Abstract
Signal transducer and activator of transcription 5 (STAT5) regulates normal lympho-myeloid development through activation downstream of early-acting cytokines, their receptors, and Janus kinases (JAKs). Despite a general understanding of the role of STAT5 in hematopoietic stem cell (HSC) proliferation, survival, and self-renewal, the transcriptional targets and mechanisms of gene regulation that control multi-lineage engraftment following transplantation for the most part remain to be understood. In this review, we focus on the role of STAT5 in HSC transplantation and recent developments toward identifying the relevant downstream target genes and their role as part of a pleiotropic STAT5 mediated signaling response.
Keywords: cytokine signaling, JAK-STAT, hematopoiesis, engraftment, quiescence, self-renewal, transplantation, stem cell
STAT5 Activation in Hematopoietic Stem and Progenitor Cells
Hematopoiesis is a finely tuned process with intricate feedback mechanisms for regulating production of mature blood cells. Many of the lineage commitment and differentiation decisions of hematopoietic cells are dictated by the delicate balance of transcription factors. Most basal transcription factors are constitutively active, but some require activation in order to translocate to the nucleus and regulate gene expression. JAK-STAT pathway activation provides a rapid way for extracellular signals to rapidly transduce signals resulting in STAT activation within minutes and gene expression within hours of the initial cytokine exposure. Once tyrosine phosphorylated, transcriptionally activated STATs accumulate in the nucleus.
Signal transducer and activator of transcription-5 (STAT5) comprises two separate genes,1 STAT5A and STAT5B, which collectively are major regulators of normal hematopoiesis with pleiotropic roles in hematopoietic stem cell (HSC),1-7 hematopoietic progenitor cell (HPC),8-10 and mature cell populations.11-14 Although the differences in phenotype of mice lacking STAT5A vs. STAT5B15,16 were primarily assumed to be due to differences in tissue specific gene expression, functional differences have been more recently discovered. For example, ERβ regulation appears to be mediated only by STAT5B,17 and STAT5B-deficient patients have been discovered that have reduced numbers of regulatory T cells and short stature.18-20 These phenotypes are consistent with major non-redundant roles for STAT5B in regulation of target genes such as FoxP3 and IGF1. Knockdown studies of STAT5A and STAT5B in human CD4+ T cells confirmed this specificity for FoxP321 and knockdown studies in IL-3 stimulated murine BaF3 cells identified overlapping and unique sets of STAT5 target genes by chromatin immunoprecipitation.22
Interestingly, the ability for STAT5 binding to modulate gene expression appears to be under additional levels of regulation such as serine phosphorylation23-25 and glycosylation26 which may influence interactions with CREB-binding protein. Serine phosphorylation has been described for most STATs27,28 although the positive or negative influence of this modification has been debated.29 The normal role of STAT5 serine phosphorylation in hematopoiesis has not been tested and the effect may be very cell type and cell context specific. Serine phosphorylation could facilitate interaction of STAT5 with critical accessory proteins required for nuclear localization of tyrosine phosphorylated STAT5. Cooperative signals mediated by serine and tyrosine phosphorylation could also be lineage-specific and further studies of serine phosphorylation deficient STAT5 mutants are needed to fully understand this level of regulation in HSC and throughout hematopoiesis.
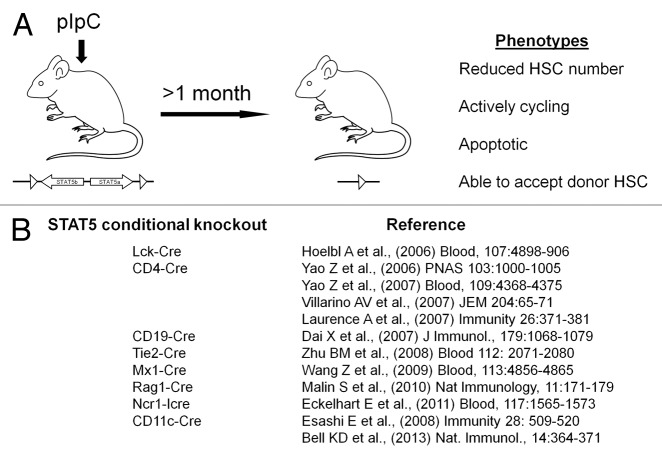
We have reported that STAT5 is required for HSC “fitness” with its deficiency resulting in greatly impaired long-term multilineage competitive repopulation capacity of fetal liver4,10 and bone marrow2-7 HSC in lethally-irradiated transplant recipients and a reciprocal permissiveness for wild-type donor HSC to engraft when transplanted into STAT5-deficient hosts in the absence of myeloablation.2,7,10 In our study, using interferon-induced Cre-mediated deletion, STAT5-deficient HSC were found to be persistently more actively cycling and become apoptotic, resulting in a reduction of long-term HSC number7 (Fig. 1A). However in this model the interferon response could result in an initial stimulation of the HSC pool and contribute to the loss of quiescence phenotype. Although our analyses were done many months post interferon induction, the role of STAT5 in the complete absence of induced interferon requires additional HSC-specific deletion approaches. Figure 1B summarizes the current state-of-the-art of STAT5 conditional knockout in hematopoietic lineages.
Figure 1. Conditional knockout strategies for STAT5 deletion in the hematopoietic system. (A) We have previously studied HSC with interferon-induced deletion of STAT5 using the Mx1-Cre system. This approach gives conditional deletion of STAT5 in adult mice. The limitation of the approach is the need to wait for the interferon response to subside before subsequent analysis. (B) Since 2006 various other groups have conditionally knocked out STAT5 floxed alleles of STAT5A and STAT5B that were generated by Lothar Hennighausen (NIDDK). These knockouts include expression of Cre recombinase in early hematopoietic stem cells (and vascular progenitors), as well as T cells, B cells, NK cells, and dendritic cell lineages.
STAT5 is activated by several early acting cytokines such as IL-3,30-32 thrombopoietin (TPO),33-37 granulocyte-colony stimulating factor (G-CSF),38,39 and granulocyte-macrophage colony stimulating factor (GM-CSF).38,40-42 Although initially all of these factors were believed to act on committee myeloid lineages, intracellular flow cytometry data from several groups using HSC/HPC fractions shows marked STAT5 activation.38,43,44 In HSCs, Mpl synergizes with early acting cytokines45-48 and regulates multilineage progenitors and HSC self-renewal.49-51 Interestingly, Mpl also promotes HSC quiescence.52 Our prior studies have found close parallels between Mpl deficiency and STAT5 deficiency2,3,7 (Fig. 2). These phenotypes include loss of multi-lineage engraftment ability, thrombocytopenia, decreased HSC self-renewal, loss of HSC quiescence, and decreased expression of Tie2 and p57. Although STAT5 may not be a major signaling partner utilized by TPO/Mpl in megakaryocytes and platelets, a direct functional link between Mpl/STAT5 is likely. Mpl promoter constructs driving STAT5A expression or generation of an inducible STAT5A expression vector crossed into a Mpl-deficient genetic background could address the role of STAT5 by testing rescue of function. Reciprocally, testing Mpl agonists in STAT5-deficient hosts might give important insight into whether STAT5 is essential for the hematopoiesis-promoting effects of these new clinically relevant agents.
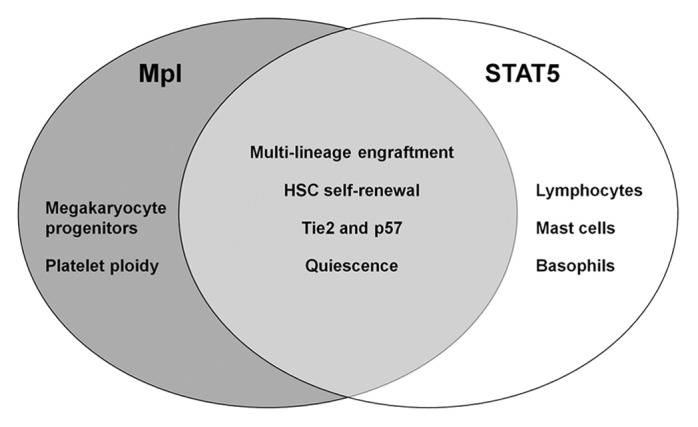
Figure 2. Comparison between STAT5 and Mpl knockout phenotypes. While c-Mpl deficiency causes severe megakaryocyte and platelet production defects, most other lymphoid and myeloid lineages are unaffected. Interestingly, Mpl knockout also selectively impairs early HSC/HPC function as determined by analysis of peripheral blood chimerism following transplantation. STAT5 knockout mice share the common HSC/HPC level defects, do not have intrinsic megakaryocyte/platelet defects, but have severe combined immunodeficiency that is unique to the role of STAT5 downstream of the common gamma chain and JAK3.
Interestingly, in hematopoiesis, most of the classical bifurcation points in lineage development are regulated by at least two transcription factors that have antagonistic functions. Examples include Notch/Pax5 and C/EBPα/GATA-1 where progressive changes in the dosage of these factors results in a shift of lineage potential. Comparable antagonism may exist at the hematopoietic stem cell/progenitor cell commitment decision. Mpl appears to control myeloid commitment and its downregulation is associated with increased lymphoid priming in HSC.53 It will be interesting to determine whether STAT5 similarly controls HSC fate decisions, perhaps downstream of Mpl. Recent studies of MPP heterogeneity, highlight downregulation of c-Mpl marking the transition to the lymphoid-primed multipotent progenitor (LMPP)53 and we have previously linked c-Mpl to STAT5 in HSC maintenance and self-renewal3,7 but have not examined lineage commitment. IL-7 is a major activator of STAT5 in the common lymphoid progenitor and the IL-7, common gamma chain, JAK3, STAT5 signaling axis is required for normal T and B lymphocyte development.54 Expression of the IL-7 receptor marks the transition toward the common lymphoid progenitor (CLP). Additionally CD150low HSC are functionally B-lymphoid-biased55-57 and express flk2 and CD86.55,58 Further, normal long-term repopulating HSC (LT-HSC) are CD150+48− but become B-lymphoid biased when localized to the central bone marrow microenvironment.59 Understanding the heterogeneity at all levels of hematopoiesis is currently a major area of study and new information is being obtained through techniques such as single cell PCR and lineage tracing with knock-in transgenic mouse models.
STAT5 is potently activated by IL-3/IL-5/GM-CSF all of which utilize the common β chain in the myeloid lineage to produce mast cells, basophils, eosinophils, granulocytes, and monocytes, including the basophil/mast cell progenitor.14,60 It is clear that STAT5 deletion causes significant development defects in both lymphoid and myeloid lineages. Interestingly, when STAT5 is deleted in fetal liver HSC, we have observed increased myelopoiesis and decreased lymphopoiesis following transplant into lethally-irradiated adult hosts.6 However, STAT5 is required at later stages for normal granulopoiesis.42 Therefore, STAT5 acts at many levels of hematopoiesis and the overall output in the peripheral blood lineages is a composite of the effects on HSC, primitive HPC, and the lineage committed progenitors. EPO is also a major activator of STAT5 in the erythroid lineage61-63 and it can synergize with SCF to activate STAT5,64 whereas activation of STAT5 by SCF alone in HSC/HPC still remains unclear.13,38,65,66 In addition to the canonical receptor/JAK interactions, cytokine mediated functional activation of STAT5 may depend on additional factors such as lipid rafts, Src family kinase activation, or recruitment by Grb2-associated binding proteins (Gabs). Prospective isolation of committed progenitor pools has not yet been tested, so it is difficult to separate the role of STAT5 in hematopoietic stem cells from the various progenitor cells and the frequency and output of progenitor populations must be considered when interpreting the outcome of long-term multilineage competitive repopulation assays.
STAT5 Regulation of Hematopoietic Stem Cell Transplantation
Recent advances in understanding HSC heterogeneity67,68 have defined a gradient of high CD150 expression regulating self-renewal potential, quiescence in long-term repopulating HSC (LT-HSC), and myeloid lineage bias and low CD150 expression associated with loss of self-renewal potential, loss of quiescence, and lymphoid-biased differentiation in short-term repopulating HSC (ST-HSC).55-57,69,70 While intermediate term HSC are beginning to be identified, their genetic control is not well characterized. Regulation of quiescence by STAT5 may be secondary to other defects in hematopoietic stem cell potential. Greater understanding of the impact of STAT5 on HSC heterogeneity and commitment may lead to improved methods for mobilization and stem cell engraftment in the absence of genotoxic myeloablative conditioning. It still remains to be determined whether enforced quiescence alone would be sufficient to correct function. Recently a method for induced quiescence was described that was based on pharmacologic stabilization of Hif-1α causing increased quiescence of HSC and accelerated recovery from myelosuppression.71
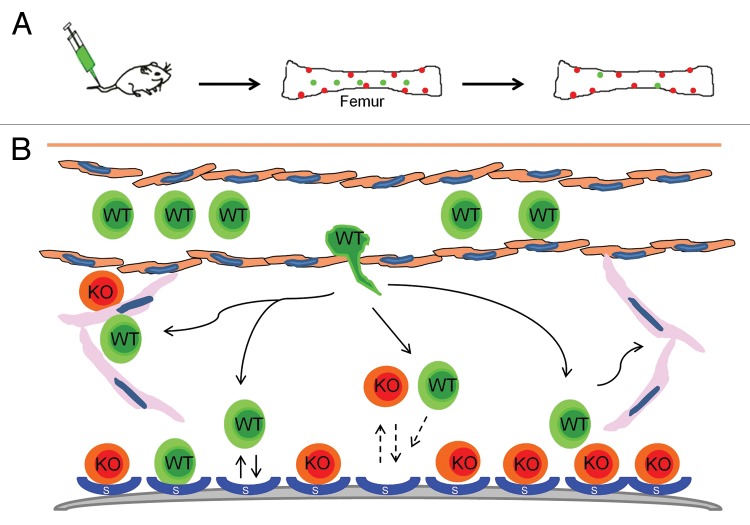
HSC niche occupancy is an essential ingredient for successful stem cell transplantation. In mice lacking STAT5, it is unclear why some niches are open and available to be filled by a donor graft. Interestingly, a percentage of the filled niches remain occupied by host cells and donor grafts do not eventually take over. Instead there is equilibrium between donor and host that is maintained long-term in the KLS compartment, although several peripheral blood lineages are dominated by wild-type grafts which selectively out-compete the STAT5-deficient host cells (Fig. 3). HSC could leave but not be able to compete for the niche once a specific set of HSC niches are filled. There are likely unique HSC and HPC niches and only the HPC niches may be fully open and able to accept wild-type donor HSC. HSC lacking STAT5 may also never leave the niche and instead act as a “dominant negative” by blocking niche space from the wild-type cells and continuing to produce blood cells, albeit at a reduced rate and without the production of mature T/B lymphocytes.
Figure 3. Possible hematopoietic stem/progenitor fates in the absence of STAT5. We have observed high levels of HSC/HPC engraftment in mice lacking STAT5. However, due to the strong multilineage selective growth advantage of the wild-type donor cells vs. the STAT5 knockout cells, it has been difficult to assess the impact in HSC vs. HPC compartments of the bone marrow. It is possible that only a subset of niches are vacated in the absence of STAT5 and that competition for these niches is not selectively impaired (homing, engraftment). STAT5 deficient HSC may not leave the niches as part of normal trafficking but rather prevent donor HSC from occupying niches. STAT5 deficient HPC may be vacated from putative HPC niches resulting in greater multilineage engraftment from multipotent progenitors.
Mechanistic studies have provided insight into how STAT5 modulates HSC engraftment and self-renewal. STAT5 transcriptional targets differ between human HSC and progenitor compartments.72 We observed that STAT5 promotes HSC quiescence and self-renewal but promotes proliferation in more differentiated cell types. More detailed molecular understanding of this dichotomy of function remains to be discovered. Roles for phosphorylated STAT5 in HSC vs. HPC might relate to the levels of phosphorylation. This is important because changes in phosphorylation might directly affect gene expression of distinct genes.73 STAT5 transcriptional function in cultured human cells has been described to involve Gata-dependent and independent pathways in control of megakaryocyte–erythrocyte development.74 STAT5 also controls key target genes such as Hif-2α to regulate HSC expansion in vitro.72 While constitutively active STAT5 cannot confer self-renewal on progenitors,75 STAT5 targets differ between HSC and progenitors in human cells.72 STAT5 is required for efficient lympho-myeloid repopulation and HSC quiescence7 and the hypoxia inducible gene Cited2 (STAT5 target gene) plays similar roles in fetal76 and adult HSC.77
Perspective on Targeting STAT5 in Normal and Leukemic Hematopoiesis
Recycling of STATs between nucleus and cytoplasm and retention in the nucleus only occurring following activation has been described.78,79 It is now clear that molecules such as Rac-GAP are important for STAT5 nuclear import.80 The negative regulation of this pathway is also critically important and much is known about the inducible negative regulators Cis and Socs. Hyper-activation of STAT5 is a validated leukemia driver and is currently a high value target for therapy. In chronic myeloid leukemia, unique roles for STAT5A and STAT5B in drug resistance downstream of BCR-ABL have been described.81 Activating mutations of STAT5 (STAT5B Y665F and N642H) in leukemia are also found in large granular lymphocyte (LGL) leukemia.82 With STAT5, it is clear that phosphorylation by JAKs is the major mechanism of activation; however, other mechanisms appear to play important roles in non-canonical activation of STAT5. These mechanisms include activation by Src family kinases downstream of Flt3 mutations, although conflicting results have been reported.83,84 Roles for integrin signaling and the spleen tyrosine kinase (Syk) in activation of STAT5 in AML blasts, including MLL-AF9 positive AML have also been reported.85,86
Interestingly, retention of phosphorylated STAT5 in the cytoplasm has been observed in leukemias and the cytoplasmic functions for STAT5 will be important to define. STAT5 nuclear accumulation occurs predominately in Flt3-ITD+ AML87 where other mutations and pathways could provide complimentary signals. Constitutively active STAT5 via Socs1 is capable of p53 activation and induction of an oncogene-induced cell senescence response.88,89 Therefore, negative regulatory mechanisms may be overcome in AML through alternative pathway activation. In contrast, BCR-ABL driven leukemogenesis in CML and ALL may require engagement of mechanisms to restrain STAT5 activation because of the principal requirement of STAT5 for leukemic progression and the absence of other compensating pathways. Notably, CML is associated with pSTAT5 localization in the cytoplasm at an unusually high level compared with normal cells.90 Since nuclear pSTAT5 might not be well tolerated in CML, understanding mechanisms controlling nuclear and cytoplasmic distribution of STAT5 has emerged as an important area.
It has been estimated that about half of STAT5 binding sites are in introns and half are in the promoter region.91,92 Nelson et al. propose about 200 highly conserved tandem STAT5 binding sites.91 Perhaps providing clues to the activity of STAT5 in HSC, transcriptional activation or repression determined by gene expression array has been completed in mouse embryonic fibroblasts lacking STAT5.93 Although STAT5 targets in various tissues such as liver94 may have some likely conservation with hematopoietic cells, the microenvironment of these diverse cell types is different and the intrinsic genetic programs and co-activators are different. Importantly, it has recently been shown that STAT binding patterns are cell type specific.95 Perhaps better understanding the HSC and HPC niches will elucidate novel mechanisms of gene regulation that are niche-dependent. Furthermore, better understanding of how STAT5 switches from activation to repression and on what sites in HSC that STAT5 is active, will be important for moving the field forward. Recent work has suggested that in lymphocyte development STAT5 tetramers play an important role in normal cytokine responses and normal immune function.96
Therapeutics for improving the transplantation into non-ablated hosts is the “Holy Grail” for many biological disorders. Also, the same principles may apply for leukemic HSC and their targeted therapy. STAT5 targeted small molecules have been recently described but more sophisticated efforts may also help such as targeting protein:protein interactions with the N-domain or by targeting STAT5 activation signatures. STAT5 serine phosphorylation also represents a potential new target.24 Identification of the serine kinase(s) could yield druggable targets capable of eradicating pSTAT5 in leukemias. Due to the prime role of STAT5 at the intersection of normal and leukemic hematopoiesis, innovative approaches for modulating STAT5 activation hold much promise for future therapeutic applications.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by NIH R01DK059380, The Cure Childhood Cancer Foundation, The Rally Foundation for Childhood Cancer Research, and the Aflac Cancer and Blood Disorders Center (KD Bunting).
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/27159
References
- 1.Grimley PM, Dong F, Rui H. Stat5a and Stat5b: fraternal twins of signal transduction and transcriptional activation. Cytokine Growth Factor Rev. 1999;10:131–57. doi: 10.1016/S1359-6101(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 2.Bradley HL, Hawley TS, Bunting KD. Cell intrinsic defects in cytokine responsiveness of STAT5-deficient hematopoietic stem cells. Blood. 2002;100:3983–9. doi: 10.1182/blood-2002-05-1602. [DOI] [PubMed] [Google Scholar]
- 3.Bradley HL, Couldrey C, Bunting KD. Hematopoietic-repopulating defects from STAT5-deficient bone marrow are not fully accounted for by loss of thrombopoietin responsiveness. Blood. 2004;103:2965–72. doi: 10.1182/blood-2003-08-2963. [DOI] [PubMed] [Google Scholar]
- 4.Bunting KD, Bradley HL, Hawley TS, Moriggl R, Sorrentino BP, Ihle JN. Reduced lymphomyeloid repopulating activity from adult bone marrow and fetal liver of mice lacking expression of STAT5. Blood. 2002;99:479–87. doi: 10.1182/blood.V99.2.479. [DOI] [PubMed] [Google Scholar]
- 5.Couldrey C, Bradley HL, Bunting KDA. A STAT5 modifier locus on murine chromosome 7 modulates engraftment of hematopoietic stem cells during steady-state hematopoiesis. Blood. 2005;105:1476–83. doi: 10.1182/blood-2004-06-2302. [DOI] [PubMed] [Google Scholar]
- 6.Li G, Wang Z, Zhang Y, Kang Z, Haviernikova E, Cui Y, Hennighausen L, Moriggl R, Wang D, Tse W, et al. STAT5 requires the N-domain to maintain hematopoietic stem cell repopulating function and appropriate lymphoid-myeloid lineage output. Exp Hematol. 2007;35:1684–94. doi: 10.1016/j.exphem.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Li G, Tse W, Bunting KD. Conditional deletion of STAT5 in adult mouse hematopoietic stem cells causes loss of quiescence and permits efficient nonablative stem cell replacement. Blood. 2009;113:4856–65. doi: 10.1182/blood-2008-09-181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snow JW, Abraham N, Ma MC, Abbey NW, Herndier B, Goldsmith MA. STAT5 promotes multilineage hematolymphoid development in vivo through effects on early hematopoietic progenitor cells. Blood. 2002;99:95–101. doi: 10.1182/blood.V99.1.95. [DOI] [PubMed] [Google Scholar]
- 9.Dai X, Chen Y, Di L, Podd A, Li G, Bunting KD, Hennighausen L, Wen R, Wang D. Stat5 is essential for early B cell development but not for B cell maturation and function. J Immunol. 2007;179:1068–79. doi: 10.4049/jimmunol.179.2.1068. [DOI] [PubMed] [Google Scholar]
- 10.Li G, Wang Z, Miskimen KL, Zhang Y, Tse W, Bunting KD. Gab2 promotes hematopoietic stem cell maintenance and self-renewal synergistically with STAT5. PLoS One. 2010;5:e9152. doi: 10.1371/journal.pone.0009152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnstein BO, Li G, Wang Z, Kennedy S, Chalfant C, Nakajima H, Bunting KD, Ryan JJ. Stat5 expression is required for IgE-mediated mast cell function. J Immunol. 2006;177:3421–6. doi: 10.4049/jimmunol.177.5.3421. [DOI] [PubMed] [Google Scholar]
- 12.Moriggl R, Topham DJ, Teglund S, Sexl V, McKay C, Wang D, Hoffmeyer A, van Deursen J, Sangster MY, Bunting KD, et al. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity. 1999;10:249–59. doi: 10.1016/S1074-7613(00)80025-4. [DOI] [PubMed] [Google Scholar]
- 13.Shelburne CP, McCoy ME, Piekorz R, Sexl V, Roh KH, Jacobs-Helber SM, Gillespie SR, Bailey DP, Mirmonsef P, Mann MN, et al. Stat5 expression is critical for mast cell development and survival. Blood. 2003;102:1290–7. doi: 10.1182/blood-2002-11-3490. [DOI] [PubMed] [Google Scholar]
- 14.Ohmori K, Luo Y, Jia Y, Nishida J, Wang Z, Bunting KD, Wang D, Huang H. IL-3 induces basophil expansion in vivo by directing granulocyte-monocyte progenitors to differentiate into basophil lineage-restricted progenitors in the bone marrow and by increasing the number of basophil/mast cell progenitors in the spleen. J Immunol. 2009;182:2835–41. doi: 10.4049/jimmunol.0802870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–86. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 16.Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, Waxman DJ, Davey HW. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci U S A. 1997;94:7239–44. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frasor J, Park K, Byers M, Telleria C, Kitamura T, Yu-Lee LY, Djiane J, Park-Sarge OK, Gibori G. Differential roles for signal transducers and activators of transcription 5a and 5b in PRL stimulation of ERalpha and ERbeta transcription. Mol Endocrinol. 2001;15:2172–81. doi: 10.1210/me.15.12.2172. [DOI] [PubMed] [Google Scholar]
- 18.Vidarsdottir S, Walenkamp MJ, Pereira AM, Karperien M, van Doorn J, van Duyvenvoorde HA, White S, Breuning MH, Roelfsema F, Kruithof MF, et al. Clinical and biochemical characteristics of a male patient with a novel homozygous STAT5b mutation. J Clin Endocrinol Metab. 2006;91:3482–5. doi: 10.1210/jc.2006-0368. [DOI] [PubMed] [Google Scholar]
- 19.Bernasconi A, Marino R, Ribas A, Rossi J, Ciaccio M, Oleastro M, Ornani A, Paz R, Rivarola MA, Zelazko M, et al. Characterization of immunodeficiency in a patient with growth hormone insensitivity secondary to a novel STAT5b gene mutation. Pediatrics. 2006;118:e1584–92. doi: 10.1542/peds.2005-2882. [DOI] [PubMed] [Google Scholar]
- 20.Cohen AC, Nadeau KC, Tu W, Hwa V, Dionis K, Bezrodnik L, Teper A, Gaillard M, Heinrich J, Krensky AM, et al. Cutting edge: Decreased accumulation and regulatory function of CD4+ CD25(high) T cells in human STAT5b deficiency. J Immunol. 2006;177:2770–4. doi: 10.4049/jimmunol.177.5.2770. [DOI] [PubMed] [Google Scholar]
- 21.Jenks JA, Seki S, Kanai T, Huang J, Morgan AA, Scalco RC, Nath R, Bucayu R, Wit JM, Al-Herz W, et al. Differentiating the roles of STAT5B and STAT5A in human CD4+ T cells. Clin Immunol. 2013;148:227–36. doi: 10.1016/j.clim.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basham B, Sathe M, Grein J, McClanahan T, D’Andrea A, Lees E, Rascle A. In vivo identification of novel STAT5 target genes. Nucleic Acids Res. 2008;36:3802–18. doi: 10.1093/nar/gkn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beuvink I, Hess D, Flotow H, Hofsteenge J, Groner B, Hynes NE. Stat5a serine phosphorylation. Serine 779 is constitutively phosphorylated in the mammary gland, and serine 725 phosphorylation influences prolactin-stimulated in vitro DNA binding activity. J Biol Chem. 2000;275:10247–55. doi: 10.1074/jbc.275.14.10247. [DOI] [PubMed] [Google Scholar]
- 24.Friedbichler K, Kerenyi MA, Kovacic B, Li G, Hoelbl A, Yahiaoui S, Sexl V, Müllner EW, Fajmann S, Cerny-Reiterer S, et al. Stat5a serine 725 and 779 phosphorylation is a prerequisite for hematopoietic transformation. Blood. 2010;116:1548–58. doi: 10.1182/blood-2009-12-258913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue HH, Fink DW, Jr., Zhang X, Qin J, Turck CW, Leonard WJ. Serine phosphorylation of Stat5 proteins in lymphocytes stimulated with IL-2. Int Immunol. 2002;14:1263–71. doi: 10.1093/intimm/dxf101. [DOI] [PubMed] [Google Scholar]
- 26.Gewinner C, Hart G, Zachara N, Cole R, Beisenherz-Huss C, Groner B. The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5. J Biol Chem. 2004;279:3563–72. doi: 10.1074/jbc.M306449200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Blenis J, Li HC, Schindler C, Chen-Kiang S. Requirement of serine phosphorylation for formation of STAT-promoter complexes. Science. 1995;267:1990–4. doi: 10.1126/science.7701321. [DOI] [PubMed] [Google Scholar]
- 28.Wen Z, Zhong Z, Darnell JEJ., Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–50. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 29.Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–37. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 30.Nosaka T, Kawashima T, Misawa K, Ikuta K, Mui AL, Kitamura T. STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells. EMBO J. 1999;18:4754–65. doi: 10.1093/emboj/18.17.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dumon S, Santos SC, Debierre-Grockiego F, Gouilleux-Gruart V, Cocault L, Boucheron C, Mollat P, Gisselbrecht S, Gouilleux F. IL-3 dependent regulation of Bcl-xL gene expression by STAT5 in a bone marrow derived cell line. Oncogene. 1999;18:4191–9. doi: 10.1038/sj.onc.1202796. [DOI] [PubMed] [Google Scholar]
- 32.Azam M, Erdjument-Bromage H, Kreider BL, Xia M, Quelle F, Basu R, Saris C, Tempst P, Ihle JN, Schindler C. Interleukin-3 signals through multiple isoforms of Stat5. EMBO J. 1995;14:1402–11. doi: 10.1002/j.1460-2075.1995.tb07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ooi J, Tojo A, Asano S, Sato Y, Oka Y. Thrombopoietin induces tyrosine phosphorylation of a common beta subunit of GM-CSF receptor and its association with Stat5 in TF-1/TPO cells. Biochem Biophys Res Commun. 1998;246:132–6. doi: 10.1006/bbrc.1998.8588. [DOI] [PubMed] [Google Scholar]
- 34.Bacon CM, Tortolani PJ, Shimosaka A, Rees RC, Longo DL, O’Shea JJ. Thrombopoietin (TPO) induces tyrosine phosphorylation and activation of STAT5 and STAT3. FEBS Lett. 1995;370:63–8. doi: 10.1016/0014-5793(95)00796-C. [DOI] [PubMed] [Google Scholar]
- 35.Drachman JG, Sabath DF, Fox NE, Kaushansky K. Thrombopoietin signal transduction in purified murine megakaryocytes. Blood. 1997;89:483–92. [PubMed] [Google Scholar]
- 36.Schulze H, Ballmaier M, Welte K, Germeshausen M. Thrombopoietin induces the generation of distinct Stat1, Stat3, Stat5a and Stat5b homo- and heterodimeric complexes with different kinetics in human platelets. Exp Hematol. 2000;28:294–304. doi: 10.1016/S0301-472X(99)00154-X. [DOI] [PubMed] [Google Scholar]
- 37.Kirito K, Watanabe T, Sawada K, Endo H, Ozawa K, Komatsu N. Thrombopoietin regulates Bcl-xL gene expression through Stat5 and phosphatidylinositol 3-kinase activation pathways. J Biol Chem. 2002;277:8329–37. doi: 10.1074/jbc.M109824200. [DOI] [PubMed] [Google Scholar]
- 38.Han L, Wierenga AT, Rozenveld-Geugien M, van de Lande K, Vellenga E, Schuringa JJ. Single-cell STAT5 signal transduction profiling in normal and leukemic stem and progenitor cell populations reveals highly distinct cytokine responses. PLoS One. 2009;4:e7989. doi: 10.1371/journal.pone.0007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu F, Kunter G, Krem MM, Eades WC, Cain JA, Tomasson MH, Hennighausen L, Link DC. Csf3r mutations in mice confer a strong clonal HSC advantage via activation of Stat5. J Clin Invest. 2008;118:946–55. doi: 10.1172/JCI32704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mui AL, Wakao H, O’Farrell AM, Harada N, Miyajima A. Interleukin-3, granulocyte-macrophage colony stimulating factor and interleukin-5 transduce signals through two STAT5 homologs. EMBO J. 1995;14:1166–75. doi: 10.1002/j.1460-2075.1995.tb07100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ko JS, Rayman P, Ireland J, Swaidani S, Li G, Bunting KD, Rini B, Finke JH, Cohen PA. Direct and differential suppression of myeloid-derived suppressor cell subsets by sunitinib is compartmentally constrained. Cancer Res. 2010;70:3526–36. doi: 10.1158/0008-5472.CAN-09-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimura A, Rieger MA, Simone JM, Chen W, Wickre MC, Zhu BM, Hoppe PS, O’Shea JJ, Schroeder T, Hennighausen L. The transcription factors STAT5A/B regulate GM-CSF-mediated granulopoiesis. Blood. 2009;114:4721–8. doi: 10.1182/blood-2009-04-216390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibbs KD, Jr., Gilbert PM, Sachs K, Zhao F, Blau HM, Weissman IL, Nolan GP, Majeti R. Single-cell phospho-specific flow cytometric analysis demonstrates biochemical and functional heterogeneity in human hematopoietic stem and progenitor compartments. Blood. 2011;117:4226–33. doi: 10.1182/blood-2010-07-298232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalaitzidis D, Neel BG. Flow-cytometric phosphoprotein analysis reveals agonist and temporal differences in responses of murine hematopoietic stem/progenitor cells. PLoS One. 2008;3:e3776. doi: 10.1371/journal.pone.0003776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ku H, Yonemura Y, Kaushansky K, Ogawa M. Thrombopoietin, the ligand for the Mpl receptor, synergizes with steel factor and other early acting cytokines in supporting proliferation of primitive hematopoietic progenitors of mice. Blood. 1996;87:4544–51. [PubMed] [Google Scholar]
- 46.Matsunaga T, Kato T, Miyazaki H, Ogawa M. Thrombopoietin promotes the survival of murine hematopoietic long-term reconstituting cells: comparison with the effects of FLT3/FLK-2 ligand and interleukin-6. Blood. 1998;92:452–61. [PubMed] [Google Scholar]
- 47.Sitnicka E, Lin N, Priestley GV, Fox N, Broudy VC, Wolf NS, Kaushansky K. The effect of thrombopoietin on the proliferation and differentiation of murine hematopoietic stem cells. Blood. 1996;87:4998–5005. [PubMed] [Google Scholar]
- 48.Ramsfjell V, Borge OJ, Veiby OP, Cardier J, Murphy MJ, Jr., Lyman SD, Lok S, Jacobsen SE. Thrombopoietin, but not erythropoietin, directly stimulates multilineage growth of primitive murine bone marrow progenitor cells in synergy with early acting cytokines: distinct interactions with the ligands for c-kit and FLT3. Blood. 1996;88:4481–92. [PubMed] [Google Scholar]
- 49.Kimura S, Roberts AW, Metcalf D, Alexander WS. Hematopoietic stem cell deficiencies in mice lacking c-Mpl, the receptor for thrombopoietin. Proc Natl Acad Sci U S A. 1998;95:1195–200. doi: 10.1073/pnas.95.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carver-Moore K, Broxmeyer HE, Luoh SM, Cooper S, Peng J, Burstein SA, Moore MW, de Sauvage FJ. Low levels of erythroid and myeloid progenitors in thrombopoietin-and c-mpl-deficient mice. Blood. 1996;88:803–8. [PubMed] [Google Scholar]
- 51.Alexander WS, Roberts AW, Nicola NA, Li R, Metcalf D. Deficiencies in progenitor cells of multiple hematopoietic lineages and defective megakaryocytopoiesis in mice lacking the thrombopoietic receptor c-Mpl. Blood. 1996;87:2162–70. [PubMed] [Google Scholar]
- 52.Qian H, Buza-Vidas N, Hyland CD, Jensen CT, Antonchuk J, Månsson R, Thoren LA, Ekblom M, Alexander WS, Jacobsen SE. Critical role of thrombopoietin in maintaining adult quiescent hematopoietic stem cells. Cell Stem Cell. 2007;1:671–84. doi: 10.1016/j.stem.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 53.Luc S, Anderson K, Kharazi S, Buza-Vidas N, Böiers C, Jensen CT, Ma Z, Wittmann L, Jacobsen SE. Down-regulation of Mpl marks the transition to lymphoid-primed multipotent progenitors with gradual loss of granulocyte-monocyte potential. Blood. 2008;111:3424–34. doi: 10.1182/blood-2007-08-108324. [DOI] [PubMed] [Google Scholar]
- 54.Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang Q, Bhandoola A, et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci U S A. 2006;103:1000–5. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Challen GA, Boles NC, Chambers SM, Goodell MA. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell. 2010;6:265–78. doi: 10.1016/j.stem.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med. 2010;207:1173–82. doi: 10.1084/jem.20091318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beerman I, Bhattacharya D, Zandi S, Sigvardsson M, Weissman IL, Bryder D, Rossi DJ. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc Natl Acad Sci U S A. 2010;107:5465–70. doi: 10.1073/pnas.1000834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimazu T, Iida R, Zhang Q, Welner RS, Medina KL, Alberola-Lla J, Kincade PW. CD86 is expressed on murine hematopoietic stem cells and denotes lymphopoietic potential. Blood. 2012;119:4889–97. doi: 10.1182/blood-2011-10-388736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grassinger J, Haylock DN, Williams B, Olsen GH, Nilsson SK. Phenotypically identical hemopoietic stem cells isolated from different regions of bone marrow have different biologic potential. Blood. 2010;116:3185–96. doi: 10.1182/blood-2009-12-260703. [DOI] [PubMed] [Google Scholar]
- 60.Qi X, Hong J, Chaves L, Zhuang Y, Chen Y, Wang D, Chabon J, Graham B, Ohmori K, Li Y, et al. Antagonistic regulation by the transcription factors C/EBPα and MITF specifies basophil and mast cell fates. Immunity. 2013;39:97–110. doi: 10.1016/j.immuni.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Socolovsky M, Fallon AE, Wang S, Brugnara C, Lodish HF. Fetal anemia and apoptosis of red cell progenitors in Stat5a-/-5b-/- mice: a direct role for Stat5 in Bcl-X(L) induction. Cell. 1999;98:181–91. doi: 10.1016/S0092-8674(00)81013-2. [DOI] [PubMed] [Google Scholar]
- 62.Silva M, Benito A, Sanz C, Prosper F, Ekhterae D, Nuñez G, Fernandez-Luna JL. Erythropoietin can induce the expression of bcl-x(L) through Stat5 in erythropoietin-dependent progenitor cell lines. J Biol Chem. 1999;274:22165–9. doi: 10.1074/jbc.274.32.22165. [DOI] [PubMed] [Google Scholar]
- 63.Oda A, Sawada K, Druker BJ, Ozaki K, Takano H, Koizumi K, Fukada Y, Handa M, Koike T, Ikeda Y. Erythropoietin induces tyrosine phosphorylation of Jak2, STAT5A, and STAT5B in primary cultured human erythroid precursors. Blood. 1998;92:443–51. [PubMed] [Google Scholar]
- 64.Boer AK, Drayer AL, Vellenga E. Stem cell factor enhances erythropoietin-mediated transactivation of signal transducer and activator of transcription 5 (STAT5) via the PKA/CREB pathway. Exp Hematol. 2003;31:512–20. doi: 10.1016/S0301-472X(03)00075-4. [DOI] [PubMed] [Google Scholar]
- 65.Harir N, Boudot C, Friedbichler K, Sonneck K, Kondo R, Martin-Lannerée S, Kenner L, Kerenyi M, Yahiaoui S, Gouilleux-Gruart V, et al. Oncogenic Kit controls neoplastic mast cell growth through a Stat5/PI3-kinase signaling cascade. Blood. 2008;112:2463–73. doi: 10.1182/blood-2007-09-115477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ryan JJ, Huang H, McReynolds LJ, Shelburne C, Hu-Li J, Huff TF, Paul WE. Stem cell factor activates STAT-5 DNA binding in IL-3-derived bone marrow mast cells. Exp Hematol. 1997;25:357–62. [PubMed] [Google Scholar]
- 67.Schroeder T. Hematopoietic stem cell heterogeneity: subtypes, not unpredictable behavior. Cell Stem Cell. 2010;6:203–7. doi: 10.1016/j.stem.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 68.Hock H. Some hematopoietic stem cells are more equal than others. J Exp Med. 2010;207:1127–30. doi: 10.1084/jem.20100950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kent DG, Copley MR, Benz C, Wöhrer S, Dykstra BJ, Ma E, Cheyne J, Zhao Y, Bowie MB, Zhao Y, et al. Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood. 2009;113:6342–50. doi: 10.1182/blood-2008-12-192054. [DOI] [PubMed] [Google Scholar]
- 70.Benveniste P, Frelin C, Janmohamed S, Barbara M, Herrington R, Hyam D, Iscove NN. Intermediate-term hematopoietic stem cells with extended but time-limited reconstitution potential. Cell Stem Cell. 2010;6:48–58. doi: 10.1016/j.stem.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 71.Forristal CE, Winkler IG, Nowlan B, Barbier V, Walkinshaw G, Levesque JP. Pharmacologic stabilization of HIF-1α increases hematopoietic stem cell quiescence in vivo and accelerates blood recovery after severe irradiation. Blood. 2013;121:759–69. doi: 10.1182/blood-2012-02-408419. [DOI] [PubMed] [Google Scholar]
- 72.Fatrai S, Wierenga AT, Daenen SM, Vellenga E, Schuringa JJ. Identification of HIF2alpha as an important STAT5 target gene in human hematopoietic stem cells. Blood. 2011;117:3320–30. doi: 10.1182/blood-2010-08-303669. [DOI] [PubMed] [Google Scholar]
- 73.Wierenga AT, Vellenga E, Schuringa JJ. Maximal STAT5-induced proliferation and self-renewal at intermediate STAT5 activity levels. Mol Cell Biol. 2008;28:6668–80. doi: 10.1128/MCB.01025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wierenga AT, Vellenga E, Schuringa JJ. Down-regulation of GATA1 uncouples STAT5-induced erythroid differentiation from stem/progenitor cell proliferation. Blood. 2010;115:4367–76. doi: 10.1182/blood-2009-10-250894. [DOI] [PubMed] [Google Scholar]
- 75.Kato Y, Iwama A, Tadokoro Y, Shimoda K, Minoguchi M, Akira S, Tanaka M, Miyajima A, Kitamura T, Nakauchi H. Selective activation of STAT5 unveils its role in stem cell self-renewal in normal and leukemic hematopoiesis. J Exp Med. 2005;202:169–79. doi: 10.1084/jem.20042541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Y, Haviernik P, Bunting KD, Yang YC. Cited2 is required for normal hematopoiesis in the murine fetal liver. Blood. 2007;110:2889–98. doi: 10.1182/blood-2007-01-066316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Du J, Chen Y, Li Q, Han X, Cheng C, Wang Z, Danielpour D, Dunwoodie SL, Bunting KD, Yang YC. HIF-1α deletion partially rescues defects of hematopoietic stem cell quiescence caused by Cited2 deficiency. Blood. 2012;119:2789–98. doi: 10.1182/blood-2011-10-387902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Swameye I, Muller TG, Timmer J, Sandra O, Klingmuller U. Identification of nucleocytoplasmic cycling as a remote sensor in cellular signaling by databased modeling. Proc Natl Acad Sci U S A. 2003;100:1028–33. doi: 10.1073/pnas.0237333100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iyer J, Reich NC. Constitutive nuclear import of latent and activated STAT5a by its coiled coil domain. FASEB J. 2008;22:391–400. doi: 10.1096/fj.07-8965com. [DOI] [PubMed] [Google Scholar]
- 80.Kawashima T, Bao YC, Nomura Y, Moon Y, Tonozuka Y, Minoshima Y, Hatori T, Tsuchiya A, Kiyono M, Nosaka T, et al. Rac1 and a GTPase-activating protein, MgcRacGAP, are required for nuclear translocation of STAT transcription factors. J Cell Biol. 2006;175:937–46. doi: 10.1083/jcb.200604073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Casetti L, Martin-Lannerée S, Najjar I, Plo I, Augé S, Roy L, Chomel JC, Lauret E, Turhan AG, Dusanter-Fourt I. Differential contributions of STAT5A and STAT5B to stress protection and tyrosine kinase inhibitor resistance of chronic myeloid leukemia stem/progenitor cells. Cancer Res. 2013;73:2052–8. doi: 10.1158/0008-5472.CAN-12-3955. [DOI] [PubMed] [Google Scholar]
- 82.Rajala HL, Eldfors S, Kuusanmäki H, van Adrichem AJ, Olson T, Lagström S, Andersson EI, Jerez A, Clemente MJ, Yan Y, et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013;121:4541–50. doi: 10.1182/blood-2012-12-474577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leischner H, Albers C, Grundler R, Razumovskaya E, Spiekermann K, Bohlander S, Rönnstrand L, Götze K, Peschel C, Duyster J. SRC is a signaling mediator in FLT3-ITD- but not in FLT3-TKD-positive AML. Blood. 2012;119:4026–33. doi: 10.1182/blood-2011-07-365726. [DOI] [PubMed] [Google Scholar]
- 84.Choudhary C, Brandts C, Schwable J, Tickenbrock L, Sargin B, Ueker A, Böhmer FD, Berdel WE, Müller-Tidow C, Serve H. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood. 2007;110:370–4. doi: 10.1182/blood-2006-05-024018. [DOI] [PubMed] [Google Scholar]
- 85.Miller PG, Al-Shahrour F, Hartwell KA, Chu LP, Järås M, Puram RV, Puissant A, Callahan KP, Ashton J, McConkey ME, et al. In Vivo RNAi screening identifies a leukemia-specific dependence on integrin beta 3 signaling. Cancer Cell. 2013;24:45–58. doi: 10.1016/j.ccr.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oellerich T, Oellerich MF, Engelke M, Münch S, Mohr S, Nimz M, Hsiao HH, Corso J, Zhang J, Bohnenberger H, et al. β2 integrin-derived signals induce cell survival and proliferation of AML blasts by activating a Syk/STAT signaling axis. Blood. 2013;121:3889–99, S1-66. doi: 10.1182/blood-2012-09-457887. [DOI] [PubMed] [Google Scholar]
- 87.Bunting KD, Xie XY, Warshawsky I, Hsi ED. Cytoplasmic localization of phosphorylated STAT5 in human acute myeloid leukemia is inversely correlated with Flt3-ITD. Blood. 2007;110:2775–6. doi: 10.1182/blood-2007-05-090969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–8. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Calabrese V, Mallette FA, Deschênes-Simard X, Ramanathan S, Gagnon J, Moores A, Ilangumaran S, Ferbeyre G. SOCS1 links cytokine signaling to p53 and senescence. Mol Cell. 2009;36:754–67. doi: 10.1016/j.molcel.2009.09.044. [DOI] [PubMed] [Google Scholar]
- 90.Harir N, Pecquet C, Kerenyi M, Sonneck K, Kovacic B, Nyga R, Brevet M, Dhennin I, Gouilleux-Gruart V, Beug H, et al. Constitutive activation of Stat5 promotes its cytoplasmic localization and association with PI3-kinase in myeloid leukemias. Blood. 2007;109:1678–86. doi: 10.1182/blood-2006-01-029918. [DOI] [PubMed] [Google Scholar]
- 91.Nelson EA, Walker SR, Li W, Liu XS, Frank DA. Identification of human STAT5-dependent gene regulatory elements based on interspecies homology. J Biol Chem. 2006;281:26216–24. doi: 10.1074/jbc.M605001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nelson EA, Walker SR, Alvarez JV, Frank DA. Isolation of unique STAT5 targets by chromatin immunoprecipitation-based gene identification. J Biol Chem. 2004;279:54724–30. doi: 10.1074/jbc.M408464200. [DOI] [PubMed] [Google Scholar]
- 93.Zhu BM, Kang K, Yu JH, Chen W, Smith HE, Lee D, Sun HW, Wei L, Hennighausen L. Genome-wide analyses reveal the extent of opportunistic STAT5 binding that does not yield transcriptional activation of neighboring genes. Nucleic Acids Res. 2012;40:4461–72. doi: 10.1093/nar/gks056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hosui A, Hennighausen L. Genomic dissection of the cytokine-controlled STAT5 signaling network in liver. Physiol Genomics. 2008;34:135–43. doi: 10.1152/physiolgenomics.00048.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kang K, Robinson GW, Hennighausen L. Comprehensive meta-analysis of Signal Transducers and Activators of Transcription (STAT) genomic binding patterns discerns cell-specific cis-regulatory modules. BMC Genomics. 2013;14:4. doi: 10.1186/1471-2164-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lin JX, Li P, Liu D, Jin HT, He J, Ata Ur Rasheed M, Rochman Y, Wang L, Cui K, Liu C, et al. Critical Role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function. Immunity. 2012;36:586–99. doi: 10.1016/j.immuni.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]