Abstract
Emerging evidence suggests that metformin, a widely used anti-diabetic drug, may be useful in the prevention and treatment of different cancers. In the present study, we demonstrate that metformin directly inhibits the enzymatic function of hexokinase (HK) I and II in a cell line of triple-negative breast cancer (MDA-MB-231). The inhibition is selective for these isoforms, as documented by experiments with purified HK I and II as well as with cell lysates. Measurements of 18F-fluoro-deoxyglycose uptake document that it is dose- and time-dependent and powerful enough to virtually abolish glucose consumption despite unchanged availability of membrane glucose transporters. The profound energetic imbalance activates phosphorylation and is subsequently followed by cell death. More importantly, the “in vivo” relevance of this effect is confirmed by studies of orthotopic xenografts of MDA-MB-231 cells in athymic (nu/nu) mice. Administration of high drug doses after tumor development caused an evident tumor necrosis in a time as short as 48 h. On the other hand, 1 mo metformin treatment markedly reduced cancer glucose consumption and growth. Taken together, our results strongly suggest that HK inhibition contributes to metformin therapeutic and preventive potential in breast cancer.
Keywords: breast cancer, metformin, hexokinases, glucose metabolism, orthotopic xenografts, in vivo imaging
Introduction
Metformin, the drug most widely used to treat type 2 diabetes, recently emerged as a potential anticancer agent. Epidemiological, clinical and preclinical evidence supports its use in the treatment of many tumors and particularly of breast cancer.1-7 This potential has been first attributed to indirect mechanisms characterized by the drug capability to reduce serum levels of glucose, insulin, and insulin-like growth factor.8,9 However, experimental evidence pointed out that it can also directly reduce cancer growth by triggering phosphorylation of 5′ AMP-activated protein kinase (AMPk).10-12
How metformin activates this energy sensor pathway is not fully understood. Actually, it selectively inhibits respiratory chain complex I, resulting in ATP depletion.13,14 However, the consequences of mitochondrial impairment should be of limited relevance in cancer in which the fraction of ATP produced by glycolysis is more than 50 times higher than in normal tissues.15 Moreover, while p-AMPK activation should increase glucose consumption,16 studies in mouse models of colon cancer reported that metformin can decrease uptake of 18F-fluorodeoxyglycose (FDG).17 Finally, the primary role of mitochondrial action was observed in hepatocytes and β cells of pancreatic islets characterized by a peculiar glucose processing machinery with a preferential expression of hexokinase (HK) IV.18
Recently, we demonstrated that metformin impairs cancer energy asset in vitro via a direct and selective enzymatic inhibition of HK isoforms I and II.19 This observation might thus indicate that metformin might directly hamper a pivotal aspect of cancer biology and hinder cell energy asset beyond the consequences of mitochondrial impairment.18,20,21 Accordingly, we hypothesized that HK I and II inhibition by metformin could modify glucose metabolism in triple-negative breast cancer both in cultured cells and xenograft models. Obtained data confirm this hypothesis, documenting that the drug actually inhibits HK function, resulting in an immediate cytotoxic effect both in vitro and in vivo as well as in a significant reduction of cancer growth rate under chronic treatment.
Results
HKI and HKII as molecular targets of metformin effect on cancer metabolism and biology
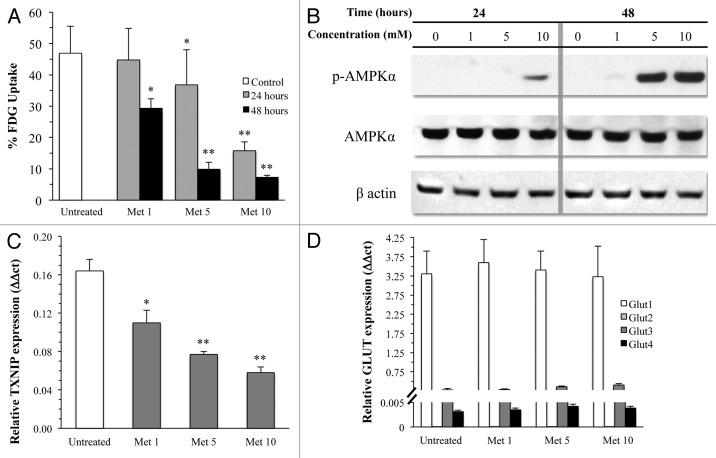
Metformin strikingly impaired glucose consumption of MDA-MB-231 in a dose- and time-dependent manner. Maximal effect occurred with exposure to 10 mM drug concentration that progressively reduced FDG uptake down to its minimum values after 48 h (Fig. 1A). The reduction in glycolytic rate throughout the whole 48 h experiment duration was paralleled by a progressive increase in AMPK phosphorylation and by a progressive reduction (Fig. 1B) in TXNIP gene expression22 (Fig. 1C).
Figure 1. (A) FDG uptake in cultured cells under control conditions (white columns) or after 24 (gray columns) and 48 (black columns) hours exposure to different concentrations of metformin. The drug caused a dose- and time-dependent reduction in tracer retention and thus in glucose consumption (*P < 0.05, ** P < 0.01 vs. control, respectively). This metabolic response was associated with a dose-dependent decrease in TXNIP gene expression (B), ruling out a possible accumulation of G6P caused by a block in downstream glycolytic chain. This metabolic derangement resulted in an activation of the energy sensor pathway leading to an increased phosphorylation of AMPK at higher drug doses. On the other hand, levels of mRNA encoding for the different GLUT carriers (D) did not report any effect on glucose transport system characterized by a high expression of GLUT1 that was not altered by metformin treatment. This neutral response was duplicated when the markedly lower expression of GLUT 2–4 were tested.
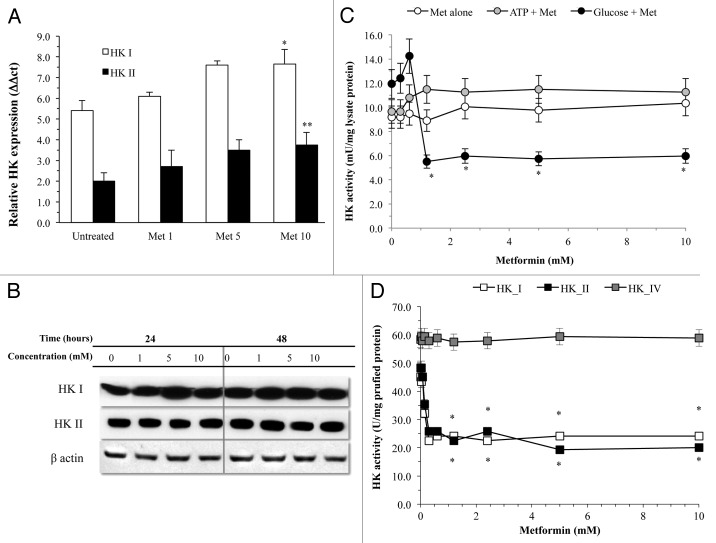
In agreement with the malignant phenotype,23,24 MDA-MB-231 expression profile for glucose carriers was limited to mRNA encoding for GLUT1, with a low prevalence of GLUT3 and the virtual absence of GLUT2 and GLUT4 (Fig. 1D). Metformin did not affect this pattern, thus excluding drug effects on trans-membrane glucose transport (Fig. 1D). A similar finding was obtained for HK expression profile. As expected,25-27 this cell line mostly expressed HKI and HKII, while isoforms III and IV were almost undetectable (Fig. 2A). Treatment slightly increased gene expression for HKI and II (Fig. 2A), ruling out any effect on enzyme availability, as also confirmed by western blot analysis (Fig. 2B).
Figure 2. HK response to metformin. Biguanide treatment did not reduce expression for isoforms I and II of HK that, rather, showed a slight but significant increase at the highest drug level. Similarly, protein availability of these enzymes was not modified by treatment even at prolonged times (B). (C) displays glucose phosphorylating activity of MDA-MB-231 cell lysate that was not altered by incubation with metformin alone (white circles) nor by metformin and ATP (black circles). On the contrary, it was almost halved by the incubation with metformin and glucose, confirming preliminary experience in CALU1 cells derived by non-small cell lung carcinoma (*P < 0.05 vs. control condition). (D) displays the selectivity of metformin interference purified human HK isoform I (white squares) and II (black squares) with absent response by HK IV (gray squares) (*P < 0.05 vs. control condition).
On the contrary, metformin inhibited glucose-phosphorylating activity of cell lysates in a dose-dependent fashion (Fig. 2C). In agreement with our previous experience,19 experiments with purified proteins showed that the interference selectively affected enzymatic function of HK I and II; it did not modify activity of HK IV and requested the presence of glucose during incubation (Fig. 2D).
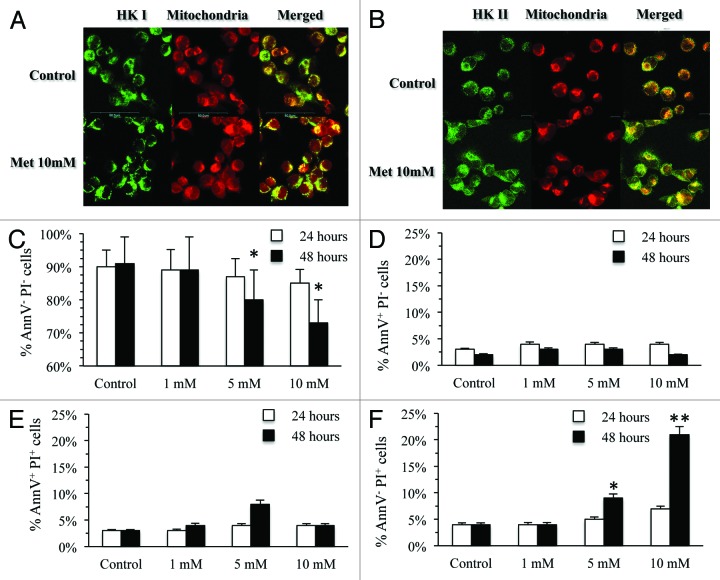
At immunofluorescence analysis, metformin exposure was associated with a preferential cytosolic localization of isoform II (Fig. 3A and B) that preceded the expected cell damage. Apoptosis, as evidenced by annexin V, was already detectable after 24 h of exposure to a low dose of metformin (1 mM) and remained relatively stable regardless drug concentrations or exposure time (Fig. 3D and E). Cell death was evident only after incubation with metformin 10 mM for 24 h and significantly increased after 48 h of treatment (Fig. 3F). Accordingly, the metabolic alteration preceded cell death and largely exceeded its degree, with the virtual abolition of glucose consumption at 48 h, facing a reduction in the number living cells of only 20%.
Figure 3. (A and B) Display the confocal microscopy for HK I and HK II, respectively, and mitochondria in MDA-MB-231 cells untreated and after 24 h incubation with metformin 10 mM. Mitochondria were labeled by MitoTracker Far Red; HKI and II were stained by indirect immunofluorescence, using a FITC-conjugated secondary antibody. Left, right, and central panels show staining for HKI/II, mitochondria and both, respectively. Merged images document that metformin causes a significant and selective dislocation of HK II isoform from mitochondrial membrane to the cytosol. (C) displays effect of different metformin concentrations cell viability at 24 (white columns) and 48 h (black columns). Number of AnnV- PI- signficantly decreased only at the highest dose and at 48 h (*P < 0.05): Number of cells in early (AnnV+ PI−, D) or late (AnnV+ PI+, E) apoptosis remained relatively low throughout the whole study period. On the contrary, number of dead cells (AnnV− PI+, F) progressively increased for highest doses at 48 h of exposure (**P < 0.01).
Metformin effect on glucose metabolism of cancer and whole body
In vivo experiments confirmed the relevance of the metabolic effect of metformin on cancer metabolism and growth rate. The experiment was completed in all animals, and no side effects occurred at the drug dosage used. As shown in Table 1, body weight was not significantly different in the 3 groups of animals at the time of imaging. On the contrary, serum glucose level was left unaltered by acute (48 h) treatment, while it was slightly, though not significantly, reduced in animals treated for the whole experiment duration (Table 1).
Table 1. Metabolic findings.
| Untreated | Pulsed | Prolonged | |||||||
|---|---|---|---|---|---|---|---|---|---|
| (n = 10) | (n = 10) | (n = 10) | |||||||
| Whole body | |||||||||
| Animal weight (grams) | 24.21 | ± | 2.12 | 23.48 | ± | 2.61 | 22.89 | ± | 2.4 |
| Serum glucose level (mM L−1) | 7.1 | ± | 2.22 | 7.5 | ± | 1.72 | 6.93 | ± | 2.34 |
| Whole body FDG clearance (mL X min−1) | 0.24 | ± | 0.08 | 0.27 | ± | 0.10 | 0.14 | ± | 0.07* |
| Whole body glucose consumption (µM X min−1) | 1.73 | ± | 0.93 | 1.78 | ± | 0.78 | 1.22 | ± | 0.23* |
| Cancer lesion | |||||||||
| Animals with palpable tumor | 10 | 9 | 7 | ||||||
| Tumor weight (mg) | 366 | ± | 133 | 353 | ± | 146 | 233 | ± | 105* |
| Tumor volume at PET (µL) | 306 | ± | 93 | 306 | ± | 125 | 187 | ± | 89* |
| Tumor glucose consumption (nM X min−1 X g−1) | 79.5 | ± | 14 | 74.9 | ± | 29 | 41.64 | ± | 5.5* |
| Total lesion glucose consumption (nM) | 24.33 | ± | 4.87 | 22.92 | ± | 5.21 | 7.79 | ± | 1.87 |
| FDG uptake (SUV mean) | 1.16 | ± | 0.09 | 1.14 | ± | 0.08 | 1.09 | ± | 0.04 |
| FDG uptake (SUV max) | 2.32 | ± | 0.48 | 2.26 | ± | 0.76 | 2.11 | ± | 0.71 |
More importantly, metformin treatment had a relevant influence on cancer growth. In fact, weight of explanted lesions was almost halved in “prolonged” mice treated with metformin for the whole month of study duration, while it was obviously not affected by pulsed treatment (Table 1).
In vivo imaging confirmed the drug effect on tumor progression. In fact, analysis of PET visible lesions reproduced the data obtained by evaluation of cancer weight as tumor volume was markedly lower in “prolonged” animals than in the remaining 2 groups (Table 1).
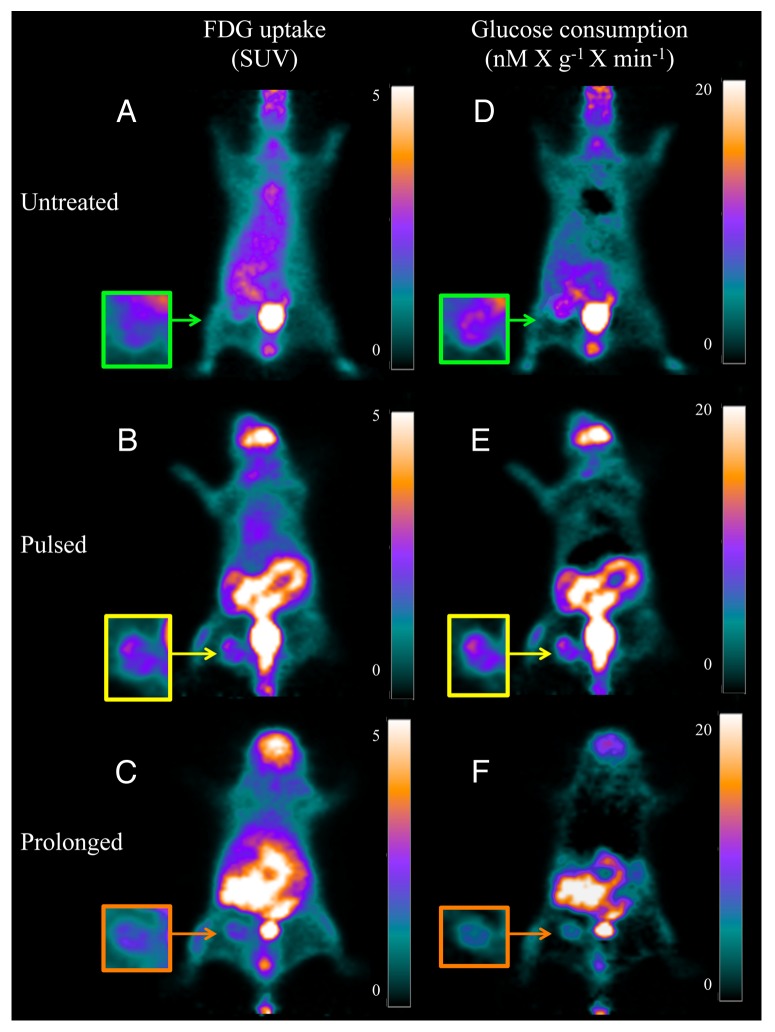
Actually, analysis of FDG uptake did not show significant differences in both average and maximal SUV within cancer lesions (Table 1 and Fig. 4A–C). However, this finding was intriguingly contradicted by the quantitative analysis of tumor glucose metabolism. In fact, glucose consumption was 79.5 ± 14 and 74.9 ± 29 nM × min−1 × g−1 in untreated and pulsed mice, respectively (P = ns) (Table 1 and Fig. 4D and E). By contrast, prolonged metformin administration markedly decreased cancer glucose utilization to 41.6 ± 5.5 nM × min−1 × g−1 (P < 0.01 vs. untreated and pulsed animals, Table 1 and Fig. 4D–F).
Figure 4. Coronal slices and enlarged tumor image of PET/FDG scans. (A–C) display the FDG uptake, documented in the last acquisition frame (from 40–50 min after injection), reproducing the usual representation of PET used in the clinical setting. Cancer uptake is visible and similar in all animals, regardless treatment protocol, as documented by the similarity of SUV reported in the color scale on the right side of each panel. (D–F) display the data of the same PET studies represented as glucose consumption (in nanomol × min-1-X g-1) as measured in each voxel by the product between the slope Patlak regression line and serum glucose concentration. This analysis shows reduced glucose consumption in cancer of the mouse treated for the whole study period.
Response of whole-body glucose metabolism explained the apparent disagreement between the 2 different analyses of the same FDG images. In fact, FDG clearance was 0.24 ± 0.05 ml × min−1 in untreated mice and remained relatively stable after 2 days treatment (Table 1), while it markedly fell after 1 month metformin treatment in the prolonged group (0.14 ± 0.07 ml × min−1, P < 0.01 vs. both untreated and pulsed animals). Obviously, this finding was largely reproduced when normalization of glycaemia was applied. In fact, whole-body glucose consumption was similar in untreated and pulsed animals, while it reached the lowest values in those treated for prolonged time (Table 1).
Metabolic correlates of tumor xenografts molecular pathology
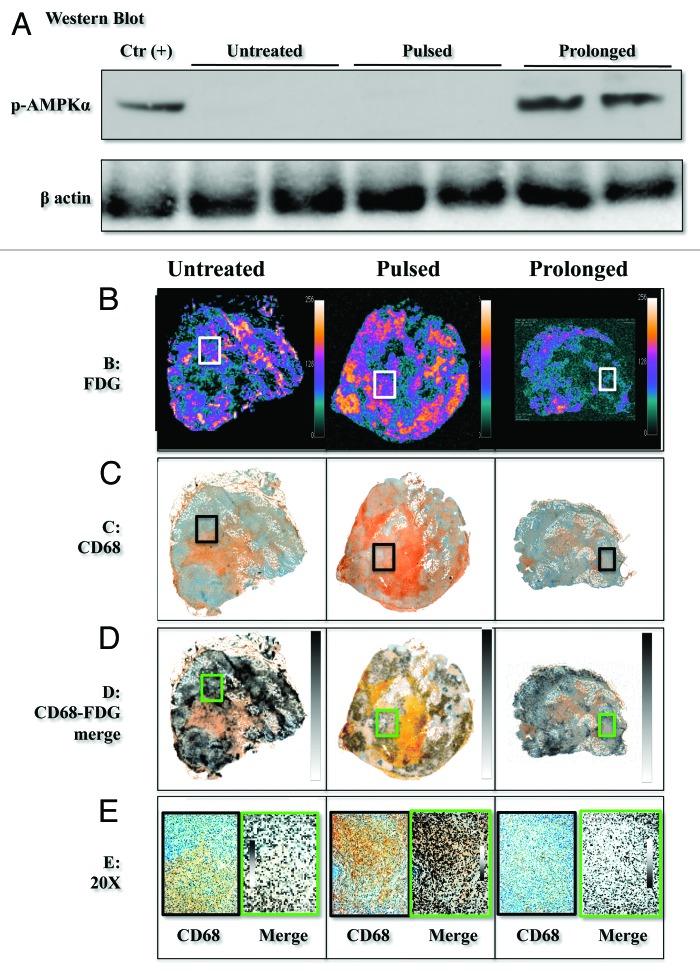
In agreement with in vitro data, treatment did not significantly alter gene expression of GLUT 1 and GLUT 3 or HK I and II in harvested lesions. On the other hand, cancer mRNA levels encoding for TXNIP were slightly, though not significantly, reduced in treated animals, regardless of the duration of metformin exposure, for 48 h or the whole month. On the contrary, AMPK phosphorylation was only activated in tumors harvested from mice submitted to prolonged treatment, while it did not occur after pulsed therapy (Fig. 5A).
Figure 5. Pictures from harvested tumors. (A) displays western blot data showing that increased phosphorylation of AMPK only occurred in animals exposed to prolonged treatment. (B) display a large macrophage infiltration in pulsed group as detected by double staining immunohistochemistry for CD68 antigen (brown) and cytokeratin CK19 (purple). (C) display autoradiography of FDG uptake, while coregistration of metabolic and immunohistochemical data are reported in (D) and a magnified area (20×) in (E). Co-localization of tracer retention and CD68+ cells can be best appreciated after pulsed treatment, while it is less evident in the remaining specimens.
Quantitative analysis of autoradiography confirmed in vivo imaging data: radioactivity content was similar in cancers harvested in pulsed (4401 ± 961 PSL × 103 × square mm−1 × min−1) and untreated mice (4918 ± 568 PSL × 103 × square mm−1 × min−1, P = ns), while prolonged treatment significantly reduced FDG uptake (2510 ± 784 × 103 PSL × square mm−1 × min−1, P < 0.01 vs. all other groups) (Fig. 5B).
At hematoxylin/eosin staining, all cancers showed a visible necrotic core whose extension, however, was peculiar: it was largest in lesions collected in “pulsed” animals (76 ± 8% of total section), intermediate in untreated mice (38 ± 11%, P < 0.01 vs. pulsed), and lowest in prolonged ones (13 ± 5%, P < 0.01 vs. both pulsed and untreated). As expected, this pattern was paralleled by a differential macrophage infiltration: CD68+ cells were largely represented in cancers harvested from pulsed models and almost absent in those exposed to prolonged treatment (Fig. 5C).
Interestingly, coregistration of autoradiography and immuno-histochemistry documented that different cell types accounted for FDG uptake in the different models (Fig. 5D). In particular, a high co-localization of radioactivity and CD68+ positivity was consistently observed in pulsed models (Fig. 5E). On the contrary, tracer distribution and thus glucose consumption was more homogeneously distributed in prolonged animals.
Discussion
The present study documents that metformin reduces cancer metabolism and growth at least partially via a direct and selective inhibition of HK I and II enzymatic function. In vitro, the profound energetic impairment caused by G6P depletion triggered the apoptotic process and eventually resulted in cell death.28,29 In vivo, administration of high drug doses after lesion development caused a large tumor necrosis in a similar interval. By contrast, prolonged treatment (from tumor implantation to harvesting) markedly reduced cancer growth and glucose consumption. Accordingly, these data extend to the xenograft model of triple-negative breast cancer our previous observations in cell cultures of non-small cell lung carcinoma19 and document the relevance of direct action on cancer metabolism in anticancer potential of chronic metformin treatment reported in the clinical setting.1-7
Metformin effect on glucose metabolism
As expected,10,11 high drug doses activated AMPK phosphorylation both in vitro and in vivo. Nevertheless, they also markedly reduced FDG uptake both in cultured cells and in cancers implanted in living animals, indicating that metformin might directly impair glycolytic flux with mechanisms independent from this energy sensor pathway.
As a probe of glucose consumption, FDG uptake enters the cell via the same facilitative transporters of glucose;30-32 it is then phosphorylated to FDG6P and remains trapped within cytosol, being a false substrate for all further reactions channeling G6P to alternative pathways. Obviously, a block in downstream glycolytic chain might reduce tracer retention, since any G6P accumulation would inevitably limit the HK-mediated glucose trapping mechanism. However, this hypothesis was ruled out by the evaluation of TXNIP, whose expression is powerfully triggered by G6P itself:22 the marked and dose-dependent reduction in FDG uptake was paralleled by a decrease in mRNA levels encoding for this protein, indicating a relative reduction in G6P availability.
Treatment did not modify gene expression for GLUT and HK. On the contrary, metformin directly inhibited glucose phosphorylating activity of cell lysates. Experiments with purified enzymes documented that this effect is selective on isoforms I and II and thus on HK asset of MDA-MB-231 cells. Thus, present data confirm that interference with HK I and II at least partially explains metformin potential in cancers characterized by this enzymatic asset,19 being probably less relevant in cells also expressing other HK isoforms.33
The selective nature of metformin activity on HK I and II is further and independently confirmed by immunofluorescence analysis. Actually, both these isoforms are closely placed on the mitochondrial outer membrane.25-27 However, this link is at least partially dependent on G6P for HK II.27 Metformin displaced this isoform away from mitochondrial membrane to the cytosol, limiting its preferential access to ATP for glucose phosphorylation and, thus, hampering a major mechanism of cancer growth and immortality28,34,35 as documented in pioneering experiments by Bustamante et al.36
HK-2 metabolic step can be used for monitoring clinical cancers via PET analysis
MicroPET-derived indexes of cancer glucose consumption were markedly reduced by prolonged drug administration.37 Again, this finding was paralleled by AMPK activation38,39 and eventually resulted in a significant reduction in both weight of harvested lesions and tumor volume at PET analysis. By contrast, and differently from cultured cells, no evident change in cancer glucose metabolism was observed after 48 h therapy in pulsed mice. However, this paradox is explained by the micrometric analysis of FDG uptake offered by autoradiography. In fact, lesion radioactivity was heterogeneously distributed in pulsed animals, with peripheral regions characterized by an intense metabolism surrounding a metabolically inert necrotic core. At immunohistochemistry, peripheral ring displayed a large number of CD68+ cells whose metabolism largely accounted for FDG uptake. This pattern closely agrees with the limitations of PET FDG imaging in the early evaluation of chemotherapy effect, as infiltrating macrophages and newly formed granulation tissue most often show greater FDG uptake than viable tumor cells.40
The reduction in tumor glucose consumption under “chronic” metformin treatment was not paralleled by a corresponding decrease in cancer FDG uptake confirming the acknowledged limitations of clinical evaluation of cancer metabolism. Actually both average and maximal SUV represent the most largely used indexes of lesion glucose consumption, since they can be easily and accurately measured with only one image of late FDG distribution. However, their value is also dependent upon tracer disposal, which, in turn, reflects systemic metabolic regulation.41 In agreement with previous observations,42 chronic metformin treatment reduced whole-body glucose consumption and blood FDG clearance in non-diabetic nude mice. This systemic response delayed FDG removal from blood and thus increased its availability for cancer uptake. Accordingly, this observation indicates that SUV data should be carefully interpreted, particularly in experimental models implying the use of drugs active on whole body metabolic pattern. On the other hand, on a clinical ground, it might also explain the limited influence of chronic metformin treatment on the diagnostic accuracy of FDG imaging in cancer diabetic patients.
Limitations
The experimental study implies a dynamic acquisition to correlate blood time concentration curves of FDG with tissue glucose metabolism. Tracer injection is particularly relevant to this purpose, since its extravasation may hamper the analysis of tracer kinetics. To overcome this problem, we already developed a procedure for tail vein catheterization using 26G cannula, and maintaining its stability as confirmed by late imaging of animal tail that ruled out any extravasation in the site of injection.
As a second limitation, non-diabetic animals were studied. However, our main purpose was to document the direct drug effect on cancer: this task would have been hampered by the presence of diabetes, due to the contamination of tracer still present in the blood both at imaging and at lesion harvesting.
Only MDA-MB-231 cell line was tested in the present study. This model of triple-negative breast cancer is particularly relevant due to the high incidence of this disease in obese, insulin-resistant, and diabetic women who are candidates to chronic biguanides treatment.
Finally, HK interference could not be directly documented in xenograft models. Accordingly, in vivo action of metformin might imply further mechanisms able to modify tumor biology and growth. However, the metabolic response of cancer agreed well with HK inhibition, suggesting that this enzymatic interference might play a relevant role in metformin anticancer potential and should be carefully considered in both experimental and clinical settings.
Conclusion
The present study indicates that metformin impairs HK enzymatic function, and its interaction with mitochondria hampering a crucial aspect for tumor immortality. Actually, this metabolic “target point” could only be reached with high drug concentrations in vitro. Similarly, the doses administered to mouse models largely exceeded clinical guidelines. This points out that interference with mitochondrial-bound HK might represent a potential target for cancer therapy. However, it also implies the need for more aggressive dosing of metformin in clinical trials or for a new class of molecules able to interfere with this step of glucose consumption.
The effect of metformin on tumor biology was strongly dependent upon treatment duration. Prolonged therapy caused a marked reduction in cancer growth without any visible necrosis. On the contrary, acute drug administration caused a profound cytotoxic effect. This observation suggests that biguanides therapy actually selected a population of drug resistant cells probably characterized by a relatively slow proliferation rate. Elucidating the mechanisms underlying this resistance might be of great interest. This task was far beyond the scopes of our study. However, this finding indicates that type of experimental protocol and duration of treatment should be carefully considered whenever data about metformin therapy are evaluated.
Materials and Methods
Chemicals
Metformin was provided by Sigma-Aldrich. FDG was produced according to standard methodology. Daily quality controls always documented adequate standards and, in particular, a radiochemical purity ≥98%.
Cell line and culture conditions
Certified human breast cancer cell line MDA-MB-231 was obtained from the Italian Cell Line Collection and cultured in DMEM medium (Gibco-BRL) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine and 100 U/ml penicillin/streptomycin at 37 °C in a humidified atmosphere of 5% CO2. Medium was changed each other day.
FDG uptake evaluation
Labeling was performed by incubating 106 cells with FDG according to a procedure validated in our laboratory.19,33 Immediately before the experiment, glucose-free medium was added with FBS and FDG at a concentration of 37KBq/mL. Tracer exposure was maintained for 60 min at 37 °C. Thereafter, uptake process was stopped by adding 4 ml of PBS before centrifugation at 450× g for 10 min. Supernatant was removed and cell pellet re-suspended in 1 ml of saline buffer. Free and bound activities were thus simultaneously counted using a Packard Cobra II gamma counter (Packard) with a 10% energy window centered at 511KeV. FDG retention was measured as the ratio between bound and total radioactivity. In all cases, labeling procedure did not affect cell viability as documented by trypan blue staining.
Real-time PCR analysis
Expression analysis was performed for glucose transporters (GLUT 1, 2, 3, and 4), HK I to IV and Thioredoxin interacting protein (TXNIP). RNA was isolated from cell culture pellets by homogenization in Qiazol followed by further purifcation on RNeasy columns (Qiagen) and DNase1 treatment to remove contaminating genomic DNA. Reverse transcription was performed with oligodT primers in a final volume of 20 µL using the SuperScript reverse transcription kit (Invitrogen). Primers were designed using Primer3 software (http://bioinfo.ut.ee/primer3/) and checked for secondary structures of the amplicon using mfold (http://mfold.rna.albany.edu/?q=mfold). Real-time polymerase chain reaction was performed on a LightCycler 480 (Roche) using Platinum SYBR Green qPCR SuperMix UDG (Invitrogen) supplemented with 2 µl of cDNA, 10 nM each sense and antisense primers, and 20× bovine serum albumin (1mg/ml) (Invitrogen) in a final volume of 20 µl. An initial denaturation step of 2 min, during which the well factor was measured, was followed by 50 cycles of 5 s at 94 °C, 10 s at 55 °C, and 10 s at 72 °C. Fluorescence measurements were performed during the annealing step in each cycle. After amplifcation, melting curves with 80 steps of 15 s and 0.5 °C temperature increase were obtained to monitor unspecific amplification products. Expression data were normalized on the mean of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), tubulin-β3, and β2-microglobulin gene expression data obtained in parallel. Relative expression values were obtained using QGene software.43
Western blot analysis
MDA-MB-231 cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) containing protease inhibitors. Alternatively, proteins were obtained from tumor tissues by AllPrep® DNA/RNA/Proteinkit (Qiagen). Proteins were resolved by 10% SDS-PAGE and transferred to nitrocellulose membranes. Membranes were incubated overnight at 4 °C and probed with the following antibodies: anti human HKI (C35C4), HKII (C64G5), total AMPK (23A3), p-AMPK (Thr172-40H9), and anti β-actin (1801) All reagents were purchased from Cell Signaling Technologies, while anti-human Actin was provided by Abcam. Primary antibodies were detected using a goat anti-mouse or goat anti-rabbit IgG HRP conjugated, and developed with ECL substrate (Thermo Fisher Scientific).
Hexokinase activity assay
HK activity was assayed spectrophotometrically, through a coupled reaction with glucose-6-phosphate dehydrogenase (G6PD), following the NADP reduction at 340 nm. The mix contained: 100 mM Tris HCl pH 8, 5 mM MgCl2, 100 Mm glucose, 0.8 mM ATP, 1 mM NADP, 3 units of G6PD.44 The assay was made on human purified HK isoforms (ProSpec-Tany TechnoGene Ltd) as well as on untreated MDA-MB-231 cell lysates. Different metformin (0.3 to 10 mM) concentrations were used. The enzyme was incubated with metformin for 10 min before the assay.
Immunofluorescence analysis
MDA-MB-231 cells were incubated with MitoTracker probe (Life Technologies Ltd) and treated as previously described.19 Coverslips were incubated overnight with rabbit anti-HKI (C35C4) or anti-HKII (C64G5) primary antibodies (Cell Signaling Technologies). Specific staining was visualized with a goat anti-Rabbit Alexa Fluor 488 secondary antibody (Molecular Probes) washed and mounted using Prolong Gold antifade reagent (Life Technologies Ltd). Results were analyzed using an Olympus (Olympus Optical) laser-scanning microscope FV500 equipped with an Olympus IX81 inverted microscope and Argon ion 488 nm, He-Ne 543 nm, and He-Ne 633 nm lasers. Digital images were acquired through a PLAPO 60× objective, with the Fluoview 4.3b software program. Images were acquired sequentially as single trans-cellular optical sections. Spatial co-localization was analyzed by ImageJ 1.34f software (NIH).
Apoptosis was determined by annexin V-FITC and propidium iodide (PI) double staining (eBioscience) according to the manufacturer's instructions. Analysis was performed using FACScan (BD Pharmigen).
Animal models
All animal experiments were reviewed and approved by the Licensing and Ethical Committee of our Institute and by the Italian Ministry of Health. The study included 30 4-wk-old female athymic (nu/nu) mice, purchased from Charles River Laboratories and housed under specific pathogen-free conditions. Mice were anaesthetized with ketamine/xylazine (100 and 10 mg/kg) and 106 MDA-MB-231 cells were orthotopically implanted in a mammary fat pad. Animals were then submitted to 3 different treatments: “untreated” mice (n = 10) did not receive any treatment and were kept under standard conditions for the whole duration of the study; “pulsed” animals (n = 10) received metformin treatment only for the 48 h before PET imaging, and “prolonged” (n = 10) were treated for the entire duration of the study. In all groups, imaging and tumor harvesting were performed 1 mo after tumor implantation.
The caloric content of the normal chow was distributed as 58% carbohydrate, 12% fat, and 30% protein, and normal food consumption was maintained during the whole experiments for all groups. All animals were allowed free access to water. Metformin was orally administered by diluting the drug in autoclaved drinking water at a concentration of 3 mg/mL according to a procedure approximately accounting for a dose of 750 mg/Kg/die.45
Experimental micro-PET scanning protocol
Before each PET scan, mice were kept under fasting conditions for 6 h. Mice were weighted, and anesthesia was induced by intraperitoneal administration of ketamine/xylazine (100 and 10 mg/kg). Serum glucose level was tested, and animals were positioned on the bed of a dedicated micro-PET system (Albira, Carestream) whose two-ring configuration permits to cover the whole animal body in a single bed position. A dose of 3–4 MBq of FDG was then injected through a tail vein, soon after start of a list mode acquisition lasting 50 min.
Image processing
Acquisition was reconstructed using the following framing rate: 10 × 15 s, 5 × 30 s, 2 × 150 s, 6 × 300 s, 1 × 600 s). PET data were reconstructed using a maximal likelihood expectation maximization method (MLEM). An experienced observer, unaware of the experimental type of analyzed mouse, identified a volume of interest (VOI) in the left ventricular chamber. Then, the computer was asked to plot the time–concentration curve within this VOI throughout the whole acquisition to define tracer input function. Whole-body FDG clearance (in ml × min−1) was calculated using the conventional stochastic approach as the ratio between injected dose and integral of input function from 0 to infinity, fitting the last 20 min with a monoexponential function.30-42 Whole-body glucose consumption (in μMol x min−1) was thus obtained multiplying clearance data for the corresponding glucose blood concentration. A further VOI was drawn over the cancer lesion to measure maximal standardized uptake values (SUV), i.e., the most commonly accepted indexes of tissue FDG uptake, expressed as the fraction of injected tracer dose normalized for body weight.
Tumor glucose consumption (in nM X min−1 × g−1) was estimated in this last VOI according to Gjedde-Patlak31 graphical analysis by using the routine of a dedicated software (PMOD). Briefly, the software utilizes the time–concentration curve in the left ventricle as the input function. The algorithm then transforms the original tissue activity measurements by fitting the data in each voxel with the slope of the regression line defined by the model. In all cases, lumped constant value was set at 1.
Autoradiographic image analysis
Immediately after imaging study, cancer lesions were surgically harvested and divided into block A for autoradiography and block B for immunohistochemistry. Block A was frozen in isopentane chilled with dry ice for sectioning with the cryo-microtome, obtaining a minimum of 3 sequential sections of 5 µm. All slices were placed onto a microscope slide to be exposed for 60 min to an imaging plate (Cyclone, Perkin Elmer Analysis Facilities) that provides an image resolution of 50 µm.
Images were analyzed for count densities (photo-stimulated luminescence per unit area, PSL/square mm) with the dedicated software. In order to analyze the whole cancer lesion, ROIs were manually drawn on each image. FDG uptake in all samples was estimated by comparing the number of PSD (counts) normalized for 18F physical decay, according to conventional formulations.46
Block B was fixed in 10% formalin solution and embedded in paraffin. Thereafter, 5 μm-thick paraffin sections were obtained, starting from cutting border facing toward block A. Besides hematoxylin–eosin staining, immunohistochemistry was performed to identify cytokeratin CK19 and CD68R expression,46 to identify neoplastic cells (revealed in fast red) and infiltrating macrophages (revealed in brown).
Both histological and autoradiographic images were saved in TIFF format. Their co-registration was performed using a dedicated software (Osirix, http://www.osirix-viewer.com/).
Statistical analysis
The data are presented as mean ± standard deviation (SD). For comparison between different groups, the null hypothesis was tested by a single factor analysis of variance (ANOVA) for multiple groups. Statistical significance was considered for P values P < 0.05.
Acknowledgments
Financial support: AA is recipient of a fellowship PO CRO Fondo Sociale Europeo Regione Liguria 2007–2013 Asse IV “Capitale Umano,” AIE is recipient of a doctoral fellowship of the University of Genoa. This work was supported by the following grants: Associazione Italiana per la Ricerca sul Cancro (UP); Fondazione Cassa di Risparmio di Genova (GS); Italian Ministry of Health (PRIN 2010JS3PMZ_009).
Glossary
Abbreviations:
- HK
hexokinase
- AMPK
5′ AMP-activated protein kinase
- PET
Positron Emission Tomography
- FDG
18F-fluorodeoxyglycose
- TXNIP
Thioredoxin interacting protein
- GLUT
glucose transporter
- PSL
photo-stimulated luminescence
- G6P
Glucose-6-Phosphate
- SUV
Standardized uptake value
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- G6PD
glucose-6-phosphate dehydrogenase
- MLEM
maximal likelihood expectation maximization
- VOI
volume of interest
- ROI
region of interest)
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26461
References
- 1.Jalving M, Gietema JA, Lefrandt JD, de Jong S, Reyners AKL, Gans ROB, de Vries EG. Metformin: taking away the candy for cancer? Eur J Cancer. 2010;46:2369–80. doi: 10.1016/j.ejca.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Dowling RJ, Goodwin PJ, Stambolic V. Understanding the benefit of metformin use in cancer treatment. BMC Med. 2011;9:33–8. doi: 10.1186/1741-7015-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. BMJ. 2005;330:1304–5. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez-Angulo AM, Meric-Bernstam F. Metformin: a therapeutic opportunity in breast cancer. Clin Cancer Res. 2010;16:1695–700. doi: 10.1158/1078-0432.CCR-09-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiralerspong S, Palla SL, Giordano SH, Meric-Bernstam F, Liedtke C, Barnett CM, Hsu L, Hung MC, Hortobagyi GN, Gonzalez-Angulo AM. Metformin and pathologic complete responses to neoadjuvant chemotherapy in diabetic patients with breast cancer. J Clin Oncol. 2009;27:3297–302. doi: 10.1200/JCO.2009.19.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cufi S, Corominas-Faja B, Vazquez-Martin A, Oliveras-Ferraros C, Dorca J, Bosch-Barrera J, Martin-Castillo B, Menendez JA. Metformin-induced preferential killing of breast cancer initiating CD44+CD24-/low cells is sufficient to overcome primary resistance to trastuzumab in HER2+ human breast cancer xenografts. Oncotarget. 2012;3:395–8. doi: 10.18632/oncotarget.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Del Barco S, Vazquez-Martin A, Cufí S, Oliveras-Ferraros C, Bosch-Barrera J, Joven J, Martin-Castillo B, Menendez JA. Metformin: multi-faceted protection against cancer. Oncotarget. 2011;2:896–917. doi: 10.18632/oncotarget.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viollet B, Guigas B, Sanz Garcia N, Leclerc J, Foretz M, Andreelli F. Cellular and molecular mechanisms of metformin: an overview. Clin Sci (Lond) 2012;122:253–70. doi: 10.1042/CS20110386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodwin PJ, Pritchard KI, Ennis M, Clemons M, Graham M, Fantus IG. Insulin-lowering effects of metformin in women with early breast cancer. Clin Breast Cancer. 2008;8:501–5. doi: 10.3816/CBC.2008.n.060. [DOI] [PubMed] [Google Scholar]
- 10.Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–74. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zakikhani M, Dowling R, Fantus IG, Sonenberg N, Pollak M. Metformin is an AMP kinase-dependent growth inhibitor for breast cancer cells. Cancer Res. 2006;66:10269–73. doi: 10.1158/0008-5472.CAN-06-1500. [DOI] [PubMed] [Google Scholar]
- 12.Vakana E, Platanias LC. AMPK in BCR-ABL expressing leukemias. Regulatory effects and therapeutic implications. Oncotarget. 2011;2:1322–8. doi: 10.18632/oncotarget.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Mir MY, Nogueira V, Fontaine E, Avéret N, Rigoulet M, Leverve X. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem. 2000;275:223–8. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- 14.Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J. 2000;348:607–14. doi: 10.1042/0264-6021:3480607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zu XL, Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochem Biophys Res Commun. 2004;313:459–65. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 16.Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Mashhedi H, Blouin MJ, Zakikhani M, David S, Zhao Y, Bazile M, Birman E, Algire C, Aliaga A, Bedell BJ, et al. Metformin abolishes increased tumor (18)F-2-fluoro-2-deoxy-D-glucose uptake associated with a high energy diet. Cell Cycle. 2011;10:2770–8. doi: 10.4161/cc.10.16.16219. [DOI] [PubMed] [Google Scholar]
- 18.Wilson JE. Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol. 2003;206:2049–57. doi: 10.1242/jeb.00241. [DOI] [PubMed] [Google Scholar]
- 19.Salani B, Marini C, Rio AD, Ravera S, Massollo M, Orengo AM, Amaro A, Passalacqua M, Maffioli S, Pfeffer U, et al. Metformin impairs glucose consumption and survival in Calu-1 cells by direct inhibition of hexokinase-II. Sci Rep. 2013;3:2070. doi: 10.1038/srep02070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn KJ, Hwang HS, Park JH, Bang SH, Kang WJ, Yun M, Lee JD. Evaluation of the role of hexokinase type II in cellular proliferation and apoptosis using human hepatocellular carcinoma cell lines. J Nucl Med. 2009;50:1525–32. doi: 10.2967/jnumed.108.060780. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez-Alvarez R, Martinez-Outschoorn UE, Lamb R, Hulit J, Howell A, Gandara R, Sartini M, Rubin E, Lisanti MP, Sotgia F. Mitochondrial dysfunction in breast cancer cells prevents tumor growth: understanding chemoprevention with metformin. Cell Cycle. 2013;12:172–82. doi: 10.4161/cc.23058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoltzman CA, Peterson CW, Breen KT, Muoio DM, Billin AN, Ayer DE. Glucose sensing by MondoA:Mlx complexes: a role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc Natl Acad Sci U S A. 2008;105:6912–7. doi: 10.1073/pnas.0712199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macheda ML, Rogers S, Best JD. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J Cell Physiol. 2005;202:654–62. doi: 10.1002/jcp.20166. [DOI] [PubMed] [Google Scholar]
- 24.Murakami T, Nishiyama T, Shirotani T, Shinohara Y, Kan M, Ishii K, Kanai F, Nakazuru S, Ebina Y. Identification of two enhancer elements in the gene encoding the type 1 glucose transporter from the mouse which are responsive to serum, growth factor, and oncogenes. J Biol Chem. 1992;267:9300–6. [PubMed] [Google Scholar]
- 25.Mathupala SP, Ko YH, Pedersen PL, Hexokinase II. Hexokinase II: cancer’s double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25:4777–86. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathupala SP, Ko YH, Pedersen PL. Hexokinase-2 bound to mitochondria: cancer’s stygian link to the “Warburg Effect” and a pivotal target for effective therapy. Semin Cancer Biol. 2009;19:17–24. doi: 10.1016/j.semcancer.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.John S, Weiss JN, Ribalet B. Subcellular localization of hexokinases I and II directs the metabolic fate of glucose. PLoS One. 2011;6:e17674. doi: 10.1371/journal.pone.0017674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim W, Yoon JH, Jeong JM, Cheon GJ, Lee TS, Yang JI, Park SC, Lee HS. Apoptosis-inducing antitumor efficacy of hexokinase II inhibitor in hepatocellular carcinoma. Mol Cancer Ther. 2007;6:2554–62. doi: 10.1158/1535-7163.MCT-07-0115. [DOI] [PubMed] [Google Scholar]
- 29.Menendez JA, Oliveras-Ferraros C, Cufí S, Corominas-Faja B, Joven J, Martin-Castillo B, Vazquez-Martin A. Metformin is synthetically lethal with glucose withdrawal in cancer cells. Cell Cycle. 2012;11:2782–92. doi: 10.4161/cc.20948. [DOI] [PubMed] [Google Scholar]
- 30.Iozzo P, Gastaldelli A, Järvisalo MJ, Kiss J, Borra R, Buzzigoli E, Viljanen A, Naum G, Viljanen T, Oikonen V, et al. 18F-FDG assessment of glucose disposal and production rates during fasting and insulin stimulation: a validation study. J Nucl Med. 2006;47:1016–22. [PubMed] [Google Scholar]
- 31.Patlak CS, Blasberg RG, Fenstermacher JD. Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. J Cereb Blood Flow Metab. 1983;3:1–7. doi: 10.1038/jcbfm.1983.1. [DOI] [PubMed] [Google Scholar]
- 32.Aloj L, Caracó C, Jagoda E, Eckelman WC, Neumann RD. Glut-1 and hexokinase expression: relationship with 2-fluoro-2-deoxy-D-glucose uptake in A431 and T47D cells in culture. Cancer Res. 1999;59:4709–14. [PubMed] [Google Scholar]
- 33.Würth R, Pattarozzi A, Gatti M, Bajetto A, Corsaro A, Parodi A, Sirito R, Massollo M, Marini C, Zona G, et al. Metformin selectively affects human glioblastoma tumor-initiating cell viability: A role for metformin-induced inhibition of Akt. Cell Cycle. 2013;12:145–56. doi: 10.4161/cc.23050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alimova IN, Liu B, Fan Z, Edgerton SM, Dillon T, Lind SE, Thor AD. Metformin inhibits breast cancer cell growth, colony formation and induces cell cycle arrest in vitro. Cell Cycle. 2009;8:909–15. doi: 10.4161/cc.8.6.7933. [DOI] [PubMed] [Google Scholar]
- 35.Song CW, Lee H, Dings RP, Williams B, Powers J, Santos TD, Choi BH, Park HJ. Metformin kills and radiosensitizes cancer cells and preferentially kills cancer stem cells. Sci Rep. 2012;2:362. doi: 10.1038/srep00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bustamante E, Pedersen PL. High aerobic glycolysis of rat hepatoma cells in culture: role of mitochondrial hexokinase. Proc Natl Acad Sci U S A. 1977;74:3735–9. doi: 10.1073/pnas.74.9.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su H, Bodenstein C, Dumont RA, Seimbille Y, Dubinett S, Phelps ME, Herschman H, Czernin J, Weber W. Monitoring tumor glucose utilization by positron emission tomography for the prediction of treatment response to epidermal growth factor receptor kinase inhibitors. Clin Cancer Res. 2006;12:5659–67. doi: 10.1158/1078-0432.CCR-06-0368. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L, He H, Balschi JA. Metformin and phenformin activate AMP-activated protein kinase in the heart by increasing cytosolic AMP concentration. Am J Physiol Heart Circ Physiol. 2007;293:H457–66. doi: 10.1152/ajpheart.00002.2007. [DOI] [PubMed] [Google Scholar]
- 39.Hadad SM, Fleming S, Thompson AM. Targeting AMPK: a new therapeutic opportunity in breast cancer. Crit Rev Oncol Hematol. 2008;67:1–7. doi: 10.1016/j.critrevonc.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 40.Kubota R, Yamada S, Kubota K, Ishiwata K, Tamahashi N, Ido T. Intratumoral distribution of fluorine-18-fluorodeoxyglucose in vivo: high accumulation in macrophages and granulation tissues studied by microautoradiography. J Nucl Med. 1992;33:1972–80. [PubMed] [Google Scholar]
- 41.Wong CY, Thie J, Parling-Lynch KJ, Zakalik D, Margolis JH, Gaskill M, Hill J, Qing F, Fink-Bennett D, Nagle C. Glucose-normalized standardized uptake value from (18)F-FDG PET in classifying lymphomas. J Nucl Med. 2005;46:1659–63. [PubMed] [Google Scholar]
- 42.Massollo M, Marini C, Brignone M, Emionite L, Salani B, Riondato M, Capitanio S, Fiz F, Democrito A, Amaro A, et al. Metformin temporal and localized effects on gut glucose metabolism assessed using 18F-FDG PET in mice. J Nucl Med. 2013;54:259–66. doi: 10.2967/jnumed.112.106666. [DOI] [PubMed] [Google Scholar]
- 43.Simon P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics. 2003;19:1439–40. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- 44.Lamprecht W, Trautschold I. Determination with Hexokinase and glucose-6-phosphate dehydrogenase. In: Bergmeyer HU, ed. Methods of Enzymatic Analysis. New York: Academic Press. 1974:1127–1131. [Google Scholar]
- 45.Phoenix KN, Vumbaca F, Claffey KP. Therapeutic metformin/AMPK activation promotes the angiogenic phenotype in the ERalpha negative MDA-MB-435 breast cancer model. Breast Cancer Res Treat. 2009;113:101–11. doi: 10.1007/s10549-008-9916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marini C, Morbelli S, Armonino R, Spinella G, Riondato M, Massollo M, Sarocchi F, Pane B, Augeri C, Abete L, et al. Direct relationship between cell density and FDG uptake in asymptomatic aortic aneurysm close to surgical threshold: an in vivo and in vitro study. Eur J Nucl Med Mol Imaging. 2012;39:91–101. doi: 10.1007/s00259-011-1955-1. [DOI] [PubMed] [Google Scholar]