Abstract
JAK-STAT signaling pathway plays an important role in the cells’ development and homeostasis. Over the past decades, the studies have identified the role of the JAK-STAT pathway in cell proliferation and apoptosis. Here, we want to discuss that whether and how the JAK-STAT pathway affects the lipid metabolism of adipose tissue. A host of cytokines and hormones can regulate lipid metabolism through activating the JAK-STAT signaling pathway. Activated STATs can regulate lipid metabolism directly by influencing the expression of enzymes. We have summarized the relevant research and articles of JAK-STAT during the recent years. Within this review, we will introduce you the recent research and highlight the unresolved problems in understanding how JAK-STAT signaling pathway contribute to the lipid metabolism in mature adipocytes and preadipocytes. Dysregulation of the JAK-STAT pathway would lead to a multiple metabolism disorders and medicines for this signaling pathway maybe become a new idea for diseases such as metabolic syndrome, especially in children.
Keywords: JAK-STAT, lipid metabolism, adipocytes, preadipocytes, obesity
Introduction
JAK-STAT signaling pathway
The Janus kinase–signal transducers and activators of transcription (JAK-STAT) signaling pathway is a pleiotropic cascade used to transduce a multitude of signals for development and homeostasis in animals.1 These cellular signals involve in immunity, cancerogenesis,2 ontogenesis,3 inflammation,4,5 stem cell maintenance,6 neuron function7,8, and lipid metabolism.9 The dysregulation of the JAK-STAT pathway would cause disease, such as immunodeficiency, cancer, allergy,10 renal disease,11 hepatic disease,12,13 and so on.
To date, 4 members have been identified in JAK kinase family, including JAK1, JAK2, JAK3, and TYK2. JAK1 and JAK2 have been detected in adipocytes. They are critical to the role of JAK-STAT in fat tissue. JAK3 and TYK2 express in adipose tissue, but no evidence indicates that they are expressed in adipocytes.14 Each JAKs contains 4 mainly domains, including kinase domain, non-catalytic kinase-like domain (a dual-specificity protein kinase that negatively regulates cytokine signaling), phosphotyrosine binding domain and receptor binding domain.1,15,16 In humans, JAK1 maps to chromosome 1p32.3–p31.3; JAK2 maps to chromosome 9p24; JAK3 maps to chromosome 19p13.1 and TYK2 maps to chromosome 19p13.2.
The STAT proteins family contains 7 members (STATs 1, 2, 3, 4, 5A, 5B, and 6). STATs 1, 3, 5A, and 5B have been detected in adipocytes.17 STAT6 may be involved in the differentiation of preadipocytes. So far, there is no evidence to support the presence of STATs 2 and 4 in fat cells. These STAT genes all have been identified in three chromosomal clusters. In mouse, STATs 1 and 4 map to a region of chromosome 1 (equivalent to human 2q12–q33); STATs 3, 5A, and 5B map to a region of chromosome 11 (human 12q13–q14.1); and STATs 2 and 6 map to a region of chromosome 10 (human 17q11.1–q22).18 In contrast, another review shows that STATs 3, 5A, and 5B map to a region of chromosome 11 (human 17q11.2–q22); and STATs 2 and 6 map to a region of chromosome 10 (human 12 q13–q14.1).19 All STATs contain 5 domains, including oligomerization domain, coiled coil (protein interaction), DNA binding domain, phosphotyrosine binding domain, and transcriptional activation domain.1
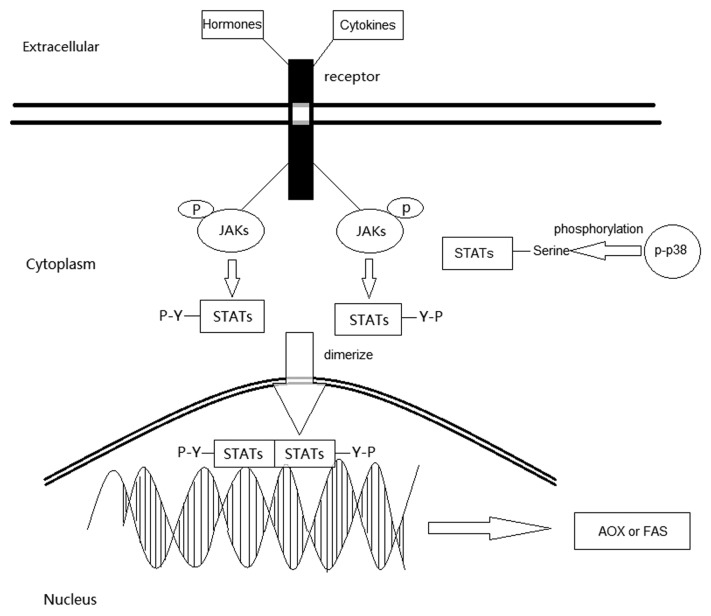
There are several forms of JAKs-STATs, including JAK1/3-STAT6, JAK1/2-STAT1/3/5, and so on.20,21 The activated JAK-STAT pathway begins with the combination of cytokine and its receptor. The receptor activates the associated JAKs, which in turn phosphorylate the receptor cytoplasmic domain to allow recruitment of a STAT. Several STATs, such as STAT, may also be directly activated by the cytokine-receptors. The STAT is phosphorylated, dimerizes, and moves to the nucleus to bind specific sequences in the genome and activate gene expression.22 There are 10 forms of STATs–STATs after phosphorylation. pSTATs 1, 2, 3, 4, 5 (5A and 5B interact in a manner as heterodimers), and 6 forms of 6 homodimers and 4 heterodimers. Few STAT2–STAT2 dimers form in the absence of STAT1 and bind target DNA sequence weakly as do STAT2:3. Other two heterodimers are STAT1:2(need p48) and STAT1:3 which is primarily involved in the cells’ apoptosis and inflammation.18 What we will review here is the homodimers of STATs-STATs. STATs could be posttranslationally modified by phosphorylation, methylation, acetylation, ubiquitylation, ISGylation, and SUMOylation. The tyrosine phosphorylation of STATs is necessary for its dimerization, nuclear translocation, and DNA binding.15 STATs can also be serine phosphorylated by MAPKs (mitogen-activated protein kinases), such as p38. The serine phosphorylation of the 727-site of STAT1 and STAT3 has been widely studied. Ser727 is of particular interest as a potential MAPK site, thus bringing STATs and MAPK together and allowing “cross-talk” among different cytokines receptor systems. An increase in transcriptional activation upon phosphorylation of Ser727 of STATs has been predicted from the prevailing (cross-talk) model. However, in cell lines, mutation of Ser727 in STAT1 displays a repressed effect and results in only 20% of wild-type activity.23 So, the effect of the serine phosphorylation remains to be clarified. It could promote or inhibit the tyrosine phospholyration.23-26 And the unphosphorylated STATs also mediates non-canonical function, such as angiotensin II-induced cardiac hypertrophy.27 In mitochondria, u-STAT3 participates in cellular respiration and mitosis.28,29 So far, Studies by independent groups have revealed that STATs 1, 3, 5A, 5B, and 6 express in fat cells.30
Lipid metabolism
Adipocytes play critical roles in lipid storage, energy homeostasis, and whole body insulin sensitivity. Approximately 90% of the adipocyte volume is TAG (triglycerides) which is located in an only lipid droplet that dislocates the nucleus to the periphery, resulting in limited cytosolic space.31 In normal individuals, the adipogenesis and lipolysis are in balance. And obesity results from an imbalance between energy intake and expenditure.32 The newest data show that the average prevalence of overweight and obesity of adults in China is 21.0% and 2.5%.33 With obesity, the individuals are in state of chronic inflammation and adipose tissue dysfunction.9 The expandability of adipose tissue determines the onset of metabolic syndrome, such as insulin resistance and diabetes type 2.34
Fat cells have two types, white adipocytes and brown adipocytes. White adipose tissue (WAT) plays important roles in energy storage through lipid release and glucose uptake. Utilizing the CM (chylomicron), VLDL, and glucose, WAT could synthetize TAG. Excess accumulation of white adipose tissue causes obesity. It can also release hormones and cytokines which contribute whole-body metabolism and insulin resistance as an endocrine tissue.35 Brown adipose tissue (BAT) contributes to energy expenditure because of its high content of mitochondria containing uncoupling protein-1 (UCP-1).9 Evidences have been provided to support the presence of BAT in adult humans except newborns and rodents. One study shows that as little as 50 g of maximally stimulated brown adipose tissue could account for up to 20% of daily energy expenditure in an adult human.36 Until recently, hormone-sensitive lipase (HSL) has been considered to be the key rate-limiting enzyme responsible for regulating TAG mobilization and a novel lipase named adipose triglyceride lipase/desnutrin (ATGL) has been identified as an important effector in the control of fat cell lipolysis.37 During physical training, TAG is first mobilized and hydrolyzed by HSL, ATGL, and monoacylglycerol lipase (MGL), each of which possesses a distinct regulatory mechanism. Eventually, it becomes FFA (free fatty acid). A great deal of energy is released during the oxygenolysis of TG. The FFA could be delivered to and used by the function organs such as heart and muscle with the help of plasma albumin.37 It is important to note that the developmental origin of white and brown fat is distinct and different precursor cells are involved in the generation of these different types of adipose tissue.38 The classical pathway of lipolysis activation in adipose tissue is cAMP-dependent. The production of cAMP is modulated by G-protein-coupled receptors of the Gs/Gi family and cAMP degradation is regulated by phosphodiesterase. Several other pathways involved in the lipolysis such as MAPK, JAK-STAT have been identified in recent years.39 As we know, leptin is a well-known hormone marker for obesity which can promote TAG hydrolysis. It has confirmed that leptin can upregulate ATGL mRNA expression but downregulate ATGL protein expression.40 With specific inhibitors of JAK2 and p38-MAPK, the ATGL mRNA expression is decreased and ATGL protein expression is increased significantly. It demonstrates that leptin-mediated regulation of ATGL expression is regulated by the JAK-STAT and MAPK signaling pathways (Fig. 1).40
Figure 1. The signaling pathway of JAK-STAT.
JAK-STAT and Lipid Metabolism
In mature adipocytes
STAT1
IFN-γ has been identified as an activator of STAT1. IFNγ induces robust STAT1 phosphorylation and SOCS1 mRNA expression, with modest, transient STAT3 phosphorylation and SOCS3 induction. Specific inhibition of JAK2 or JAK3 failed to block the effects of IFN-γ, suggesting a predominant role for JAK1–STAT1.41 The activated STAT1 proteins bind to the promoter of PPAR-γ2 (peroxisome proliferator-activated receptor-gamma 2) and lipoprotein lipase (LPL). It induces a repress action on the expression of PPAR-γ2 and LPL. PPAR-γ2 is an activator of adipogenesis42 while LPL is the rate-limiting enzyme that catalyzes the hydrolysis of serum triglycerides from lipoproteins into free fatty acids for uptake and storage in adipose tissue. Therefore, it indicates that STAT1 possibly promotes adipogenesis and inhibits lipolysis in mature fat cells.30 The other identified activators include LIF, OSM, CT-1, GH, and IL-11.14
STAT3
Many cytokines can generate their signal via the STAT3 signaling pathway which mediates a potent anti-inflammation response dependent on its target gene, suppressor of cytokine signaling-3 (SOCS3).43-45 STAT3 is involved in distinct functions in different cells.46 The ASKO mice which was created with an adipocyte-specific disruption of the STAT3 gene, had a higher weight and more adipose tissue mass than their littermate controls, associated with adipocyte hypertrophy but without adipocyte hyperplasia, hyperphagia, or reduced energy expenditure.47 This observation indicates that STAT3 promotes lipolysis and inhibits adipogenesis. As described previously, with the inhibitor of JAK2, JAK2’s downstream target, STAT3 is inhibited as a result. So, STAT3 maybe contribute to the expression of ATGL.40,48 Likewise, the ability of STAT3 inhibitor, STATTIC, to attenuate lipolysis and ATGL protein abundance in bovine adipocytes indicates leptin induced lipolysis may be regulated by a STAT3 mediated increasing in ATGL protein abundance.49 Whereas JAK-STAT has no effect on the HSL expression.50 But the STAT3-induced promotion of lipolysis is inconsistent with the fact that STAT3 inhibits inflammation which promotes lipolysis and reduces lipid storage.51,52 TNF-α, a factor of inflammation, induces the lipolysis and inhibits adipogenesis in WAT.53 Numerous of studies have indicated that pSTAT3 activated by cytokines, such as IL-10, can inhibit the LPS-induced expression of TNF-α.54,55 Moreover, in hepatocyte, forced expression of STAT3 reduces abundance of mRNAs for fatty acid synthase and acetyl-CoA carboxylase. And the amount of mRNA for acyl-CoA oxidase (AOX), which contributes to β-oxidation, is also decreased by overexpression of STAT3.12 The activators include LIF, OSM, CNTF, CT-1, IFN-γ, NP, GH, Leptin, IL-6, IL-10, and IL-11.56-59
STAT5
In adipocytes, AOX and fatty acid synthase (FAS) are identified to be the target genes of STAT5.60,61 A STAT5A binding site in FAS and AOX promoter mediates the repression of FAS expression while promotes the expression of AOX.9,30 Therefore STAT5 possibly induces an effect on the lipid metabolism like STAT3. Studies have shown that the adiogenic effect of GH could be suppressed by overexpression of STAT5A mutant (STAT5A–Y694F) with the reduction of PPARγ2 expression. In this research, STAT5A/5B stimulated C/EBPβ (C/AAAT enhancer binding protein β)- and C/EBPδ-induced adipogenesis with enhancement of PPARγ2 expression. In addition, STAT5A/5B enhanced the transcriptional activity of C/EBPβ/δ in the PPARγ gene promoter. Furthermore, STAT5A/5B stimulated PPARγ-induced adipogenesis and enhanced the transcriptional activity of PPARγ. These results suggest that the STAT5A/5B signaling pathway could stimulate adipogenesis cooperated with C/EBPβ/δ and PPARγ in the GH-induced lipid metabolism.62 STAT5 may be involved in the expression of ATGL like STAT3.39,48 And they may play a dual role in the lipid metabolism according to the activator. The activators of STAT5 (STAT5A and STAT5B) include GH and PRL.14
STAT6
Studies have indicated that only IL-4 can activate STAT6, and it has been shown to activate this transcription factor in 3T3-L1 preadipocytes but not in adipocytes.63
In preadipocytes
STAT1
On one hand, STATs tend to promote lipolysis in mature adipocyte. On the other hand, STATs tend to induce proliferation and promote adipogenesis of preadipocytes. But in cultured murine and human adipocytes, the protein level of STAT1 is downregulated during differentiation. It indicates that STAT1 may not promote adipogenesis during the differentiation of adipocytes.14 Consistently, another study shows that IFN-γ may suppress differentiation in human adipocytes via JAK-STAT1.64 Treatment with IFNγ could block differentiation of pre-adipocytes to the mature phenotype.41 Therefore, STAT1 in preadipocytes tends to promote lipolysis and repress adipogenesis.
STAT3
In preadipocytes, the target genes of STAT3 has been identified as C/EBPβ. Studies have showed that JAK2–STAT3 pathway is involved in the early stage of 3T3-L1 adipocyte differentiation though regulating the C/EBPβ transcription.65 pSTAT3 binds to its target gene, C/EBPβ, and promote the proteins level which in turn promote adipogenesis.14 C/EBP transcription factors (including C/EBP α, β, and δ) are the first family of transcription factors shown to play a critical role in the differentiation of fat cells in vitro. Mice lacking both C/EBPβ and δ or α alone exhibits a defective differentiation.66With the inhibiting factors of STAT3, such as AG490, dimethylfumarate and STAT3 siRNA, adipocyte differentiation would be suppressed. And PPAR-γ agonist may abolish the inhibitor-STAT3-induced suppression of adipogenesis, suggesting that STAT3 regulates adipocyte differentiation through PPAR-γ.67,68 Additional researches support that STAT3 promote the differentiation of preadipocytes. They have shown that pSTAT3 may promote fat cell differentiation via modulating the progress of mitotic clonal expansion, a proliferative phase that occurs immediately following induction of adipogenesis.69
STAT5
In preadipocytes, the target genes of STAT5 have been identified as C/EBPα and PPAR-γ. STAT5 induces adipogenesis of preadipocytes. STAT5, especially STAT5A, may bind to the promoter of C/EBPα and PPARγ3 and promote the expression of C/EBPα and PPARγ, which in turn induce adipogenesis of preadipocytes during its differentiation. STAT5B may enhance the STAT5A-induced preadipocytes differentiation.9 STAT5A−/− or STAT5B−/− or STAT5−/− mice exhibits reduced fat pad sizes compared with wild-type mice.66 And the athymic mice injected with Swiss 3T3 cells expressing STAT5A may result in fat pad formation at the site of injection.70 Another research also showed that virus-mediated gene transfer of the constitutively active form of both STAT5A and STAT5B resulted in enhanced adipocyte differentiation in the absence of fetal bovine serum, which was judged by the expression of proadipogenic factors as well as accumulation of fat droplets in the cell line.71 STAT5 may also modulate the proteins level of adiponectin which contributes to the lipid metabolism of adipocytes.
STAT6
STAT6 can be activated by IL-4, but its expression is not regulated during fat cell development and is uniform in preadipocytes.14 So the effect of STAT6 in lipid metabolism remains to be clarified.
Conclusion and Outlook
In conclusion, adipose tissue is the only tissue capable of hydrolyzing its stores of TAG and of mobilizing fatty acids and glycerol in the bloodstream so that they can be used by other tissues, such as heart and muscle. Disorder of lipid metabolism may result in metabolic syndrome including obesity, fatty liver, and insulin resistance and so on. Many agents and pathways involved in the regulation of lipolysis and adipogenesis in adipose tissue have been clarified. A host of cytokines and hormones can regulate lipid metabolism through activating the JAK-STAT signaling pathway. Modulation of the JAKs–STATs pathway can regulate lipid metabolism in adipocyte. Activated STATs can regulate lipid metabolism directly by influencing the expression of enzymes, such as AOX and FAS. Meanwhile, STATs also play roles in inflammation and anti-inflammation, which could impact on the lipid metabolism. But a host of problems remain to be resolved. Further studies are necessary to identify the other target genes of STATs in adipocyte and to determine how modifications such as serine phosphorylation, acetylation, methylation and sumoylation impact on STATs. How JAKs-STATs impact on the expression of ATGL? Whether JAKs–STATs participate in the HSL or MGL-induced lipolysis? And the JAKs-STATs pathway in BAT remains to be elucidated. Although the roles of different JAKs-STATs in the lipid metabolism of adipocytes and preadipocytes have been studied with cell lines and animal models as reviewed, it will be important to continue to explore the roles and mechanisms of JAKs-STATs in human adipose tissue because of the impact of species variation. The dysregulation of JAKs-STATs pathway would lead to a multiple metabolism disorders and medicines aimed at this signaling pathway maybe become a new idea for metabolic syndrome.72
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/27203
References
- 1.Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117:1281–3. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 2.O’Shea JJ, Holland SM, Staudt LM. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med. 2013;368:161–70. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaposhnikov AV, Komar’kov IF, Lebedeva LA, Shidlovskii YV. Molecular components of JAK/STAT signaling pathway and its interaction with transcription machinery. Mol Biol. 2013;47:343–51. doi: 10.1134/S0026893313030126. [DOI] [PubMed] [Google Scholar]
- 4.Stepkowski SM, Chen W, Ross JA, Nagy ZS, Kirken RA. STAT3: an important regulator of multiple cytokine functions. Transplantation. 2008;85:1372–7. doi: 10.1097/TP.0b013e3181739d25. [DOI] [PubMed] [Google Scholar]
- 5.Kaplan MH. STAT signaling in inflammation. JAKSTAT. 2013;2:e24198. doi: 10.4161/jkst.24198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stine R, Matunis E. JAK-STAT Signaling in Stem Cells. In: Hime G, Abud H, eds. Transcriptional and Translational Regulation of Stem Cells: Springer Netherlands, 2013:247-67. [Google Scholar]
- 7.Nicolas CS, Amici M, Bortolotto ZA, Doherty A, Csaba Z, Fafouri A, Dournaud P, Gressens P, Collingridge GL, Peineau S. The role of JAK-STAT signaling within the CNS. JAKSTAT. 2013;2:e22925. doi: 10.4161/jkst.22925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicolas CS, Peineau S, Amici M, Csaba Z, Fafouri A, Javalet C, Collett VJ, Hildebrandt L, Seaton G, Choi SL, et al. The Jak/STAT pathway is involved in synaptic plasticity. Neuron. 2012;73:374–90. doi: 10.1016/j.neuron.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richard AJ, Stephens JM. The role of JAK-STAT signaling in adipose tissue function. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbadis.2013.05.030. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morales JK, Falanga YT, Depcrynski A, Fernando J, Ryan JJ. Mast cell homeostasis and the JAK-STAT pathway. Genes Immun. 2010;11:599–608. doi: 10.1038/gene.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuang PY, He JC. JAK/STAT signaling in renal diseases. Kidney Int. 2010;78:231–4. doi: 10.1038/ki.2010.158. [DOI] [PubMed] [Google Scholar]
- 12.Kinoshita S, Ogawa W, Okamoto Y, Takashima M, Inoue H, Matsuki Y, Watanabe E, Hiramatsu R, Kasuga M. Role of hepatic STAT3 in the regulation of lipid metabolism. Kobe J Med Sci. 2008;54:E200–8. [PubMed] [Google Scholar]
- 13.Mair M, Blaas L, Österreicher CH, Casanova E, Eferl R. JAK-STAT signaling in hepatic fibrosis. Front Biosci (Landmark Ed) 2011;16:2794–811. doi: 10.2741/3886. [Landmark Ed] [DOI] [PubMed] [Google Scholar]
- 14.Richard AJ, Stephens JM. Emerging roles of JAK-STAT signaling pathways in adipocytes. Trends Endocrinol Metab. 2011;22:325–32. doi: 10.1016/j.tem.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–11. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 16.Ungureanu D, Wu J, Pekkala T, Niranjan Y, Young C, Jensen ON, Xu CF, Neubert TA, Skoda RC, Hubbard SR, et al. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat Struct Mol Biol. 2011;18:971–6. doi: 10.1038/nsmb.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephens JM, Morrison RF, Pilch PF. The expression and regulation of STATs during 3T3-L1 adipocyte differentiation. J Biol Chem. 1996;271:10441–4. doi: 10.1074/jbc.271.18.10441. [DOI] [PubMed] [Google Scholar]
- 18.Darnell JE., Jr. STATs and gene regulation. Science. 1997;277:1630–5. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 19.Ihle JN. The Stat family in cytokine signaling. Curr Opin Cell Biol. 2001;13:211–7. doi: 10.1016/S0955-0674(00)00199-X. [DOI] [PubMed] [Google Scholar]
- 20.Meier C, Hoeller S, Bourgau C, Hirschmann P, Schwaller J, Went P, Pileri SA, Reiter A, Dirnhofer S, Tzankov A. Recurrent numerical aberrations of JAK2 and deregulation of the JAK2-STAT cascade in lymphomas. Mod Pathol. 2009;22:476–87. doi: 10.1038/modpathol.2008.207. [DOI] [PubMed] [Google Scholar]
- 21.Heinrich T, Rengstl B, Muik A, Petkova M, Schmid F, Wistinghausen R, Warner K, Crispatzu G, Hansmann ML, Herling M, et al. Mature T-cell lymphomagenesis induced by retroviral insertional activation of Janus kinase 1. Mol Ther. 2013;21:1160–8. doi: 10.1038/mt.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray PJ. The JAK-STAT signaling pathway: input and output integration. J Immunol. 2007;178:2623–9. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- 23.Wen Z, Zhong Z, Darnell JE., Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–50. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 24.Aaronson DS, Horvath CM. A road map for those who don’t know JAK-STAT. Science. 2002;296:1653–5. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 25.Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–37. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 26.Chung J, Uchida E, Grammer TC, Blenis J. STAT3 serine phosphorylation by ERK-dependent and -independent pathways negatively modulates its tyrosine phosphorylation. Mol Cell Biol. 1997;17:6508–16. doi: 10.1128/mcb.17.11.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yue H, Li W, Desnoyer R, Karnik SS. Role of nuclear unphosphorylated STAT3 in angiotensin II type 1 receptor-induced cardiac hypertrophy. Cardiovasc Res. 2010;85:90–9. doi: 10.1093/cvr/cvp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohr A, Chatain N, Domoszlai T, Rinis N, Sommerauer M, Vogt M, Müller-Newen G. Dynamics and non-canonical aspects of JAK/STAT signalling. Eur J Cell Biol. 2012;91:524–32. doi: 10.1016/j.ejcb.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Stark GR, Darnell JE., Jr. The JAK-STAT pathway at twenty. Immunity. 2012;36:503–14. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao P, Stephens JM. Identification of STAT target genes in adipocytes. JAKSTAT. 2013;2:e23092. doi: 10.4161/jkst.23092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson BR, Lobo S, Bernlohr DA. Fatty acid flux in adipocytes: the in’s and out’s of fat cell lipid trafficking. Mol Cell Endocrinol. 2010;318:24–33. doi: 10.1016/j.mce.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hossain P, Kawar B, El Nahas M. Obesity and diabetes in the developing world--a growing challenge. N Engl J Med. 2007;356:213–5. doi: 10.1056/NEJMp068177. [DOI] [PubMed] [Google Scholar]
- 33.Tian X, Zhao G, Li Y, Wang L, Shi Y. Overweight and Obesity Difference of Chinese Population Between Different Urbanization Levels. J Rural Health. 2013 doi: 10.1111/jrh.12041. Forthcoming. [DOI] [PubMed] [Google Scholar]
- 34.Mlinar B, Marc J. New insights into adipose tissue dysfunction in insulin resistance. Clin Chem Lab Med. 2011;49:1925–35. doi: 10.1515/CCLM.2011.697. [DOI] [PubMed] [Google Scholar]
- 35.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–53. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–17. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lafontan M, Langin D. Lipolysis and lipid mobilization in human adipose tissue. Prog Lipid Res. 2009;48:275–97. doi: 10.1016/j.plipres.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Lefterova MI, Lazar MA. New developments in adipogenesis. Trends Endocrinol Metab. 2009;20:107–14. doi: 10.1016/j.tem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Chaves VE, Frasson D, Kawashita NH. Several agents and pathways regulate lipolysis in adipocytes. Biochimie. 2011;93:1631–40. doi: 10.1016/j.biochi.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 40.Li YC, Zheng XL, Liu BT, Yang GS. Regulation of ATGL expression mediated by leptin in vitro in porcine adipocyte lipolysis. Mol Cell Biochem. 2010;333:121–8. doi: 10.1007/s11010-009-0212-4. [DOI] [PubMed] [Google Scholar]
- 41.McGillicuddy FC, Chiquoine EH, Hinkle CC, Kim RJ, Shah R, Roche HM, Smyth EM, Reilly MP. Interferon γ attenuates insulin signaling, lipid storage, and differentiation in human adipocytes via activation of the JAK/STAT pathway. J Biol Chem. 2009;284:31936–44. doi: 10.1074/jbc.M109.061655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J-J, Toy WC, Liu S, Cheng A, Lim BK, Subramaniam T, Sum CF, Lim SC. Acetyl-keto-β-boswellic acid induces lipolysis in mature adipocytes. Biochem Biophys Res Commun. 2013;431:192–6. doi: 10.1016/j.bbrc.2012.12.136. [DOI] [PubMed] [Google Scholar]
- 43.El Kasmi KC, Holst J, Coffre M, Mielke L, de Pauw A, Lhocine N, Smith AM, Rutschman R, Kaushal D, Shen Y, et al. General nature of the STAT3-activated anti-inflammatory response. J Immunol. 2006;177:7880–8. doi: 10.4049/jimmunol.177.11.7880. [DOI] [PubMed] [Google Scholar]
- 44.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–65. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 45.Murray PJ. STAT3-mediated anti-inflammatory signalling. Biochem Soc Trans. 2006;34:1028–31. doi: 10.1042/BST0341028. [DOI] [PubMed] [Google Scholar]
- 46.Levy DE, Lee CK. What does Stat3 do? J Clin Invest. 2002;109:1143–8. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cernkovich ER, Deng J, Bond MC, Combs TP, Harp JB. Adipose-specific disruption of signal transducer and activator of transcription 3 increases body weight and adiposity. Endocrinology. 2008;149:1581–90. doi: 10.1210/en.2007-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frühbeck G. Intracellular signalling pathways activated by leptin. Biochem J. 2006;393:7–20. doi: 10.1042/BJ20051578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koltes DA, Spurlock DM. Appendix A. Translocation of adipose triglyceride lipase to the lipid droplet increased with leptin treatment in bovine adipocytes. Novel mechanisms involved with lipid metabolism in adipose tissue of dairy cows 2013:150. [Google Scholar]
- 50.Bergan HE, Kittilson JD, Sheridan MA. PKC and ERK mediate GH-stimulated lipolysis. J Mol Endocrinol. 2013;51:213–24. doi: 10.1530/JME-13-0039. [DOI] [PubMed] [Google Scholar]
- 51.Langin D, Mouisel E. Adipose Tissue Lipolysis. In: Bastard J-P, Fève B, eds. Physiology and Physiopathology of Adipose Tissue: Springer Paris, 2013:141-57. [Google Scholar]
- 52.Wang Y, Wang H, Hegde V, Dubuisson O, Gao Z, Dhurandhar NV, Ye J. Interplay of pro- and anti-inflammatory cytokines to determine lipid accretion in adipocytes. Int J Obes (Lond) 2013;37:1490–8. doi: 10.1038/ijo.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torres-Leal FL, Fonseca-Alaniz MH, Rogero MM, Tirapegui J. The role of inflamed adipose tissue in the insulin resistance. Cell Biochem Funct. 2010;28:623–31. doi: 10.1002/cbf.1706. [DOI] [PubMed] [Google Scholar]
- 54.Kontoyiannis D, Kotlyarov A, Carballo E, Alexopoulou L, Blackshear PJ, Gaestel M, Davis R, Flavell R, Kollias G. Interleukin-10 targets p38 MAPK to modulate ARE-dependent TNF mRNA translation and limit intestinal pathology. EMBO J. 2001;20:3760–70. doi: 10.1093/emboj/20.14.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niemand C, Nimmesgern A, Haan S, Fischer P, Schaper F, Rossaint R, Heinrich PC, Müller-Newen G. Activation of STAT3 by IL-6 and IL-10 in primary human macrophages is differentially modulated by suppressor of cytokine signaling 3. J Immunol. 2003;170:3263–72. doi: 10.4049/jimmunol.170.6.3263. [DOI] [PubMed] [Google Scholar]
- 56.Wolsk E, Mygind H, Grøndahl TS, Pedersen BK, van Hall G. IL-6 selectively stimulates fat metabolism in human skeletal muscle. Am J Physiol Endocrinol Metab. 2010;299:E832–40. doi: 10.1152/ajpendo.00328.2010. [DOI] [PubMed] [Google Scholar]
- 57.Wehinger J, Gouilleux F, Groner B, Finke J, Mertelsmann R, Weber-Nordt RM. IL-10 induces DNA binding activity of three STAT proteins (Stat1, Stat3, and Stat5) and their distinct combinatorial assembly in the promoters of selected genes. FEBS Lett. 1996;394:365–70. doi: 10.1016/0014-5793(96)00990-8. [DOI] [PubMed] [Google Scholar]
- 58.Agrawal S, Gollapudi S, Su H, Gupta S. Leptin activates human B cells to secrete TNF-α, IL-6, and IL-10 via JAK2/STAT3 and p38MAPK/ERK1/2 signaling pathway. J Clin Immunol. 2011;31:472–8. doi: 10.1007/s10875-010-9507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murray PJ. Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response. Curr Opin Pharmacol. 2006;6:379–86. doi: 10.1016/j.coph.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 60.Hogan JC, Stephens JM. The regulation of fatty acid synthase by STAT5A. Diabetes. 2005;54:1968–75. doi: 10.2337/diabetes.54.7.1968. [DOI] [PubMed] [Google Scholar]
- 61.Coulter AA, Stephens JM. STAT5 activators modulate acyl CoA oxidase (AOX) expression in adipocytes and STAT5A binds to the AOX promoter in vitro. Biochem Biophys Res Commun. 2006;344:1342–5. doi: 10.1016/j.bbrc.2006.04.071. [DOI] [PubMed] [Google Scholar]
- 62.Kawai M, Namba N, Mushiake S, Etani Y, Nishimura R, Makishima M, Ozono K. Growth hormone stimulates adipogenesis of 3T3-L1 cells through activation of the Stat5A/5B-PPARgamma pathway. J Mol Endocrinol. 2007;38:19–34. doi: 10.1677/jme.1.02154. [DOI] [PubMed] [Google Scholar]
- 63.Deng J, Hua K, Lesser SS, Greiner AH, Walter AW, Marrero MB, Harp JB. Interleukin-4 mediates STAT6 activation in 3T3-L1 preadipocytes but not adipocytes. Biochem Biophys Res Commun. 2000;267:516–20. doi: 10.1006/bbrc.1999.1993. [DOI] [PubMed] [Google Scholar]
- 64.McGillicuddy FC, Chiquoine EH, Hinkle CC, Kim RJ, Shah R, Roche HM, Smyth EM, Reilly MP. Interferon gamma attenuates insulin signaling, lipid storage, and differentiation in human adipocytes via activation of the JAK/STAT pathway. J Biol Chem. 2009;284:31936–44. doi: 10.1074/jbc.M109.061655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang K, Guo W, Yang Y, Wu J. JAK2/STAT3 pathway is involved in the early stage of adipogenesis through regulating C/EBPβ transcription. J Cell Biochem. 2011;112:488–97. doi: 10.1002/jcb.22936. [DOI] [PubMed] [Google Scholar]
- 66.White UA, Stephens JM. Transcriptional factors that promote formation of white adipose tissue. Mol Cell Endocrinol. 2010;318:10–4. doi: 10.1016/j.mce.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang D, Zhou Y, Lei W, Zhang K, Shi J, Hu Y, Shu G, Song J. Signal transducer and activator of transcription 3 (STAT3) regulates adipocyte differentiation via peroxisome-proliferator-activated receptor γ (PPARgamma) Biol Cell. 2010;102:1–12. doi: 10.1042/BC20090070. [DOI] [PubMed] [Google Scholar]
- 68.Kang HJ, Seo HA, Go Y, Oh CJ, Jeoung NH, Park KG, Lee IK. Dimethylfumarate suppresses adipogenic differentiation in 3T3-L1 preadipocytes through inhibition of STAT3 activity. PLoS One. 2013;8:e61411. doi: 10.1371/journal.pone.0061411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cernkovich ER, Deng J, Hua K, Harp JB. Midkine is an autocrine activator of signal transducer and activator of transcription 3 in 3T3-L1 cells. Endocrinology. 2007;148:1598–604. doi: 10.1210/en.2006-1106. [DOI] [PubMed] [Google Scholar]
- 70.Stewart WC, Pearcy LA, Floyd ZE, Stephens JM. STAT5A expression in Swiss 3T3 cells promotes adipogenesis in vivo in an athymic mice model system. Obesity (Silver Spring) 2011;19:1731–4. doi: 10.1038/oby.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wakao H, Wakao R, Oda A, Fujita H. Constitutively active Stat5A and Stat5B promote adipogenesis. Environ Health Prev Med. 2011;16:247–52. doi: 10.1007/s12199-010-0193-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kiu H, Nicholson SE. Biology and significance of the JAK/STAT signalling pathways. Growth Factors. 2012;30:88–106. doi: 10.3109/08977194.2012.660936. [DOI] [PMC free article] [PubMed] [Google Scholar]