Abstract
We previously demonstrated that DNA and protein co-administration induced differentiation of immature dendritic cells (iDCs) into CD11c+CD40lowIL-10+ regulatory DCs (DCregs) via the caveolin-1 (Cav-1) -mediated signal pathway. Here, we demonstrate that production of IL-10 and the low expression of CD40 play a critical role in the subsequent induction of regulatory T cells (Tregs) by the DCregs. We observed that DNA and protein were co-localized with DC-SIGN in caveolae and early lysosomes in the treated DCs, as indicated by co-localization with Cav-1 and EEA-1 compartment markers. DNA and protein also co-localized with LAMP-2. Gene-array analysis of gene expression showed that more than a thousand genes were significantly changed by the DC co-treatment with DNA + protein compared with controls. Notably, the level of DC-SIGN expression was dramatically upregulated in pOVA + OVA co-treated DCs. The expression levels of Rho and Rho GNEF, the down-stream molecules of DC-SIGN mediated signal pathway, were also greatly upregulated. Further, the level of TLR9, the traditional DNA receptor, was significantly downregulated. These results suggest that DC-SIGN as the potential receptor for DNA and protein might trigger the negative pathway to contribute the induction of DCreg combining with Cav-1 mediated negative signal pathway.
Keywords: DNA and protein co-administration, cavelin-1, DC-SIGN, iTreg, tolerance
Introduction
Regulatory T cells (Tregs) play a central role in the maintenance of immunological self-tolerance and immune homeostasis.1 Tregs suppress activation and expansion of self-reactive lymphocytes via both cell contact-dependent and cytokine-dependent mechanisms.2-6 Tregs can be induced by immature DCs (iDCs)7,8 or by DCs that are modified by cytokines such as TGF-β and GM-CSF.9,10 Further, T cells can be induced into anergy or become Tregs if co-stimulatory signals of DCs are not fully expressed,11,12 suggesting an important influence of the state of DCs maturation during Treg induction.
Recently, we demonstrated that iTregs can be induced from naïve T cells by a co-immunization with DNA and protein vaccines and then have preventive or therapeutic functions against autoimmune diseases.13-17 Furthermore, both in vitro and in vivo, co-administration of DNA and protein induces DC differentiation into an IL-10-producing regulatory DC form (DCregs) that has a CD11c+CD40low phenotype. These DCregs in turn could convert naïve CD4+ T cells into antigen specific CD4+CD25-Foxp3+ iTregs in vitro and in vivo.17 The induction of DCregs was mediated by caveolae-dependent endocytosis and caveolin-1 (Cav-1) signal pathway.18 Nevertheless, the distribution of vaccine DNA and protein in the DCregs and the role of the CD40low and IL-10 producing phenotype of the DCregs in the induction of Tregs remained to be determined.
In this study, we demonstrated that CD40low and IL-10 producing form of the DCregs plays a critical role in the induction of iTregs. DNA and protein were found to co-localize with caveolae and lysosomes. Microarray analysis showed that more than a thousand genes were significantly changed at their transcriptional levels after DNA and protein co-treatment. These genes were associated with the TLR signal pathways, kinase activation, NO production, transcriptional factors and cytokines. These results provide new insight into understanding the mechanism of the induction of DCregs by DNA and protein co-administration.
Results
Induction of Tregs by DCregs depends on IL-10 and low expression of CD40
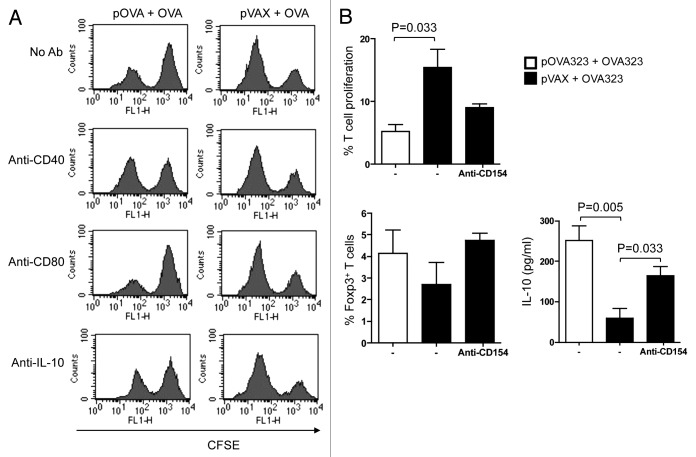
Our previous studies showed that DCs isolated from spleen of mice that had been co-treated with DNA and protein, or DC that had been co-treated in vitro, could convert naïve or antigen-primed T cells into Tregs.17,18 To investigate roles of CD40 and IL-10 in such modified DCs (DCregs) in the induction of iTregs, we used DCs isolated from the spleens of mice 48 h after the co-administration of pOVA and OVA. Naïve CD4+ T cells were isolated from naïve syngeneic mice and labeled with CFSE. The freshly-isolated DCs and the labeled T cells were then co-cultured in the presence or absence of various mAbs that included anti-CD40 (stimulatory), anti-IL-10 (a functional blocker) and anti-CD80 (a functional blocker). After seven days, the T cells were purified from the co-culture and added into a mixed-leukocyte reaction (MLR) system to examine their suppressive functions. The results showed that anti-CD40 and anti-IL-10 mAbs could partially reverse the suppressive effects of the Tregs on the MLR, whereas mAbs against CD80 did not (Fig. 1A).
Figure 1. Both CD40 and IL-10 signals of DCreg were required for Tregs development in vitro. (A) For in vivo generated DCreg, DCreg were isolated from spleen of DNA plus antigen co-administrated mice and co-cultured with CFSE labeled syngeneic naive CD4+ T cells in the presence of mAb (50 μg/ml) to CD40 (a stimulator), CD80 (a blocker) and IL-10 (a blocker) or an isotype control. T cells isolated from DC-T co-cultures were analyzed for their abilities to inhibit MLR. After 48 h, T cell proliferation was analyzed by FACS and cells were gated on the CFSE positive population. Results are representative of three experiments. (B) For in vitro generated DCreg, co-treated DC were co-cultured with CFSE-CD4+ T cells purified from spleens of mice immunized with OVA in IFA and in the presence or absence of anti-CD40L (blocker) for 5 d. T cell proliferation and the productions of Foxp3 and IL-10 were detected by FACS. P values (Mann-Whitney test) are indicated.
To investigate whether low expression of CD40 by the DCregs was essential for the induction of iTregs, we first generated DCregs by adding pOVA323 + OVA323 (test), or pVAX + OVA323 (control) to cultured JAWS II cells. The activity of the resulting cells was then tested by co-culturing the cells with syngeneic CD4+ T cells that had been purified from OVA immunized mice and labeled with CFSE. Co-culture was in the presence or absence of anti-CD154 mAb as a blocking agent. The expression of Foxp3 and IL-10 by the CFSE-T cells was analyzed after 5-d co-culture. pOVA323 + OVA323 treated DCs had increased expression of both Foxp3 and IL-10 compared with pVAX + OVA323 treated DCs. Moreover, the addition of anti-CD154 to the pVAX + OVA323 treated DCs could exert a similar, though smaller, effect compared with that seen in pOVA323 + OVA323 treated DCs (Fig. 2). These results revealed that DCreg cells that were generated by co-treatment both in vitro and in vivo could induce T cells to differentiate into regulatory iTregs. Thus, the presence of IL-10 and the low expression of CD40 play crucial roles in the induction of iTregs.
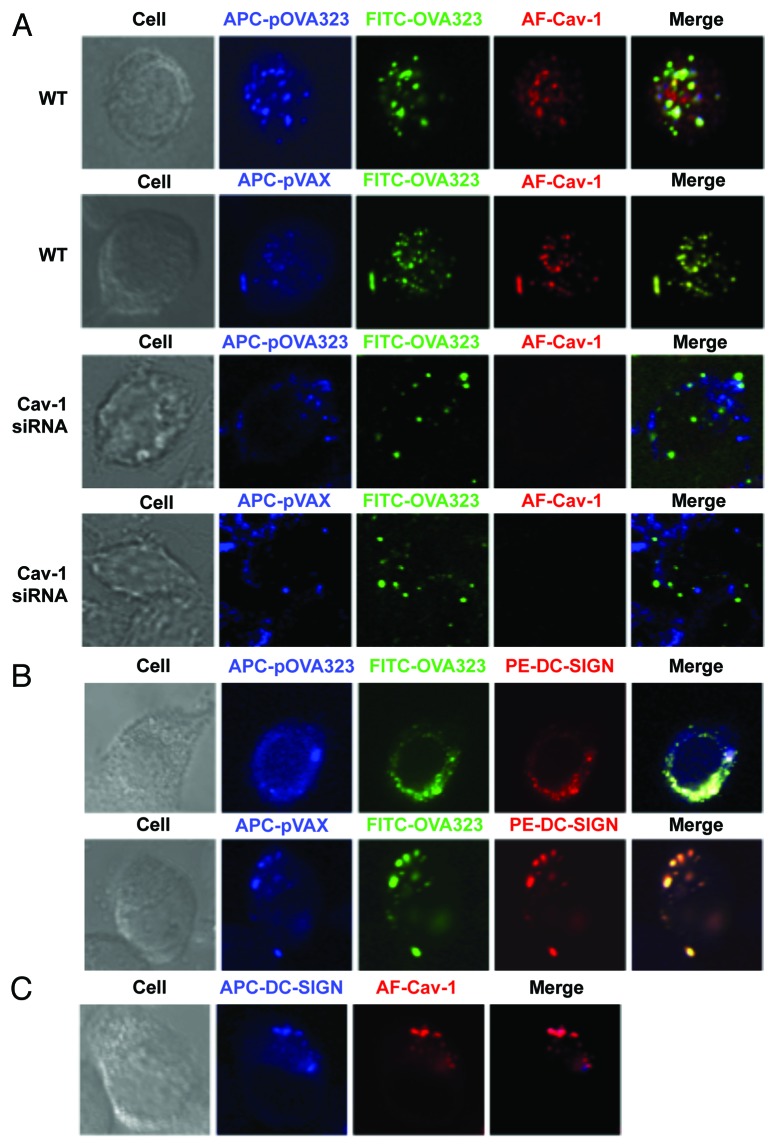
Figure 2. The distribution of DNA and protein in co-administrated DCs. (A) The co-localization of Cav-1 with DNA and protein. WT JAWS II cells (WT) or Cav-1 silenced JAWS II cells (Cav-1 siRNA) were co-treated with Cy5-pOVA323 + FITC-OVA323 or Cy5-pVAX + FITC-OVA323 for 24 h, then fixed and permeabilized with 0.1% Triton X-100 in PBS buffer. Cells were incubated with rabbit anti-Cav-1 for 1 h and subsequently reacted with the Alexa Fluor 546-labeled goat anti-rabbit IgG before the cells were observed using an inverted Nikon ECLIPSE IE2000-E confocal microscope. (B) The co-localization of DC-SIGN with DNA and protein. JAWS II cells were co-treated with Cy5-pOVA323 + FITC-OVA323 or Cy5-pVAX + FITC-OVA323 for 24 h, then fixed and permeabilized with 0.1% Triton X-100 in PBS buffer. Cells were stained with PE-DC-SIGN and observed using an inverted Nikon ECLIPSE IE2000-E confocal microscope. (C) The co-localization of DC-SIGN and Cav-1. JAWS II cells were fixed and permeabilized. Cells were inoculated with rabbit anti-Cav-1 for 1 h and subsequently reacted with the Alexa Fluor 546-labeld goat anti-rabbit IgG and APC-DC-SIGN before cells were observed using an inverted Nikon ECLIPSE IE2000-E confocal microscope.
DNA and protein localized in caveolae
Our previous study indicated that the effect of DNA and protein co-entry on the expression of CD40 was mediated via caveolae-dependent endocytosis.18 It is reasonable to propose that the DNA + protein could co-localize within the caveolae. To confirm this, the co-localization of DNA and protein with Cav-1 was analyzed. The fluorescent dye-labeled DNA and peptides were used to treat control and Cav-1-silenced JAWSII cells. After 5 h-stimulation, control and Cav-1 silenced JAWSII cells were incubated with rabbit anti-Cav-1 antibodies. In control WT JAWSII cells, DNA and protein were observed to be co-localized with Cav-1, after either pOVA323 + OVA323 or pVAX + OVA323 treatment (Fig. 2A). However, in Cav-1-silenced JAWSII cells DNA did not co-localize with protein (Fig. 2A).
A large array of pathogens can interact with C-type lectin DC-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN) to modulate DC-mediated immune responses, including induction of Tregs and upregulation of IL-10 expression.19,20 We investigated whether DC-SIGN might act as the receptor in DNA and protein co-treatment. The localization of DNA, protein, and DC-SIGN was analyzed under a confocal microscope. DNA and protein were co-localized with DC-SIGN both in pOVA323 + OVA323 and pVAX + OVA323 treated JAWSII cells (Fig. 2B), indicating that DC-SIGN might be a receptor for both DNA and protein. The co-localization of Cav-1 and DC-SIGN was also observed (Fig. 2C). The result confirmed that DNA and protein were taken up by caveolae-mediated endocytosis that leads to downregulation of CD40 expression.
DNA and protein co-localized with EEA1 and LAMP-2 compartments
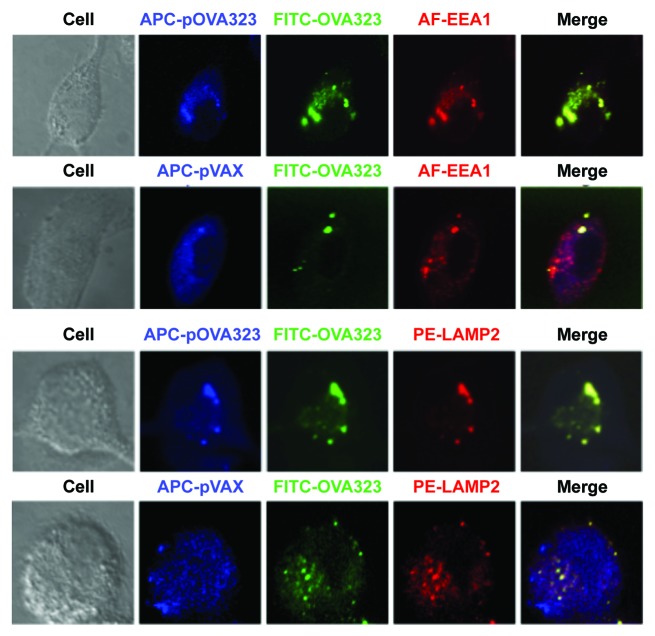
We further investigated whether this distinct antigen processing pathway caused the different signals. The co-localization in JAWSII cells of fluorescent dye-labeled DNA and peptides with early endosome antigen 1 (EEA1) and with lysosome-associated membrane protein-2 (LAMP-2) was analyzed under a confocal microscope. DNA and protein co-localized with EEA1 in both the pOVA323 + OVA323 and the pVAX + OVA323 treated JAWSII cells (Fig. 3). The co-localization of DNA and protein with LAMP-2 was observed in the pOVA323 + OVA323 treated cells but not in the pVAX + OVA323 treated cells. OVA323 co-localized with LAMP-2 in the pVAX + OVA323 treated cells but pVAX did not (Fig. 3). Previous studies have demonstrated that processed exogenous antigens are bound to MHC class II molecules in late endosomes21,22 and that the food antigen ovalbumin that was targeted to late endosomes of enterocytes after oral administration was able to induce oral tolerance.23 Our results thus indicated that the co-localization of DNA and protein with LAMP-2 might be required for the induction of antigen-specific Tregs.
Figure 3. The co-localization of DNA and protein with EEA1 or LAMP2. JAWS II cells were co-treated with Cy5-pOVA323 + FITC-OVA323 or Cy5-pVAX + FITC-OVA323 for 5 h, then fixed and permeabilized with 0.1% Triton X-100 in PBS buffer. Cells were incubated with rabbit anti-EEA1 or rat anti-LAMP2 for 1 h and subsequently reacted with the Alexa Fluor 546-labeled goat anti-rabbit IgG or PE-labeled goat anti-rat IgG. Cells were observed using an inverted Nikon ECLIPSE IE2000-E confocal microscope.
DC-SIGN co-localized with EEA1 in pOVA323 + OVA323 treated DCs
DC-SIGN co-localized with Cav-1 might also serve as a receptor for either DNA or protein. To further address this issue, the localization of DC-SIGN and EEA1 was investigated. JAWS II cells were treated with pOVA323 + FITC-OVA323 or pVAX + FITC-OVA323. After 5 h, the cells were incubated with anti-EEA1 or anti-DC-SIGN antibodies. DC-SIGN co-localized with EEA1 only in the pOVA323 + FITC-OVA323 treated DCs, but not with pVAX + FITC-OVA323 treated DCs (Fig. 4). The results suggested that DC-SIGN could also be acting as a receptor for the matched DNA + protein (pOVA323 + OVA323) co-administration but not for the mismatched control (pVAX + OVA323) treatment, and so may also contribute to triggering the negative signals in the co-treated DCs.
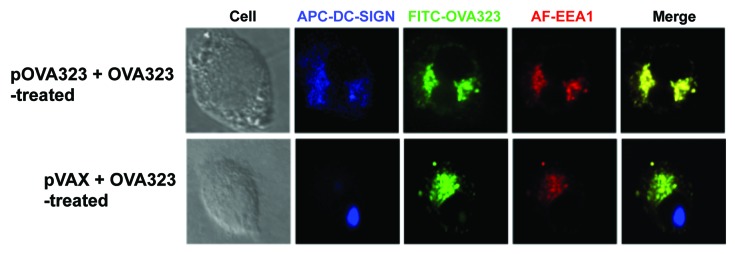
Figure 4. The co-localization of DC-SIGN and EEA1. JAWS II cells were co-treated with pOVA323 + FITC-OVA323 or pVAX + FITC-OVA323 for 5 h, then fixed and permeabilized with 0.1% Triton X-100 in PBS buffer. Cells were incubated with rabbit anti-EEA1 for 1 h and subsequently reacted with the Alexa Fluor 546-labeled goat anti-rabbit IgG and APC-DC-SIGN. Cells were observed using an inverted Nikon ECLIPSE IE2000-E confocal microscope.
Comparison of gene expression between the co-treated DCs and non-treated, DNA treated or protein treated DCs
To explore the mechanism of DCreg induction, gene expression after 24 h treatment was compared by Genechip in the non-treated DCs, DCs treated with pOVA, DCs treated with OVA, and DCs co-treated with pOVA and OVA. More than a thousand genes were significantly upregulated or downregulated (-fold difference > 2 or < 0.5; the levels of gene expression in the pOVA + OVA co-administration over that in the non-treated DCs, pOVA- or OVA-treated DCs). Some more significantly affected genes are listed in Table 1.
Table 1. Upregulated and downregulated genes.
| Accession number | Gene symbol | Gene description | Transcript level (fold) | ||
|---|---|---|---|---|---|
| pOVA + OVA/ Control | pOVA | OVA | |||
| Upregulated | |||||
| AK020411 | BMP7 | Bone morphogenetic protein 7 | 5.2 | 11.9 | 27.4 |
| NM_007617 | Caveolin-3 | Caveolin-3 | 13.9 | 7.7 | 2.7 |
| AJ131357 | CCR9 | Chemokine (C-C motif) receptor 9 | 0.4 | 23.3 | 5.2 |
| NM_009856 | CD83 | CD83 antigen | 7.3 | 1.2 | 4.7 |
| U76372 | SMDC1 | SET and MYND domain containing 1 | 6.1 | 2 | 6 |
| AF374470 | DC-SIGN | CD209a antigen | 1.1 | 3.1 | 4.5 |
| NM_010494 | ICAM-2 | Intercellular adhesion molecule 2 | 11.6 | 5.7 | 1.7 |
| NM_010505 | IFN-a5 | Interferon α 5 | 3.4 | 1.8 | 6.9 |
| K00083 | IFN-g | Interferon gamma | 1.6 | 22.3 | 16.8 |
| BB453775 | MAPK-10 | Mitogen-activated protein kinase 10 | 32.7 | 2.3 | 9.6 |
| AF228706 | NFATZ2 | Nuclear factor of activated T-cells 5 | 3.9 | 2.4 | 4 |
| AV208159 | NOSi | Nitric oxide synthase inhibitor | 1.9 | 0.8 | 11 |
| D32210 | Notch2 | Notch gene homolog 2 (Drosophila) | 1.4 | 11.5 | 2.1 |
| NM_008716 | Notch3 | Notch gene homolog 3 (Drosophila) | 1.8 | 1.3 | 2.5 |
| NM_008840 | PI3KCD | PI3K catalytic delta polypeptide | 0.9 | 5.8 | 4.6 |
| AF309564 | Rho | ras homolog gene family, member J | 2.4 | 11.4 | 16.3 |
| AK006430 | Rho binder | Rho GTPase binding | 1.9 | 8.5 | 11 |
| BG069493 | Rho GNEF | Rho guanine nucleotide exchenge factor | 1.8 | 5.3 | 4.9 |
| BF466815 | SOCS7 | Suppressor of cytokine signaling 7 | 2.7 | 0.4 | 2.1 |
| NM_009506 | VEGFC | Vascular endothelial growth factor C | 5.1 | 5.8 | 5.3 |
| NM_018865 | Wisp1 | WNT1 inducible signaling pathway protein 1 | 2.8 | 3.9 | 1.7 |
| BC005449 | Wnt1 | Wingless-related MMTV integration site 1 | 2.4 | 1.1 | 4.9 |
| U20658 | Wnt10b | Wingless related MMTV integration site 10b | 3.9 | 1 | 3.8 |
| NM_009290 | Wnt8a | Wingless related MMTV integration site 8b | 1.9 | 2.9 | 2.4 |
| Downregulated | |||||
| X52685 | A-PKC | protein kinase C, α | 0.21 | 0.71 | 0.21 |
| NM_009756 | BMP10 | Bone morphogenetic protein 10 | 0.16 | 0.76 | 0.24 |
| NM_007556 | BMP6 | Bone morphogenetic protein 6 | 0.05 | 0.1 | 0.63 |
| NM_007558 | BMP8a | Bone morphogenetic protein 8A | 0.14 | 0.09 | 0.13 |
| NM_007560 | BMPR1b | BMP receptor, type 1B | 0.24 | 0.44 | 0.3 |
| BG070002 | CREB3 | cAMP responsive element binding protein 3 | 0.37 | 0.06 | 0.41 |
| M33036 | ICAM-1 | Intercellular adhesion molecule 1 | 0.06 | 0.03 | 0.05 |
| NM_010503 | IFN-a2 | Interferon α 2 | 1.1 | 0.4 | 1.2 |
| NM_010504 | IFN-a4 | Interferon α 4 | 1 | 0.35 | 1.1 |
| NM_008333 | IFN-a11 | Interferon α 11 | 0.35 | 0.14 | 0.08 |
| NM_010510 | IFN-b1 | Interferon β 1 | 0.61 | 0.49 | 0.8 |
| L33768 | JAK3 | Janus kinase 3 | 0.38 | 0.35 | 0.6 |
| BC021640 | MAPK12 | Mitogen-activated protein kinase 12 | 0.57 | 0.12 | 0.27 |
| BF124648 | NFATC3 | RNA binding motif protein 35b | 0.2 | 0.36 | 0.48 |
| X70514 | NODA1 | Nodal | 0.46 | 0.59 | 0.1 |
| BB794641 | NOS1 | Nitric oxide synthase 1 | 1.3 | 0.29 | 0.17 |
| BI696369 | NRARP | Notch-regulated ankyrin repeat protein | 0.31 | 0.44 | 0.26 |
| NM_011084 | PI3K c2g | PI3K C2 domain containing, gamma polypeptide | 0.13 | 0.41 | 0.05 |
| AK009381 | PTRF | Polymerase I and transcript release factor | 0.47 | 0.62 | 0.16 |
| BF539472 | Rho GAP12 | Rho GTPase activating protein 12 | 0.21 | 0.37 | 0.36 |
| NM_008113 | Rho GDI | Rho GDP dissociation inhibitor (GDI) gamma | 0.21 | 0.21 | 0.38 |
| U58993 | SMAD5 | MAD homolog 5 (Drosophila) | 0.82 | 0.48 | 0.94 |
| AF348140 | TLR9 | Toll-like receptor 9 | 0.37 | 0.28 | 0.29 |
| NM_009518 | Wnt10a | Wingless related MMTV integration site 10a | 0.43 | 0.14 | 0.33 |
| NM_009522 | Wnt3a | Wingless related MMTV integration site 3a | 0.61 | 0.9 | 0.37 |
| BC018425 | Wnt5a | Wingless related MMTV integration site 5a | 0.16 | 0.14 | 0.16 |
| AK004683 | Wnt7a | Wingless related MMTV integration site 7a | 0.44 | 0.95 | 0.21 |
| NM_008324 | IDO | Indoleamine 2,3-dioxygenase 1 | 0.76 | 0.3 | 0.35 |
| BB770528 | RA-i2 | Retinoic acid induced 2 | 0.03 | 0.3 | 0.05 |
It is known that when DNA stimulates iDCs to develop into mDCs by the TLR9 signaling pathway the mDCs can secret high levels of cytokines including Type I interferon (IFN-α, IFN-β) and the IFN-α and IFN-β can then stimulate DC maturation to more effectively activate immune responses.24,25 The levels of expression of TLR9, IFN-α and IFN-β were significantly downregulated by the pOVA + OVA co-administration, which might therefore contribute to iDC development into DCregs and not into mDCs.
DC-SIGN is a prototypic member of the C-type lectin family and facilitates broad immune surveillance by DCs. Mycobacterium tuberculosis and HIV both target DC-SIGN to infect DCs and activate the small GTPase Ras protein (Rho) and Rho guanine nucleotide exchange factor (Rho GNEF) to increase the production of IL-10 and to evade immune surveillance.26 The expression levels of DC-SIGN, Rho and Rho GNEF were dramatically upregulated in the pOVA + OVA co-treated DCs, which is consistent with the increase of IL-10 production in these DCs.18
Notch pathways play important roles in the generation and expansion of Tregs.27 The expression levels of Notch2 and Notch3 were significantly increased in the pOVA + OVA co-treated DCs and this is consistent with the induction of Tregs by the pOVA + OVA co-treated DCs in the DC-T cell co-culture system.18 The suppressor of cytokine signaling (SOCS) protein family plays a central role in the negative regulation of cytokine action and a lack of SOCS7 is associated with elevated production of pro-inflammatory cytokines.28 The expression of SOCS7 was upregulated in co-administrated DCs and could be related to the decrease of pro-inflammatory cytokine production in the co-treated group.18
Nitric oxide was reported to have a selective enhancing effect on the induction and differentiation of Th1 cells in synergy with IL-12 produced by antigen presenting cells.29 The expression of Nitric oxide synthase 1 (NOS1) was decreased whereas the expression of NOS inhibitor was increased in the co-treated DCs. These changes may help to explain the decrease of IFN-γ in CD4+ T cells and the increase of Treg frequency in the co-treated group.15
Taken together, it appears that the DNA + protein co-administration triggered several negative signaling pathways and downregulated positive signaling pathways when inducing DCregs. The pathways implicate differential transcription, TLR signaling, kinase activities, NO production, and cytokine production.
Discussion
In this study, we demonstrated that DNA and protein co-localized with Cav-1, EEA1 and LAMP-2 in DCs and that CD40low and IL-10 played important roles in the subsequent induction of Tregs by DCregs. We further explored underlying mechanisms to understand how the DNA and protein together induced DCregs. The results showed that several signal pathways were affected by the co-administration, which included the TLR pathway, kinase activity, NO production, transcriptional factors and cytokine production.
Our previous study showed that DNA and protein co-administration could induce DCregs with CD11c+CD40lowIL-10+ phenotype, which triggered Cav-1 mediated negative signaling and led to the downregulation of CD40 and upregulation of IL-10.18 However, the roles of CD40 and IL-10 in the induction of Tregs were not clear. In this study, we show that the induction of Tregs by DCregs was blocked by recovery of CD40 signal via adding anti-CD40 (stimulatory) or inhibiting the function of IL-10 via adding anti-IL-10 (a functional blocker). The results directly demonstrated that the low expression of CD40 and higher expression of IL-10 in DCregs play important roles in the induction of Tregs by DCregs. We again observed that DNA and protein co-localized with Cav-1, consistent with our previous results in which DNA and protein were seen to be taken up by caveolae-mediated endocytosis to elicit negative signal.18 However, the mismatched control DNA and protein can also co-localize with Cav-1 in the pVAX + OVA323 treated DCs. But why the mismatched control DNA and protein co-treatment did not trigger the same negative signaling as seen in the matched DNA and protein co-administration is still elusive. We found that the pOVA323 and OVA323 co-localized with LAMP-2 in the pOVA323 + OVA323 co-treatment but pVAX did not in the pVAX + OVA323 co-treatment, suggesting that an endogenous antigen production process may be involved. This seems to be supported by the observation that DC-SIGN only co-localized with EEA1 in the pOVA323 + OVA323 co-administrated DCs, but not in pVAX + OVA323 treated DCs. The differences may lead to triggering of different signals. However, this should be further investigated.
Some pathogens could evade immune surveillance through DC-SIGN mediated negative signal pathway. Our results showed that DC-SIGN not only co-localized with Cav-1 for antigen uptake but also co-localized with EEA1 for antigen process in the pOVA323 + OVA323 co-administrated DCs. The results suggested that DC-SIGN might act as a receptor for DNA and protein co-administration to facilitate the activation of negative signal pathway(s). Consistently, a genechip analysis showed that the level of DC-SIGN expression was dramatically upregulated in the pOVA + OVA co-treated DCs. We further observed that the expression levels of some molecules mediated under the DC-SIGN signaling were greatly upregulated in the pOVA + OVA co-treated DCs. These molecules include Rho and Rho GNEF, which were demonstrated to be involved in the production of IL-10.26 The results further support that DC-SIGN might act as the receptor for DNA and protein.
TLRs play important roles in the recognition of pathogens and activation of immune responses. DNA could activate DCs by the TLR9 signal pathway to secret high levels of cytokines including IFN-α, IFN-β.24,25 The level of TLR9 expression was significantly downregulated in the pOVA + OVA co-treated DCs, which might reduce interactions of TLR9 and DNA to avoid the activation of TLR9 mediated signaling. This is consistent with the downregulation of expressions of IFN-α2, IFN-α4, IFN-α11, and IFN-β1, especially compared with the pOVA alone treated DCs. Combining the downregulation of TLR9 and upregulation of DC-SIGN, the pOVA and OVA have the tendency to use DC-SIGN as the receptor to trigger a negative signal pathway rather than TLR9 to activate DCs.
Our results provide a preliminary finding for the induction of DCregs by DNA + protein co-immunization. These data provide an evidence to show the association between antigen uptake, antigen process and several negative signal pathways that initiated by Cav-1 or DC-SIGN. Our further studies will focus on the investigation of functions of several critical genes related to the mechanism, especially to interactions with DC-SIGN and it’s down-stream molecules.
In conclusion, IL-10 and CD40low of DCregs play important roles in the induction of iTregs. DNA and protein co-localization with Cav-1 and DC-SIGN apparently triggers negative signal pathways to induce iDCs differentiation into DCregs. DNA and protein co-administration affected a number of genes that are involved in different signal pathways including genes of TLR pathways, kinase activity, NO production, transcriptional factors and cytokine production. The results suggested that DNA and protein co-administration exerts its regulatory effects in DCs to lead to a tolerogenic function.
Materials and Methods
Mice and reagents
Female BALB/c and C57BL/6 mice (8–10 weeks of age) were from the Animal Institute of Chinese Medical Academy. All animals received pathogen-free water and food.
Flexset IL-10 and fluorescently labeled anti-mouse monoclonal antibodies including anti-FoxP3-allophycocyanin (APC), and isotype controls were purchased from BD Biosciences. Anti-DC-SIGN (dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin)-APC and anti-DC-SIGN-phycoerythrin (PE) were purchased from eBioscience. Antibodies against caveolin-1, early endosome antigen 1 (EEA1) and lysosome-associated membrane protein-2 (LAMP-2) were purchased from Santa Cruz Biotechnology. Alexa Fluor 546 (AF)-labeled goat anti-rabbit IgG was purchased from Invitrogen. PE-labeled goat anti-rat IgG was purchased from Biolegend. CFSE was obtained from Molecular Probes.
Vaccine preparations
The DNA vaccines, pVAX-OVA (designated as pOVA) and pVAX-OVA323 (designated as pOVA323) have been described.18 OVA was purchased from Sigma-Aldrich. The OVA peptide (aa323–339, named as OVA323) or FITC-labeled OVA323 were synthesized by GL Biochem Co., Ltd.. All plasmids were purified to remove endotoxin with the EndoFree Plasmid Maxi Kit (QIAGEN) and used as the DNA vaccines after dissolving in PBS at 2 mg/ml. Recombinant proteins and peptides were dissolved in PBS at 2 mg/ml and sterilized by filtration.
Culture and stimulation of JAWS II cells or Cav-1 silenced JAWS II cells
The JAWS II mouse DC line was purchased from the American Type Culture Collection (ATCC) and maintained in complete growth medium containing minimum essential medium α with ribonucleosides, deoxyribonucleosides, 4 mM L-glutamine, and 1 mM sodium pyruvate (Invitrogen Inc), and supplemented with 20% fetal bovine serum (ATCC) and 5 ng/ml murine recombinant GM-CSF (R&D Systems, Inc.). Cav-1 silenced JAWS II cells were generated as previously described18 and cultured using the above protocol. The cells were incubated at 37 °C with 5% CO2 and treated with different antigens (10 μg/ml) such as pVAX, pOVA, OVA, pOVA323 and OVA323.
DC-T cells co-culture
For analysis of in vivo function of purified DCregs, CD11c+ cells (DCregs) were isolated from spleen of pOVA + OVA or pVAX + OVA treated mice using CD11c MicroBeads according to the manufacturer’s protocol (Miltenyi Biotec Inc.) and co-cultured with CFSE labeled syngeneic naive CD4+ T cells in the presence of mAb (50 μg/ml) to CD40 (a stimulator), CD80 (a blocker) and IL-10 (a blocker) or an isotype control. T cells isolated from DC-T co-cultures were analyzed for their abilities to inhibit MLR.
For in vitro function of DCregs, JAWS II cells were fed with 10 μg/ml pOVA323 and OVA323 or pVAX and OVA323 (added directly into the medium) for 24 h. CD4+ T cells were purified using CD4+ T Cell Isolation Kit II according to the manufacturer’s protocol (Miltenyi Biotec Inc.) from the spleen of mice that had been immunized with OVA in incomplete Freund's adjuvant (IFA) and labeled with CFSE. CFSE-CD4+ T cells were co-cultured with the treated DCs for 5 d and then T cell proliferation was measured as the degree of CFSE dilution, expression of Foxp3 was analyzed by intracellular staining, and production of IL-10 in supernatant was measured by Mouse IL-10 Flex Set from BD Biosciences.
Confocal Microscopy
To determine the localization of plasmid in DCs, pVAX and pOVA were first labeled with the Label IT® CyTM5 Intracellular Nucleic Acid Localization Kit (Mirus) and named as CyTM5-pVAX and CyTM5-pOVA. DCs were plated out into glass-bottom Petri dishes (MatTek) 4 d before the experiment. The cells were then incubated with the FITC-labeled OVA323 and the CyTM5-labeled pVAX or pOVA323 at 37°C for either 5 or 24 h at 10 mg/ml. The cells were then fixed with 4% paraformaldehyde for 10 min at 37°C, and permeabilized with 0.1% Triton X-100 in PBS buffer for 3 min. The cells were then incubated in blocking solution (PBS containing Fc-Block or 20% blocking serum for goat Fc) for 30 min. For the staining of caveolin-1, EEA1 and LAMP-2, cells were incubated with primary antibodies for 1 h. After washing three times with PBS, cells were incubated with the appropriate AF-labeled goat anti-rabbit IgG or PE-labeled goat anti-rat IgG for 30 min, and then observed using an inverted Nikon ECLIPSE IE2000-E confocal microscope. For DC-SIGN staining, cells were incubated with the appropriate APC-labeled or PE-labeled DC-SIGN antibodies for 30 min and observed using the inverted confocal microscope. Dual color images were acquired by sequential scanning, with only one laser line per scan to avoid cross-excitation. The images were processed using the software program Nikon EZ-C1 3.00 FreeViewer.
Flow cytometric (FACS) analysis
T cells were stained with the appropriate APC-Foxp3 in PBS for 30 min at 4°C, as described elsewhere.15,17 The cells were collected by flow cytometry (FACSCalibur) and analyzed with FlowJo software.
mRNA expression profiling
GeneChip Mouse genome 430 2.0 arrays (Affymetrix), containing probe sets for > 45,000 characterized genes and expressed sequence tags, were used. Sample labeling and processing, GeneChip hybridization, and scanning were performed according to Affymetrix protocols. Briefly, double-stranded cDNA was synthesized from total RNA with the SuperScript Choice System (Invitrogen), with a T7 RNA polymerase promoter site added to its 3′ end (Genset). Biotinylated cRNAs were generated from cDNAs in vitro and amplified by using the BioArray T7 RNA polymerase labeling kit (Enzo Life Science INC.). After purification of cRNAs by the RNeasy mini kit (Qiagen), 20 μg of cRNA was fragmented at 94°C for 35 min. Approximately 12.5 μg of fragmented cRNA was used in a 250-μl hybridization mixture containing herring-sperm DNA (0.1 mg/ml; Promega), plus bacterial and phage cRNA controls (1.5 pM BioB, 5 pM BioC, 25 pM BioD, and 100 pM Cre) to serve as internal controls for hybridization efficiency. Aliquots (200 μl) of the mixture were hybridized to arrays for 16 h at 45°C in a GeneChip Hybridization Oven 640 (Affymetrix). Each array was washed and stained with streptavidin–phycoerythrin (Invitrogen) and amplified with biotinylated anti-streptavidin antibody (Vector Laboratories) on the GeneChip Fluidics Station 450 (Affymetrix). Arrays were scanned with the GeneArray G7 scanner (Affymetrix) to obtain image and signal intensities.
Data analysis
After scanning, the images were processed using Affymetrix Expression Console™ Software Version 1.0 to generate gene expression intensity values. Arrays normalization was performed using MAS 5.0 algorithm. The top 2% and bottom 2% of signal intensities are excluded, then the mean is calculated. The original signal values are normalized to the same target intensity (TGT = 500).
Statistical analysis
The data were analyzed using the Mann-Whitney test. A p value of < 0.05 was considered to be statistically significant.
Disclosure of Potential Conflicts of Interest
This work was supported by grants from the Chinese National Natural Science Foundation (30930068) and Chinese High-Tech R&D Program (2012AA02A407 and 2013ZX09102041) to BW.
Glossary
Abbreviations:
- Cav-1
caveolin-1
- CD40low
low expression of CD40
- DC
dendritic cell
- DCreg
regulatory dendritic cell
- DC-SIGN
dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin
- EEA1
early endosome antigen 1
- iTreg
Ag-specific regulatory T cell
- LAMP-2
lysosome-associated membrane protein-2
- SOCS
suppressor of cytokine signaling
- Treg
regulatory T cell
- WT
wild-type
Footnotes
Previously published online: www.landesbioscience.com/journals/vaccines/article/25011
REFERENCE
- 1.Sakaguchi S. Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol. 2004;22:531–62. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- 2.Thornton AM, Shevach EM. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J Exp Med. 1998;188:287–96. doi: 10.1084/jem.188.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gondek DC, Lu L-F, Quezada SA, Sakaguchi S, Noelle RJ. Cutting edge: contact-mediated suppression by CD4+CD25+ regulatory cells involves a granzyme B-dependent, perforin-independent mechanism. J Immunol. 2005;174:1783–6. doi: 10.4049/jimmunol.174.4.1783. [DOI] [PubMed] [Google Scholar]
- 4.Cao X, Cai SF, Fehniger TA, Song J, Collins LI, Piwnica-Worms DR, et al. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity. 2007;27:635–46. doi: 10.1016/j.immuni.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 5.Roncarolo M-G, Levings MK, Traversari C. Differentiation of T regulatory cells by immature dendritic cells. J Exp Med. 2001;193:F5–9. doi: 10.1084/jem.193.2.F5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cottrez F, Hurst SD, Coffman RL, Groux H. T regulatory cells 1 inhibit a Th2-specific response in vivo. J Immunol. 2000;165:4848–53. doi: 10.4049/jimmunol.165.9.4848. [DOI] [PubMed] [Google Scholar]
- 7.Jonuleit H, Schmitt E, Schuler G, Knop J, Enk AH. Induction of interleukin 10-producing, nonproliferating CD4(+) T cells with regulatory properties by repetitive stimulation with allogeneic immature human dendritic cells. J Exp Med. 2000;192:1213–22. doi: 10.1084/jem.192.9.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levings MK, Gregori S, Tresoldi E, Cazzaniga S, Bonini C, Roncarolo MG. Differentiation of Tr1 cells by immature dendritic cells requires IL-10 but not CD25+CD4+ Tr cells. Blood. 2005;105:1162–9. doi: 10.1182/blood-2004-03-1211. [DOI] [PubMed] [Google Scholar]
- 9.Luo X, Tarbell KV, Yang H, Pothoven K, Bailey SL, Ding R, et al. Dendritic cells with TGF-β1 differentiate naive CD4+CD25- T cells into islet-protective Foxp3+ regulatory T cells. Proc Natl Acad Sci U S A. 2007;104:2821–6. doi: 10.1073/pnas.0611646104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganesh BB, Cheatem DM, Sheng JR, Vasu C, Prabhakar BS. GM-CSF-induced CD11c+CD8a--dendritic cells facilitate Foxp3+ and IL-10+ regulatory T cell expansion resulting in suppression of autoimmune thyroiditis. Int Immunol. 2009;21:269–82. doi: 10.1093/intimm/dxn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Ambrosio A, Colucci M, Pugliese O, Quintieri F, Boirivant M. Cholera toxin B subunit promotes the induction of regulatory T cells by preventing human dendritic cell maturation. J Leukoc Biol. 2008;84:661–8. doi: 10.1189/jlb.1207850. [DOI] [PubMed] [Google Scholar]
- 12.Lavelle EC, McNeela E, Armstrong ME, Leavy O, Higgins SC, Mills KHG. Cholera toxin promotes the induction of regulatory T cells specific for bystander antigens by modulating dendritic cell activation. J Immunol. 2003;171:2384–92. doi: 10.4049/jimmunol.171.5.2384. [DOI] [PubMed] [Google Scholar]
- 13.Jin H, Kang Y, Zheng G, Xie Q, Xiao C, Zhang X, et al. Induction of active immune suppression by co-immunization with DNA- and protein-based vaccines. Virology. 2005;337:183–91. doi: 10.1016/j.virol.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 14.Kang Y, Jin H, Zheng G, Du X, Xiao C, Zhang X, et al. Co-inoculation of DNA and protein vaccines induces antigen-specific T cell suppression. Biochem Biophys Res Commun. 2007;353:1034–9. doi: 10.1016/j.bbrc.2006.12.124. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Jin H, Zhang F, Du X, Zhao G, Yu Y, et al. Treatment of autoimmune ovarian disease by co-administration with mouse zona pellucida protein 3 and DNA vaccine through induction of adaptive regulatory T cells. J Gene Med. 2008;10:810–20. doi: 10.1002/jgm.1200. [DOI] [PubMed] [Google Scholar]
- 16.Jin H, Xiao C, Geng S, Hu Y, She R, Yu Y, et al. Protein/DNA vaccine-induced antigen-specific Treg confer protection against asthma. Eur J Immunol. 2008;38:2451–63. doi: 10.1002/eji.200737899. [DOI] [PubMed] [Google Scholar]
- 17.Jin H, Kang Y, Zhao L, Xiao C, Hu Y, She R, et al. Induction of adaptive T regulatory cells that suppress the allergic response by coimmunization of DNA and protein vaccines. J Immunol. 2008;180:5360–72. doi: 10.4049/jimmunol.180.8.5360. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Geng S, Xie X, Liu H, Zheng G, Sun X, et al. Caveolin-1-mediated negative signaling plays a critical role in the induction of regulatory dendritic cells by DNA and protein coimmunization. J Immunol. 2012;189:2852–9. doi: 10.4049/jimmunol.1102828. [DOI] [PubMed] [Google Scholar]
- 19.Smits HH, Engering A, van der Kleij D, de Jong EC, Schipper K, van Capel TM, et al. Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J Allergy Clin Immunol. 2005;115:1260–7. doi: 10.1016/j.jaci.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 20.Gringhuis SI, den Dunnen J, Litjens M, van Het Hof B, van Kooyk Y, Geijtenbeek TB. C-type lectin DC-SIGN modulates Toll-like receptor signaling via Raf-1 kinase-dependent acetylation of transcription factor NF-kappaB. Immunity. 2007;26:605–16. doi: 10.1016/j.immuni.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Peters PJ, Neefjes JJ, Oorschot V, Ploegh HL, Geuze HJ. Segregation of MHC class II molecules from MHC class I molecules in the Golgi complex for transport to lysosomal compartments. Nature. 1991;349:669–76. doi: 10.1038/349669a0. [DOI] [PubMed] [Google Scholar]
- 22.Harding CV, Geuze HJ. Class II MHC molecules are present in macrophage lysosomes and phagolysosomes that function in the phagocytic processing of Listeria monocytogenes for presentation to T cells. J Cell Biol. 1992;119:531–42. doi: 10.1083/jcb.119.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmer KP, Büning J, Weber P, Kaiserlian D, Strobel S. Modulation of antigen trafficking to MHC class II-positive late endosomes of enterocytes. Gastroenterology. 2000;118:128–37. doi: 10.1016/S0016-5085(00)70421-5. [DOI] [PubMed] [Google Scholar]
- 24.Krug A, Rothenfusser S, Hornung V, Jahrsdörfer B, Blackwell S, Ballas ZK, et al. Identification of CpG oligonucleotide sequences with high induction of IFN-alpha/beta in plasmacytoid dendritic cells. Eur J Immunol. 2001;31:2154–63. doi: 10.1002/1521-4141(200107)31:7<2154::AID-IMMU2154>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 25.Levy DE, Marié I, Prakash A. Ringing the interferon alarm: differential regulation of gene expression at the interface between innate and adaptive immunity. Curr Opin Immunol. 2003;15:52–8. doi: 10.1016/S0952-7915(02)00011-0. [DOI] [PubMed] [Google Scholar]
- 26.Geijtenbeek TB, Gringhuis SI. Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol. 2009;9:465–79. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenzie GJ, Khan M, Briend E, Stallwood Y, Champion BR. Notch: a unique therapeutic target for immunomodulation. Expert Opin Ther Targets. 2005;9:395–410. doi: 10.1517/14728222.9.2.395. [DOI] [PubMed] [Google Scholar]
- 28.Knisz J, Banks A, McKeag L, Metcalfe DD, Rothman PB, Brown JM. Loss of SOCS7 in mice results in severe cutaneous disease and increased mast cell activation. Clin Immunol. 2009;132:277–84. doi: 10.1016/j.clim.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Niedbala W, Cai B, Liew FY. Role of nitric oxide in the regulation of T cell functions. Ann Rheum Dis. 2006;65(Suppl 3):iii37–40. doi: 10.1136/ard.2006.058446. [DOI] [PMC free article] [PubMed] [Google Scholar]