;4:368–73. doi: 10.1038/sj.embor.embor802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steen HC, Nogusa S, Thapa RJ, Basagoudanavar SH, Gill AL, Merali S, Barrero CA, Balachandran S, Gamero AM. Identification of STAT2 serine 287 as a novel regulatory phosphorylation site in type I interferon-induced cellular responses. J Biol Chem. 2013;288:747–58. doi: 10.1074/jbc.M112.402529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tenoever BR, Ng SL, Chua MA, McWhirter SM, García-Sastre A, Maniatis T. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science. 2007;315:1274–8. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- 18.Perwitasari O, Cho H, Diamond MS, Gale M., Jr. Inhibitor of κB kinase epsilon (IKK(epsilon)), STAT1, and IFIT2 proteins define novel innate immune effector pathway against West Nile virus infection. J Biol Chem. 2011;286:44412–23. doi: 10.1074/jbc.M111.285205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumoto M, Tanaka N, Harada H, Kimura T, Yokochi T, Kitagawa M, Schindler C, Taniguchi T. Activation of the transcription factor ISGF3 by interferon-gamma. Biol Chem. 1999;380:699–703. doi: 10.1515/BC.1999.087. [DOI] [PubMed] [Google Scholar]
- 20.Zimmermann A, Trilling M, Wagner M, Wilborn M, Bubic I, Jonjic S, Koszinowski U, Hengel H. A cytomegaloviral protein reveals a dual role for STAT2 in IFN-gamma signaling and antiviral responses. J Exp Med. 2005;201:1543–53. doi: 10.1084/jem.20041401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrow AN, Schmeisser H, Tsuno T, Zoon KC. A novel role for IFN-stimulated gene factor 3II in IFN-γ signaling and induction of antiviral activity in human cells. J Immunol. 2011;186:1685–93. doi: 10.4049/jimmunol.1001359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura T, Kadokawa Y, Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Tarutani M, Tan RS, Takasugi T, Matsuyama T, et al. Essential and non-redundant roles of p48 (ISGF3 gamma) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells. 1996;1:115–24. doi: 10.1046/j.1365-2443.1996.08008.x. [DOI] [PubMed] [Google Scholar]
- 23.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–50. doi: 10.1016/S0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 24.Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–42. doi: 10.1016/S0092-8674(00)81288-X. [DOI] [PubMed] [Google Scholar]
- 25.Hogan RJ, Gao G, Rowe T, Bell P, Flieder D, Paragas J, Kobinger GP, Wivel NA, Crystal RG, Boyer J, et al. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J Virol. 2004;78:11416–21. doi: 10.1128/JVI.78.20.11416-11421.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofer MJ, Li W, Manders P, Terry R, Lim SL, King NJ, Campbell IL. Mice deficient in STAT1 but not STAT2 or IRF9 develop a lethal CD4+ T-cell-mediated disease following infection with lymphocytic choriomeningitis virus. J Virol. 2012;86:6932–46. doi: 10.1128/JVI.07147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13:795–804. doi: 10.1016/S1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 28.Shresta S, Sharar KL, Prigozhin DM, Snider HM, Beatty PR, Harris E. Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. J Immunol. 2005;175:3946–54. doi: 10.4049/jimmunol.175.6.3946. [DOI] [PubMed] [Google Scholar]
- 29.Perry ST, Buck MD, Lada SM, Schindler C, Shresta S. STAT2 mediates innate immunity to Dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog. 2011;7:e1001297. doi: 10.1371/journal.ppat.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahm B, Trifilo MJ, Zuniga EI, Oldstone MB. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity. 2005;22:247–57. doi: 10.1016/j.immuni.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Ousman SS, Wang J, Campbell IL. Differential regulation of interferon regulatory factor (IRF)-7 and IRF-9 gene expression in the central nervous system during viral infection. J Virol. 2005;79:7514–27. doi: 10.1128/JVI.79.12.7514-7527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.George CX, Das S, Samuel CE. Organization of the mouse RNA-specific adenosine deaminase Adar1 gene 5′-region and demonstration of STAT1-independent, STAT2-dependent transcriptional activation by interferon. Virology. 2008;380:338–43. doi: 10.1016/j.virol.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarkis PT, Ying S, Xu R, Yu XF. STAT1-independent cell type-specific regulation of antiviral APOBEC3G by IFN-alpha. J Immunol. 2006;177:4530–40. doi: 10.4049/jimmunol.177.7.4530. [DOI] [PubMed] [Google Scholar]
- 34.Lou YJ, Pan XR, Jia PM, Li D, Xiao S, Zhang ZL, Chen SJ, Chen Z, Tong JH. IRF-9/STAT2 [corrected] functional interaction drives retinoic acid-induced gene G expression independently of STAT1. Cancer Res. 2009;69:3673–80. doi: 10.1158/0008-5472.CAN-08-4922. [DOI] [PubMed] [Google Scholar]
- 35.Kawakami T, Matsumoto M, Sato M, Harada H, Taniguchi T, Kitagawa M. Possible involvement of the transcription factor ISGF3 gamma in virus-induced expression of the IFN-beta gene. FEBS Lett. 1995;358:225–9. doi: 10.1016/0014-5793(94)01426-2. [DOI] [PubMed] [Google Scholar]
- 36.Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Kimura T, Kitagawa M, Yokochi T, Tan RS, Takasugi T, Kadokawa Y, et al. Regulation of IFN-alpha/beta genes: evidence for a dual function of the transcription factor complex ISGF3 in the production and action of IFN-alpha/beta. Genes Cells. 1996;1:995–1005. doi: 10.1046/j.1365-2443.1996.870287.x. [DOI] [PubMed] [Google Scholar]
- 37.Majumder S, Zhou L, Chaturvedi P, Babcock G, Aras S, Ransohoff RM. p48/STAT-1alpha-containing complexes play a predominant role in induction of IFN-gamma-inducible protein, 10 kDa (IP-10) by IFN-gamma alone or in synergy with TNF-alpha. J Immunol. 1998;161:4736–44. [PubMed] [Google Scholar]
- 38.Luker KE, Pica CM, Schreiber RD, Piwnica-Worms D. Overexpression of IRF9 confers resistance to antimicrotubule agents in breast cancer cells. Cancer Res. 2001;61:6540–7. [PubMed] [Google Scholar]
- 39.Peng T, Zhu J, Hwangbo Y, Corey L, Bumgarner RE. Independent and cooperative antiviral actions of beta interferon and gamma interferon against herpes simplex virus replication in primary human fibroblasts. J Virol. 2008;82:1934–45. doi: 10.1128/JVI.01649-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mestan J, Brockhaus M, Kirchner H, Jacobsen H. Antiviral activity of tumour necrosis factor. Synergism with interferons and induction of oligo-2′,5′-adenylate synthetase. J Gen Virol. 1988;69:3113–20. doi: 10.1099/0022-1317-69-12-3113. [DOI] [PubMed] [Google Scholar]
- 41.Robinson CM, Shirey KA, Carlin JM. Synergistic transcriptional activation of indoleamine dioxygenase by IFN-gamma and tumor necrosis factor-alpha. J Interferon Cytokine Res. 2003;23:413–21. doi: 10.1089/107999003322277829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Konan KV, Taylor MW. Importance of the two interferon-stimulated response element (ISRE) sequences in the regulation of the human indoleamine 2,3-dioxygenase gene. J Biol Chem. 1996;271:19140–5. doi: 10.1074/jbc.271.32.19140. [DOI] [PubMed] [Google Scholar]
- 43.Ohmori Y, Schreiber RD, Hamilton TA. Synergy between interferon-gamma and tumor necrosis factor-alpha in transcriptional activation is mediated by cooperation between signal transducer and activator of transcription 1 and nuclear factor kappaB. J Biol Chem. 1997;272:14899–907. doi: 10.1074/jbc.272.23.14899. [DOI] [PubMed] [Google Scholar]
- 44.Bartee E, Mohamed MR, McFadden G. Tumor necrosis factor and interferon: cytokines in harmony. Curr Opin Microbiol. 2008;11:378–83. doi: 10.1016/j.mib.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yarilina A, Park-Min KH, Antoniv T, Hu X, Ivashkiv LB. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat Immunol. 2008;9:378–87. doi: 10.1038/ni1576. [DOI] [PubMed] [Google Scholar]
- 46.Bartee E, Mohamed MR, Lopez MC, Baker HV, McFadden G. The addition of tumor necrosis factor plus beta interferon induces a novel synergistic antiviral state against poxviruses in primary human fibroblasts. J Virol. 2009;83:498–511. doi: 10.1128/JVI.01376-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fink K, Martin L, Mukawera E, Chartier S, De Deken X, Brochiero E, Miot F, Grandvaux N. IFNβ/TNFα synergism induces a non-canonical STAT2/IRF9-dependent pathway triggering a novel DUOX2 NADPH oxidase-mediated airway antiviral response. Cell Res. 2013;23:673–90. doi: 10.1038/cr.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reis LF, Ho Lee T, Vilcek J. Tumor necrosis factor acts synergistically with autocrine interferon-beta and increases interferon-beta mRNA levels in human fibroblasts. J Biol Chem. 1989;264:16351–4. [PubMed] [Google Scholar]
- 49.Tliba O, Tliba S, Da Huang C, Hoffman RK, DeLong P, Panettieri RA, Jr., Amrani Y. Tumor necrosis factor alpha modulates airway smooth muscle function via the autocrine action of interferon beta. J Biol Chem. 2003;278:50615–23. doi: 10.1074/jbc.M303680200. [DOI] [PubMed] [Google Scholar]
- 50.Kimura A, Naka T, Nagata S, Kawase I, Kishimoto T. SOCS-1 suppresses TNF-alpha-induced apoptosis through the regulation of Jak activation. Int Immunol. 2004;16:991–9. doi: 10.1093/intimm/dxh102. [DOI] [PubMed] [Google Scholar]
- 51.Guo D, Dunbar JD, Yang CH, Pfeffer LM, Donner DB. Induction of Jak/STAT signaling by activation of the type 1 TNF receptor. J Immunol. 1998;160:2742–50. [PubMed] [Google Scholar]
- 52.Miscia S, Marchisio M, Grilli A, Di Valerio V, Centurione L, Sabatino G, Garaci F, Zauli G, Bonvini E, Di Baldassarre A. Tumor necrosis factor alpha (TNF-alpha) activates Jak1/Stat3-Stat5B signaling through TNFR-1 in human B cells. Cell Growth Differ. 2002;13:13–8. [PubMed] [Google Scholar]
- 53.Cheshire JL, Baldwin AS., Jr. Synergistic activation of NF-kappaB by tumor necrosis factor alpha and gamma interferon via enhanced I kappaB alpha degradation and de novo I kappaBbeta degradation. Mol Cell Biol. 1997;17:6746–54. doi: 10.1128/mcb.17.11.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Wu TR, Cai S, Welte T, Chin YE. Stat1 as a component of tumor necrosis factor alpha receptor 1-TRADD signaling complex to inhibit NF-kappaB activation. Mol Cell Biol. 2000;20:4505–12. doi: 10.1128/MCB.20.13.4505-4512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clarke DL, Clifford RL, Jindarat S, Proud D, Pang L, Belvisi M, Knox AJ. TNFα and IFNγ synergistically enhance transcriptional activation of CXCL10 in human airway smooth muscle cells via STAT-1, NF-κB, and the transcriptional coactivator CREB-binding protein. J Biol Chem. 2010;285:29101–10. doi: 10.1074/jbc.M109.0999952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hiroi M, Ohmori Y. The transcriptional coactivator CREB-binding protein cooperates with STAT1 and NF-kappa B for synergistic transcriptional activation of the CXC ligand 9/monokine induced by interferon-gamma gene. J Biol Chem. 2003;278:651–60. doi: 10.1074/jbc.M204544200. [DOI] [PubMed] [Google Scholar]
- 57.Martinez-Moczygemba M, Gutch MJ, French DL, Reich NC. Distinct STAT structure promotes interaction of STAT2 with the p48 subunit of the interferon-alpha-stimulated transcription factor ISGF3. J Biol Chem. 1997;272:20070–6. doi: 10.1074/jbc.272.32.20070. [DOI] [PubMed] [Google Scholar]
- 58.Gupta S, Jiang M, Pernis AB. IFN-alpha activates Stat6 and leads to the formation of Stat2:Stat6 complexes in B cells. J Immunol. 1999;163:3834–41. [PubMed] [Google Scholar]
- 59.Wan L, Lin CW, Lin YJ, Sheu JJ, Chen BH, Liao CC, Tsai Y, Lin WY, Lai CH, Tsai FJ. Type I IFN induced IL1-Ra expression in hepatocytes is mediated by activating STAT6 through the formation of STAT2: STAT6 heterodimer. J Cell Mol Med. 2008;12:876–88. doi: 10.1111/j.1582-4934.2008.00143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghislain JJ, Wong T, Nguyen M, Fish EN. The interferon-inducible Stat2:Stat1 heterodimer preferentially binds in vitro to a consensus element found in the promoters of a subset of interferon-stimulated genes. J Interferon Cytokine Res. 2001;21:379–88. doi: 10.1089/107999001750277853. [DOI] [PubMed] [Google Scholar]
- 61.Kraus TA, Lau JF, Parisien JP, Horvath CM. A hybrid IRF9-STAT2 protein recapitulates interferon-stimulated gene expression and antiviral response. J Biol Chem. 2003;278:13033–8. doi: 10.1074/jbc.M212972200. [DOI] [PubMed] [Google Scholar]
- 62.Brierley MM, Marchington KL, Jurisica I, Fish EN. Identification of GAS-dependent interferon-sensitive target genes whose transcription is STAT2-dependent but ISGF3-independent. FEBS J. 2006;273:1569–81. doi: 10.1111/j.1742-4658.2006.05176.x. [DOI] [PubMed] [Google Scholar]
- 63.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]