The tumor suppressor gene TP53 and its gene product—a veritable Swiss Army knife of cellular regulation—have been the targets of intense study since their discovery in 1979.1-3 Indeed, a PubMed search for the term “p53” retrieves more than 68 000 citations, and more than 100 physical or genetic interactions with TP53 have been identified to date. Its importance in cancer biology was recognized immediately upon its discovery, and Science honored p53 as the “Molecule of the Year” some 20 y ago.
Despite the intensity of this research, spanning more than 3 decades, the mechanisms by which p53 coordinates diverse cellular functions remain enigmatic. A study by DeMicco and colleagues sheds new light on the tumor suppressor roles of p53, showing that it performs different functions depending on its cellular and developmental context.4
The canonical role of TP53 is as a tumor suppressor gene that responds to DNA damage. However, diverse roles for TP53 in coordinating cellular response to a range of stresses have been discovered. It is now known that TP53 responds to oxidative stress and oxidized DNA; monitors and regulates aspects of cellular metabolism; governs proper chromosome segregation; controls cell cycle arrest in response to cellular stressors; induces apoptosis; and regulates autophagy and senescence.5
Reflecting its key role in coordinating stress responses, TP53 is the single most frequently mutated gene in human cancer, with partial or complete loss of function occurring in 60% of tumors.6,7 This observation, together with a multitude of studies in model systems, has demonstrated a role for TP53 in tumor suppression in a host of different cell and tissue types.
DeMicco et al. have used an elegant mouse modeling approach to begin unraveling the key tumor suppressor functions of TP53 in T-cell lymphomas.4 The authors use the power of conditional TP53 deletion to inactivate it at varying stages of T-cell development and to characterize the subsequent tumors. Using mice containing a Vav–Cre transgene they induce TP53 inactivation in hematopoietic stem cells (HSC); and using an Lck-Cre transgene, they induce TP53 inactivation in CD4− CD8− double-negative (DN) immature T cells. Consistent with the phenotype of mice with germline TP53 deletion (TP53−/−), they find that inactivation of TP53 in HSC strongly predisposes to T-cell lymphomagenesis, as does inactivation in immature DN thymocytes. However, they note that TP53 deletion in HSC skews the mice toward lymphomas with an earlier T-cell developmental signature, suggesting that additional transforming events—occurring after TP53 deletion but prior to early T-cell differentiation—may contribute to transformation susceptibility at early T-cell stages.
Perhaps the most fascinating finding of this study is the effect of developmental context on the karyotypic features of the T-cell lymphomas that arise after TP53 inactivation. The very well-characterized T-cell lymphomas in TP53−/− mice almost uniformly lack chromosomal translocations, but commonly exhibit anueploidy. DeMicco and colleagues now show that inactivation of TP53 in either HSC or DN thymocytes leads to T-cell tumors with clonal translocations. Interestingly, the patterns of translocations observed differed depending on the stage at which TP53 was deleted. Inactivation in HSC led to lymphomas with translocations that generally did not involve antigen receptor genes, suggesting that they originated prior to T-cell receptor rearrangement. Conversely, inactivation in DN thymocytes led to lymphomas with translocations involving the Tcrα/δ genes. While the contribution of Cre activity in these model systems cannot be fully ruled out, the authors compellingly argue that Cre-mediated DNA damage is unlikely to be a significant component in patterning the translocations.
Although the mechanistic basis for this peculiar shift in chromosomal instability profiles is not yet known, these findings clearly highlight the fact that developmental context of TP53 activity—or its inactivation—is a critical facet of its tumor-suppressive functions.
The diverse range of TP53 functions in cellular homeostastis and tumor suppression remain mysterious. It has become clear over the years that the predominant functions of TP53 can vary by tissue type. The new study by DeMicco et al. now convinces us that the roles of TP53 can also vary by developmental stage within a single tissue type. This adds to our understanding—and raises new questions—regarding the true functions of one of the most studied, yet least understood, tumor suppressor genes. (Fig. 1)
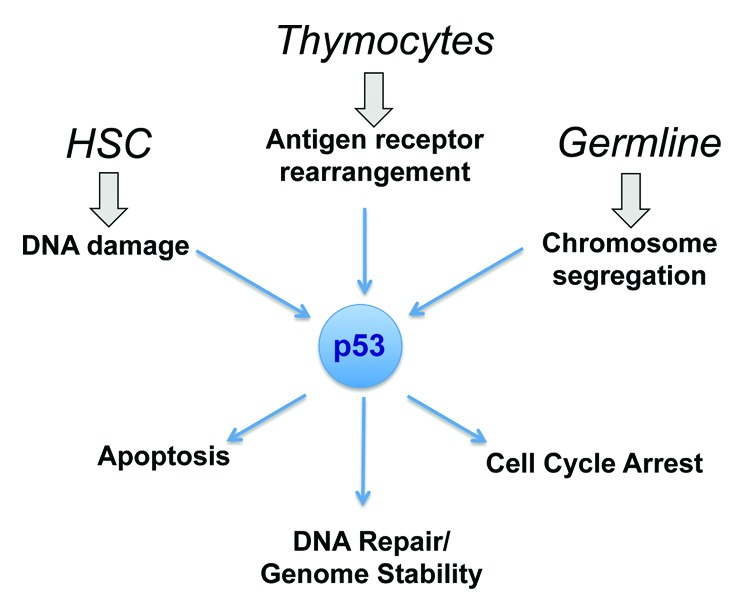
Figure 1. Context dependent integration of p53 tumor suppressor activities. The study by DeMicco and colleagues shows that the role that p53 plays in suppressing neoplastic transformation can vary by developmental stage within a single cell lineage.4 Their data show that inactivation of TP53 in HSC results in tumors with translocations that do not involve antigen receptors, while inactivation in DN thymocytes predisposes to lymphoma with antigen receptor translocations. These lymphomas contrast with the aneuploid tumors that arise after germline inactivation of TP53. This highlights the developmental context dependency of p53 functions.
DeMicco A, et al. Cell Cycle. 2013;12:3307–16. doi: 10.4161/cc.26299.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26806
References
- 1.Crawford LV, et al. Proc Natl Acad Sci U S A. 1981;78:41–5. doi: 10.1073/pnas.78.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeLeo AB, et al. Proc Natl Acad Sci U S A. 1979;76:2420–4. doi: 10.1073/pnas.76.5.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linzer DI, et al. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 4.Demicco A, et al. Cell Cycle. 2013;12 doi: 10.4161/cc.26299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nag S, et al. J Biomed Res. 2013;27:254–71. doi: 10.7555/JBR.27.20130030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung KJ, et al. Br J Haematol. 2009;146:257–69. doi: 10.1111/j.1365-2141.2009.07739.x. [DOI] [PubMed] [Google Scholar]
- 7.Meek DW. Nat Rev Cancer. 2009;9:714–23. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]


