Abstract
Sodium fluoride (NaF) is associated with embryonic and fetal development abnormalities, but the mechanism by which this occurs is unclear. DNA methylation, an important epigenetic reprogramming mechanism, is essential for normal embryonic development. Thus, we investigated the effect of NaF on DNA methylation in early mouse embryos, as well as mouse sperm and liver using bisulfite sequencing and ELISA. Data indicate that H19, a paternally imprinted gene, compared to control embryos, was less methylated in 8-cell embryos from pregnant mice treated with NaF (100 mg/l) in drinking water for 48 h. Peg3, a maternally imprinted gene, and the Line1 repeated sequence were similarly methylated in NaF-treated and control embryos. Oral ingestion of NaF for 35 days did not significantly change Line1 and genomic global DNA methylation in the liver. H19, Rasgrf1, Line1, and genomic global DNA methylation were also similar in NaF-treated and control sperm. Female mice mated with NaF-treated male mice (35 days) had less methylated H19, but Peg3 was significantly more methylated. Line1 was similarly methylated in treated 8-cell embryos, compared to control embryos. NaF treatment of male mice before copulation significantly increased the expression of H19 in blastocysts, whereas H19 expression was not detected in 8-cell embryos. Data suggest that NaF may interact directly with the embryo to disrupt the maintenance of normal gene imprinting during pregnancy. Long-term NaF exposure of males may not directly affect DNA methylation of the sperm and liver, but the sperm may signal to early embryos with abnormal gene imprinting.
Keywords: Sodium fluoride, DNA methylation, H19, Peg3, Mouse embryos
Introduction
Fluorine is one of the most abundant elements on the earth, and high fluoride concentrations can be found in soil and water; dental products; food and beverage products such as tea, soy products, and grape juice; and in fluorinated prescription drugs (Sun et al. 2010). Increased fluoride intake is reported to be associated with an increased prevalence of dental fluorosis. Likewise, excessive fluoride exposure can precipitate various pathological changes in reproductive tissues and in the liver (Gupta et al. 2007). Fluoride not only can cross the blood-testis barrier to inhibit testicular spermatogenesis and decrease sperm capacitation (Dvorakova-Hortova et al. 2008), but also it can readily cross the placental barrier to directly damage developing mammalian fetuses (Goh and Neff 2003). However, the mechanism of fluorine reproductive toxicity is not well understood.
DNA methylation is an important mammalian epigenetic mechanism thought to be involved in early embryonic development. Two major waves of genome-wide demethylation and remethylation occur during development: one occurs during germ cell development and the other occurs after fertilization. However, imprinted genes escape reprogramming after fertilization and retain methylation differences during development. Reprogramming is essential for normal embryonic development and this is inextricably linked with cell pluripotency (Seisenberger et al. 2012). Methylation of the Line1 repeated sequence (10,000–100,000 copies) presumably reflects the methylation of many different genome regions. Line1 is highly methylated overall in mature oocytes and sperm, but it is demethylated in the zygote-to-blastocyst stage (Lane et al. 2003; Okada et al. 2010).
H19, a paternally imprinted gene, is expressed on the maternal allele and repressed on the paternal allele. The H19 imprint occurs during spermatogenesis, and the differentially methylated region (DMR) of H19 is methylated in mature sperm but not in oocytes (Lucifero et al. 2002). Differential methylation of H19 is maintained during reprogramming after fertilization and is a target for environmental insults (Joss-Moore and Lane 2012; Park et al. 2004).
The DMR of Peg3, a maternally imprinted gene, is unmethylated in sperm but completely methylated in mature oocytes (Lucifero et al. 2002). In a previous study, we reported that the Peg3 methylation imprint can be erased and re-established during pre-implantation development (Liu et al. 2008). The unique methylation dynamics of Peg3 indicate that the process of methylation imprint establishment during pre-implantation development is more complex than previously thought (Imamura et al. 2005). Peg3 methylation errors can cause aberrant development. Recent research suggests that the Peg3 methylation status may be a useful molecular marker for cervical intra-epithelial neoplasia (Nye et al. 2013). Thus, Peg3 methylation also may be easily influenced by environmental factors.
This leads us to speculate that NaF may change the normal DNA methylation pattern of early embryos. Therefore, we investigated whether NaF interferes with DNA methylation, and we report that possible links exist between the DNA methylation and NaF reproductive toxicity.
Materials and methods
Materials and chemicals
Chemicals and media were purchased from Sigma-Aldrich (St. Louis, MO) unless specifically mentioned. NaF was purchased from Suyi Chemical Company (Shanghai, China). Pregnant mare serum gonadotropin (PMSG) and human chorionic gonadotropin (hCG) were obtained from Ningbo Second Hormone Factory (Ningbo, China). A DNA easy kit and RNeasy Micro Kit were procured from Qiagen (Germany). A MethyFlash Methylated DNA Quantification Kit was purchased from Epigentek (USA). Sodium metabisulfite was purchased from Merck (Germany). ExTaq hotstart polymerase, PrimeScript reverse transcriptase, and T vector were procured from TaKaRa Biotechnology Company (Dalian, China). Wizard SV Gel and the PCR Clean-Up System were purchased from Promega Biotech Company (Beijing, China). Restriction enzymes (TaqI) were purchased from New England Biolabs (USA).
Ethics statement
This study was carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Research Council. All procedures described in the present study were reviewed and approved by the Animal Care and Use Committee of Yangzhou University (approval ID: SYXK (Su) 2007-0005). Healthy ICR mice, 6–8 weeks of age, were used for all experiments. The mice were housed in an air-conditioned animal house (26 ± 2 °C) and exposed to 10–12 h of light per day. Animals were maintained on a standard diet and had free access to water.
Animal treatment
The animals were randomly divided into three groups for NaF treatment as follows:
Group I (NaF-male) was comprised of male mice given 100 mg/l NaF in water for 35 days. Group II (NaF-female) was comprised of female mice that were mated with the male mice that were not exposed to NaF. Then, only female mice with a vaginal plug were given 100 mg/l NaF in water for 48 h. Group III (Control) consisted of respective male and female animal groups, neither of which received NaF treatment.
All male mice were mated with females treated with PMSG (10 IU) and, 48 h later, with hCG (10 IU). Only female mice with vaginal plugs were determined to have successfully copulated, and this also was regarded as day 0.5 of pregnancy. Pregnant mice were bred continuously and eight-cell embryos and blastocysts were collected on days 2.5 and 3.5 of pregnancy, respectively. Liver samples and sperm were collected from five male mice from both groups (I, III).
DNA isolation and global methylation determination
DNA from sperm and livers was isolated with the DNA easy kit (Qiagen) according to the manufacturer’s protocol and finally dissolved in 25–50 μl TE buffer. The concentration and quality of DNA were tested with a spectrophotometer (NanoDrop). Eight-cell embryos were treated in lysis solution (0.5 M EDTA, 2 mg/ml proteinase K, Ph 8) at 37 °C for 0.5 h and stored at −20 °C until use. Global methylation for sperm and liver was measured using a MethyFlash Methylated DNA Quantification Kit (Epigentek), an ELISA-based colorimetric assay. The assay was performed according to the manufacturer’s instructions. Relative quantification was assessed by normalizing samples against the positive control that was provided with the kit.
Bisulfite treatment
Bisulfite treatment of genomic DNA was carried out as previously described (Zhu et al. 2008). Briefly, samples were denatured in 0.3 M NaOH at 37 °C for 15 min, mixed with 2 volumes of 2 % LMP (low melting point) agarose and pipetted into chilled mineral oil to form agarose beads. This was then treated with freshly made bisulfite solution (2.5 M sodium metabisulfite, Merck; 125 mM hydroquinone, Sigma; pH 5.0) at 50 °C for 16 h in the dark. The reaction was stopped by equilibrating the beads three times with 1 ml of TE buffer. Following desulfonation in 0.5 ml 0.3 M NaOH for 0.5 h, the beads were washed with TE buffer and H2O, respectively, and stored at −20 °C.
PCR amplification
To avoid the preferential amplification of methylated or unmethylated sequences from a mixed population of starting molecules, nested PCR was performed using bisulfite-treated DNA in the first round (12.5 μl) and 2 μl of this PCR product as a template in the second round (25 μl). All reactions contained 0.4 mM primers, 0.2 mM dNTPs, 50 mM KCl, 10 mM Tris–HCl, 1.5 mM MgCl2, and 1.25 units of ExTaq hotstart polymerase (TaKaRa). PCR was performed on an ABI-9700 using the following program. The first round: 1 time at 94 °C for 6 min, 35 times at 94 °C for 1 min, annealing temperature (AT) for 2 min, 72 °C for 3 min and 1 time at 72 °C for 5 min; the second round: 1 time at 94 °C for 5 min, 30 times at 94 °C for 40 s, AT for 45 s, 72 °C for 50 s, 1 time at 72 °C for 5 min. The primers used for bisulfite sequencing were synthesized as previously described (Li et al. 2004; Lucifero et al. 2002; Okada et al. 2010) and these are listed in Table 1.
Table 1.
Primers used for bisulfite sequencing
| Gene | Primer sequence (5′–3′) | Size (bp) | Annealing temperature |
|---|---|---|---|
| H19 | F1:GAGTATTTAGGAGGTATAAGAATT | 423 | 45 °C |
| R1:ATCAAAAACTAACATAAACCCCT | |||
| F2:GTAAGGAGATTATGTTTATTTTTGG | |||
| R2:CCTCATTAATCCCATAACTAT | |||
| Peg3 | F1:TGATAATAGTAGTTTGATTGGTAGGG | 444 | 45 °C |
| R1:TAATTCACACCTAAAACCCTAAAACC | |||
| F2:TTTTGTAGAGGATTTTGATAAGGAGG | |||
| R2:AAATACCACTTTAAATCCCTATCACC | |||
| Rasgrf1 | F1:AAGATAGTTTAGATATGGAATTTTGG | 221 | 60 °C |
| R:ATAATACAACAACAACAATAACAATC | |||
| F2:GATTTTTTAGAGAGTTTATAAAGTTAG | |||
| Line1 | F1:GTTAGAGAATTTGATAGTTTTTGGAATAGG | 250 | 56 °C |
| R1:CCAAAACAAAACCTTTCTCAAACACTATAT | |||
| F2:TAGGAAATTAGTTTGAATAGGTGAGAGGT | |||
| R2:TCAAACACTATATTACTTTAACAATTCCCA |
DNA sequencing and restriction analysis of PCR products
The PCR products from three independent amplifications were gel-purified using the Wizard SV Gel and PCR Clean-Up System (Promega). Then, purified fragments were cloned into a T vector (TaKaRa). The positive clones confirmed by PCR were sequenced using an automatic sequencer (ABI PRISM-77). A portion of the purified fragments was digested with appropriate TaqI restriction enzymes (New England Biolabs). The digested fragments were electrophoresed on 3.0 % agarose gels.
Semi-quantitative RT-PCR
Total RNA were isolated from 15 to 30 grouped embryos using the RNeasy Micro Kit (Qiagen). First-strand cDNA synthesis was carried out using PrimeScript reverse transcriptase (TaKaRa). PCR was performed on an ABI-9700 using the following program: 1 time at 94 °C for 6 min, 40 times at 94 °C for 30 s, 57 °C for 30 s, 72 °C for 40 s and 1 time at 72 °C for 5 min. The primers used for RT-PCR are given in Table 2. After PCR amplifications, each reaction mixture was separated by 2 % agarose gel electrophoresis. The intensity of each band was quantified and the relative intensity against Gapdh was calculated by using Gel Doc System (Bio-Rad).
Table 2.
Primers used for RT-PCR
| Gene | Primer sequence (5′–3′) | Size (bp) | Annealing temperature |
|---|---|---|---|
| H19 | F: GTATGCCCTAACCGCTCAGTC | 136 | 57 °C |
| R: CCAACCTCCCTCCCTAGAAAC | |||
| Peg3 | F: TCAATGACCTCACAAGCCACCAG | 261 | 57 °C |
| R: CGGGCAACAGAGCGATGAAAGC | |||
| Gapdh | F: TCTTGGGCTACACTGAGGAC | 126 | 57 °C |
| R: CATACCAGGAAATGAGCTTGA |
Statistical analysis
The methylation rates in each group were expressed as mean ± standard error from bisulfite sequencing. Relative transcripts of each gene were expressed as mean ± standard error. All data were analyzed by one-way ANOVA, and the difference between groups was determined using the Student’s T test (p < 0.05 was considered statistically significant).
Results
NaF exposure to female mice during pregnancy disrupts H19 DNA methylation in the embryo
DNA methylation of H19, Peg3, and Line1 was examined in NaF-treated male, NaF-treated female, and respective control groups. The results of bisulfite sequencing and bisulfite restriction analysis are summarized in Fig. 1. The data show that DNA methylation of H19 was almost entirely lost in the NaF-treated female group compared with the control group (Fig. 1a). Methylation of H19 in NaF-treated female and control groups were 9.12 ± 5.28 and 49.33 ± 7.60 %, respectively, and the differences between them were significant (p < 0.001) (Fig. 1b). PCR products of H19 in NaF-treated females were undigested, unlike those observed in the female untreated control group (Fig. 1c). The cleaved and uncleaved products refer to methylated and unmethylated templates, respectively.
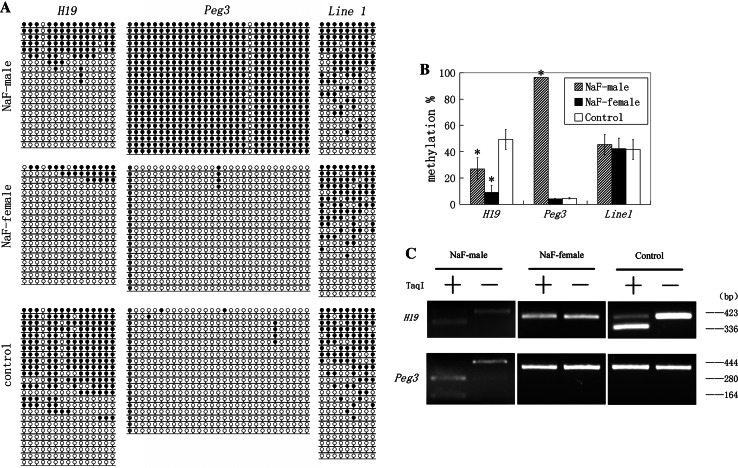
Fig. 1.
Methylation status of H19, Peg3, and Line1 in embryos in NaF-male, NaF-female, and control groups. NaF-male mice were given 100 mg/l NaF in water for 35 days. NaF-female mice with a vaginal plug were given 100 mg/l NaF in water for 48 h, after mating with male mice not exposed to NaF. Control animals did not receive NaF treatment. a Methylation profiles assayed by a bisulfite sequencing assay. Each line represents an individual clone allele. Each circle within the row represents a single CpG site (open and closed circles represent unmethylated and methylated CpGs, respectively). b Statistical analysis of methylation in three groups. Bisulfite sequencing (a) was used to plot the percent of methylated CpGs of the total number of CpG sites. Data are presented as mean ± standard error. (*p < 0.05, compared with controls). The same illustration was applied to the following figures. c Overall methylation profiles of H19 and Peg3 as revealed by bisulfite restriction analysis. The same bisulfite-treated DNA sample used for sequencing was digested with restriction enzymes. Restriction enzymes cleaved only if the recognized TaqI site is methylated. −, undigested PCR products; +, digested PCR products. Sizes of digested fragments are indicated on the right. Digestion of PCR products with TaqI enzyme produced cleaved and uncleaved products, suggesting that PCR products include methylated and unmethylated templates
The pattern and degree of methylation in Peg3 and Line1 were comparable between NaF-treated females and control females (Fig. 1a). Peg3 methylation in NaF-treated females and control females was 4.14 ± 0.32 and 4.66 ± 0.45 %, respectively. Line1 methylation in NaF-treated females and control females groups was 42.33 ± 7.99 and 41.67 ± 7.42 %, respectively. These differences were not significant (Fig. 1b). Furthermore, bisulfite restriction analysis did not clearly reveal the significant difference of Peg3 between NaF-female and control groups (Fig. 1c).
NaF treatment of male mice before copulation disrupts DNA methylation of H19 and Peg3 in the developing embryo
When female mice were mated with male mice treated with NaF in water for 35 days, DNA methylation was significantly decreased in H19 and increased in Peg3 compared to H19 and Peg3 in male mice which did not receive NaF treatment. NaF treatment of male mice did not affect methylation of Line1 in the developing embryo. The restriction pattern of H19 was similar between NaF-treated male mice and the control male group (Fig. 1c), and bisulfite sequencing revealed that the H19 methylation in the NaF-treated male group was 27.00 ± 8.54 %, which was significantly lower than that observed in the control male group (p < 0.05) (Fig. 1a, b). Peg3 methylation in NaF-treated male mice was 96.55 ± 0 %, significantly higher than that observed in the control male group (p < 0.001) (Fig. 1a, b). The majority of Peg3 PCR products in NaF-treated mice were digested, unlike those observed in the control male group (Fig. 1c).
NaF treatment of male mice before copulation increase the expression of H19 in blastocysts
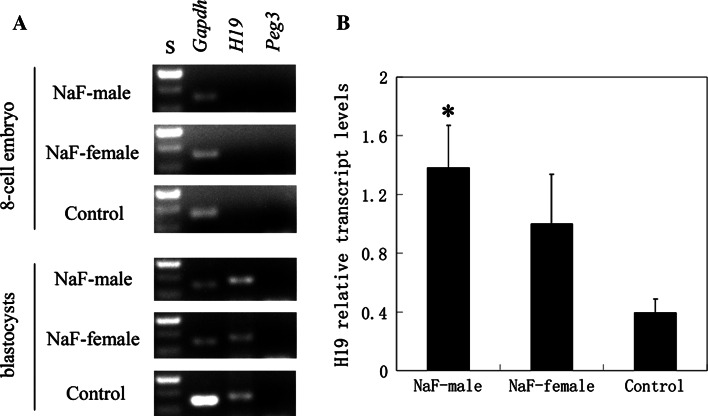
The expression of H19 and Peg3 was examined in 8-cell embryos and blastocysts from all mouse groups (See Fig. 2). H19 was expressed at the blastocyst stage, whereas Peg3 was not. NaF treatment of male mice before copulation significantly increased H19 expression in blastocysts. At the 8-cell embryo stage, H19 and Peg3 expression could not be detected by RT-PCR.
Fig. 2.
H19 and Peg3 expression in 8-cell embryo and blastocysts from NaF-treated and control mice. a Agarose gel electrophoresis. S, 100 bp ladders. b H19 relative transcript levels. Transcripts were normalized against the Gapdh control. Each column indicates the mean ± standard error (*p < 0.05, compared with controls)
NaF may not significantly affect DNA methylation of sperm and liver
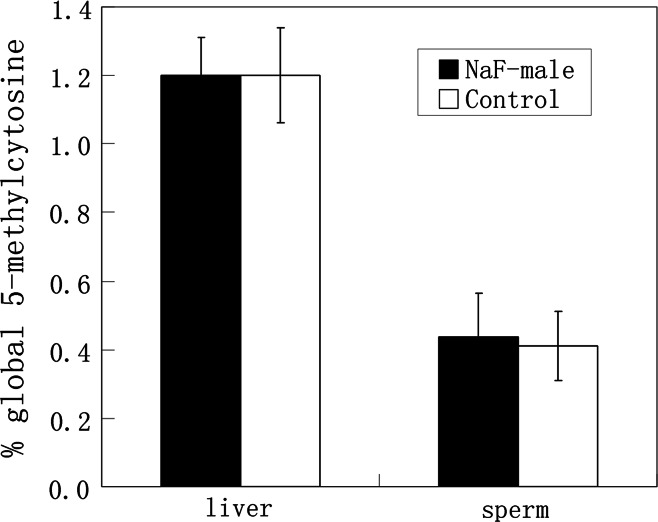
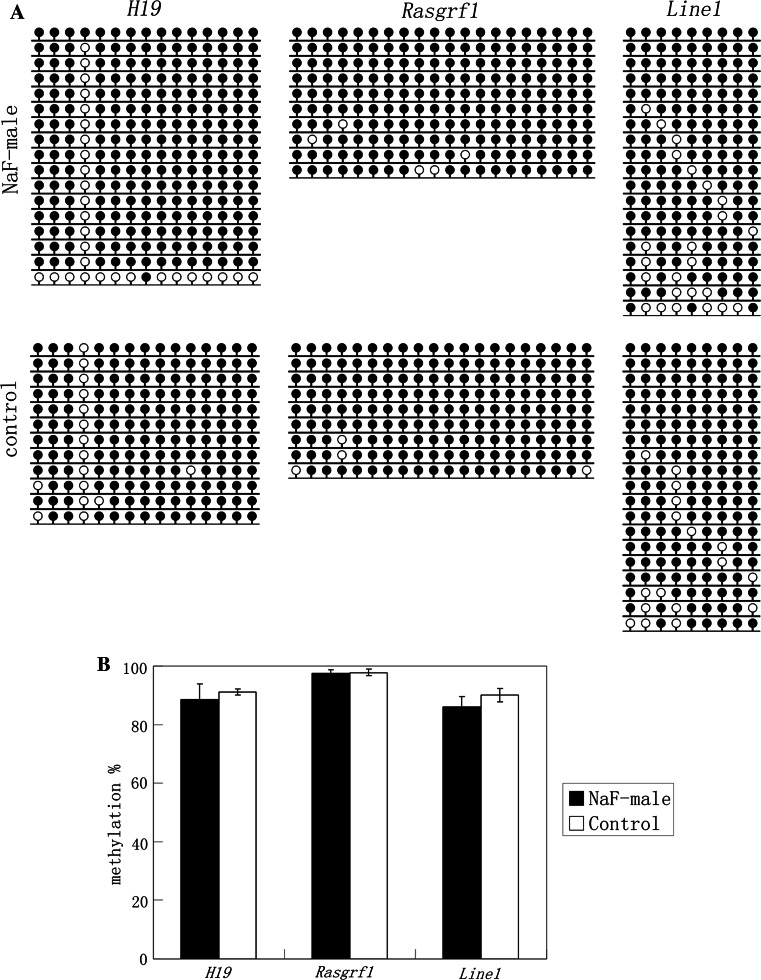
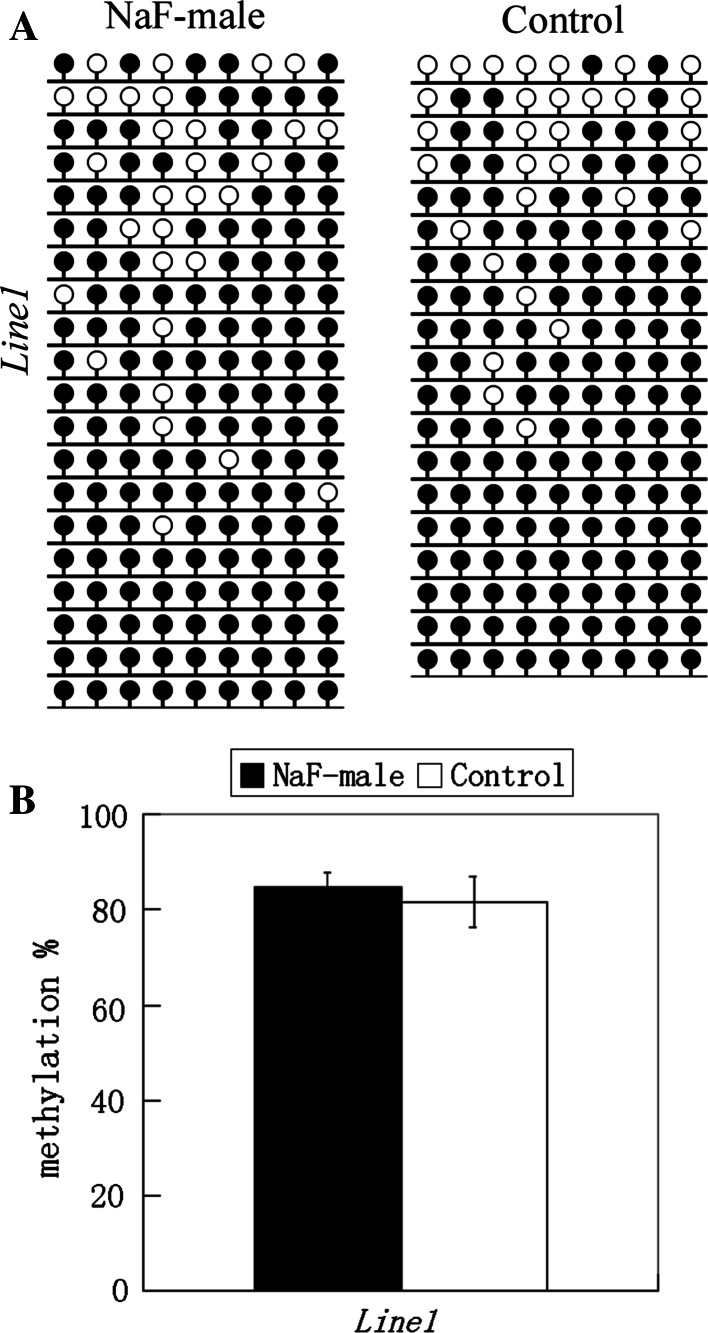
Global DNA methylation in sperm and liver were examined in NaF-treated male mice and their corresponding control group. Using an ELISA-based colorimetric assay, 5-methylcytosine in genomic DNA was measured (Fig. 3). In sperm from NaF-treated male mice and the control male group, 5-methylcytosine was 0.44 ± 0.13 and 0.41 ± 0.10 %, respectively, and 5-methylcytosine in the livers of those treatment and control groups was 1.20 ± 0.11 and 1.20 ± 0.14 %, respectively. Furthermore, DNA methylation of H19, Rasgrf1, and Line1 in sperm and Line1 methylation in liver were examined using bisulfite sequencing (Figs. 4, 5), and no significant differences were observed in all examinations. Data indicate that NaF may not significantly affect DNA methylation in the sperm and liver of male mice.
Fig. 3.
5-methylcytosine in genomic DNA of sperm and liver from NaF-treated male and control groups were measured using a MethylFlash Methylated Quantification kit
Fig. 4.
Methylation status of H19, Rasgrf1, and Line1 on sperm in NaF-male and control groups. a Methylation profiles assayed by the bisulfite sequencing assay. b Statistical analysis of methylation in two groups. Bisulfite sequencing (a) was used to plot the percent of methylated CpGs of the total number of CpG sites
Fig. 5.
Methylation status of Line1 on liver in NaF-male and control groups. a Methylation profiles assayed by the bisulfite sequencing assay. b Statistical analysis of methylation in two groups. Bisulfite sequencing (a) was used to plot the percent of methylated CpGs of the total number of CpG sites
Discussion
NaF not only cause pathological changes in a variety of tissues and organs, but also causes cancer and malformation of the reproductive system. The mechanism of NaF reproductive toxicity is not well understood, although DNA methylation is known to be a particularly important epigenetic modification mechanism. Abnormal DNA methylation is also thought to be involved in the etiology of complex diseases (Fraga et al. 2005). In the present study, the effects of NaF on DNA methylation in the embryo and in sperm and the liver were investigated. H19, Peg3, and Rasgrf1, which are imprinted genes, were selected for study because of their important biological functions and their characteristic phenotypes. The methylation of the Line1 repeated sequence was also investigated as an indicator of genome methylation. Data indicated that NaF treatment disrupted DNA methylation of H19 and Peg3 in early embryonic stages of the mouse. However, there were no significant changes in DNA methylation in sperm and liver of male mice treated with NaF. Because gene expression is affected by DNA methylation, we then investigated the effect of NaF on the expression of H19 and Peg3 in 8-cell embryos and in the blastocyst. NaF treatment of male mice before copulation significantly increased the expression of H19 in blastocysts, whereas H19 expression was not detected in 8-cell embryos. These findings indicate that NaF can disrupt DNA methylation of H19 and further affect H19 expression.
Oral ingestion of NaF in water is usually the major source of fluoride toxicosis. The mice treated with 100 mg/l NaF in water were used to model this toxicity. Pregnant female mice and male mice were treated for 48 h and 35 days, respectively. Many studies of laboratory animals indicate that adverse reproductive and developmental effects occur at high fluoride concentrations and that various injurious effects are associated with an exposure of 100 mg/l NaF in drinking water (Chlubek et al. 2003; Shanthakumari et al. 2004). The experimental period was aligned with the cycle of mouse spermatogenesis. For gonad stem cells to mature into sperm, 35 days are required. Within these 35 days, the spermiogonium stage lasts 8 days; meiosis lasts 13 days; and sperm formation needs 14 days. Finally, 5 days are needed for sperm to reach the tail of epididymis for ejaculation (Zhu et al. 2006).
In the processes of imprint erasure, establishment and maintenance are vulnerable to errors. Methylation imprints could not be re-established in the postzygotic embryos, once they were lost (Chang et al. 2006; Sato et al. 2007). Our results suggest that H19 is demethylated in embryos from NaF-treated pregnant mice, resulting in an undermethylation pattern in the embryos. This is consistent with early reports demonstrating that H19 is a target for environmental insults (Doherty et al. 2000; Joss-Moore and Lane 2012). In contrast, Peg3 imprints are not affected by NaF exposure during pregnancy. This discrepancy may be due to the fact that Peg3 methylation can be erased after fertilization (Liu et al. 2008). However, in this present study, we did not investigate whether NaF ingestion during pregnancy affected re-establishment of the Peg3 imprint in the morula or blastocyst stage. Line1 methylation, as an indicator of genome methylation, is not affected by NaF ingestion during pregnancy. Our data support the idea that NaF can cross the placental barrier and interact directly with pre-implantation embryos. NaF can disrupt maintenance of the H19 imprint during pre-implantation development, while genomic DNA methylation can be normally reprogrammed. Thus, H19 methylation is a target for NaF reproductive toxicity.
We next investigated the effects of NaF on DNA methylation of sperm and whether this effect can be transferred to early embryos. Although fluoride can cross the blood-testis barrier to inhibit testicular spermatogenesis and decrease sperm capacitation (Dvorakova-Hortova et al. 2008; Sarkar et al. 2006), NaF did not significantly affect DNA methylation in H19, Rasgrf1, Line1 and genome methylation during spermatogenesis. Of note, H19 and Peg3 were abnormally methylated in the embryo when NaF-treated male mice sperm were the source of oocyte fertilization. Data show that NaF does not directly affect DNA methylation in the sperm, but NaF can potentially affect regulation mechanisms of methylation imprints (this was beyond the scope of this investigation). Also, sperm that were not directly damaged may “carry” or transmit this insult to early embryos, giving rise to abnormal methylation imprints. In this case, Peg3 hypermethylation caused by NaF may result from early events of remethylation, because Peg3 is completely unmethylated in sperm and 2-cell embryos. In addition, we investigated the effect of NaF on DNA methylation in the liver. NaF did not appear to effect genome methylation in the liver, where DNA methylation reprogramming is not found.
In summary, we propose that NaF can harm DNA methylation of imprinted genes in embryos, but clearly, genome methylation can withstand some adverse effects of NaF and maintain balance. Our data have provided essential preliminary data for further study of potential NaF-induced reproductive toxicity. At a minimum, our data suggest that the use of fluoride in dental care products may require re-evaluation for pregnant women and men who plan to father children.
Acknowledgments
This work was supported by Natural Science Foundation of Jiangsu Province (BK2012264), Innovation Foundation of Yangzhou University (027389604035349), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
References
- Chang SC, Tucker T, Thorogood NP, Brown CJ. Mechanisms of X-chromosome inactivation. Front Biosci. 2006;11:852–866. doi: 10.2741/1842. [DOI] [PubMed] [Google Scholar]
- Chlubek D, Grucka-Mamczar E, Birkner E, Polaniak R, Stawiarska-Pieta B, Duliban H. Activity of pancreatic antioxidative enzymes and malondialdehyde concentrations in rats with hyperglycemia caused by fluoride intoxication. J Trace Elem Med Biol. 2003;17(1):57–60. doi: 10.1016/S0946-672X(03)80047-0. [DOI] [PubMed] [Google Scholar]
- Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62(6):1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- Dvorakova-Hortova K, Sandera M, Jursova M, Vasinova J, Peknicova J. The influence of fluorides on mouse sperm capacitation. Anim Reprod Sci. 2008;108(1–2):157–170. doi: 10.1016/j.anireprosci.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102(30):10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh EH, Neff AW. Effects of fluoride on Xenopus embryo development. Food Chem Toxicol. 2003;41(11):1501–1508. doi: 10.1016/S0278-6915(03)00166-2. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Khan TI, Agrawal D, Kachhawa JB. The toxic effects of sodium fluoride on the reproductive system of male rats. Toxicol Ind Health. 2007;23(9):507–513. doi: 10.1177/0748233708089041. [DOI] [PubMed] [Google Scholar]
- Imamura T, Kerjean A, Heams T, Kupiec JJ, Thenevin C, Paldi A. Dynamic CpG and non-CpG methylation of the Peg1/Mest gene in the mouse oocyte and preimplantation embryo. J Biol Chem. 2005;280(20):20171–20175. doi: 10.1074/jbc.M501749200. [DOI] [PubMed] [Google Scholar]
- Joss-Moore LA, Lane RH. Epigenetics and the developmental origins of disease: the key to unlocking the door of personalized medicine. Epigenomics. 2012;4(5):471–473. doi: 10.2217/epi.12.53. [DOI] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35(2):88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Li JY, Lees-Murdock DJ, Xu GL, Walsh CP. Timing of establishment of paternal methylation imprints in the mouse. Genomics. 2004;84(6):952–960. doi: 10.1016/j.ygeno.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Liu JH, Zhu JQ, Liang XW, et al. Diploid parthenogenetic embryos adopt a maternal-type methylation pattern on both sets of maternal chromosomes. Genomics. 2008;91(2):121–128. doi: 10.1016/j.ygeno.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM. Methylation dynamics of imprinted genes in mouse germ cells. Genomics. 2002;79(4):530–538. doi: 10.1006/geno.2002.6732. [DOI] [PubMed] [Google Scholar]
- Nye MD, Hoyo C, Huang Z, et al. Associations between methylation of paternally expressed gene 3 (PEG3), cervical intraepithelial neoplasia and invasive cervical cancer. PLoS ONE. 2013;8(2):e56325. doi: 10.1371/journal.pone.0056325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463(7280):554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KY, Sellars EA, Grinberg A, Huang SP, Pfeifer K. The H19 differentially methylated region marks the parental origin of a heterologous locus without gametic DNA methylation. Mol Cell Biol. 2004;24(9):3588–3595. doi: 10.1128/MCB.24.9.3588-3595.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar SD, Maiti R, Ghosh D. Management of fluoride induced testicular disorders by calcium and vitamin-E co-administration in the albino rat. Reprod Toxicol. 2006;22(4):606–612. doi: 10.1016/j.reprotox.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Sato A, Otsu E, Negishi H, Utsunomiya T, Arima T. Aberrant DNA methylation of imprinted loci in superovulated oocytes. Hum Reprod. 2007;22(1):26–35. doi: 10.1093/humrep/del316. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48(6):849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanthakumari D, Srinivasalu S, Subramanian S. Effect of fluoride intoxication on lipidperoxidation and antioxidant status in experimental rats. Toxicology. 2004;204(2–3):219–228. doi: 10.1016/j.tox.2004.06.058. [DOI] [PubMed] [Google Scholar]
- Sun Z, Niu R, Su K, et al. Effects of sodium fluoride on hyperactivation and Ca2 + signaling pathway in sperm from mice: an in vivo study. Arch Toxicol. 2010;84(5):353–361. doi: 10.1007/s00204-009-0508-x. [DOI] [PubMed] [Google Scholar]
- Zhu B, Huang X, Chen J, Lu Y, Chen Y, Zhao J. Methylation changes of H19 gene in sperms of X-irradiated mouse and maintenance in offspring. Biochem Biophys Res Commun. 2006;340(1):83–89. doi: 10.1016/j.bbrc.2005.11.154. [DOI] [PubMed] [Google Scholar]
- Zhu JQ, Liu JH, Liang XW, et al. Heat stress causes aberrant DNA methylation of H19 and Igf-2r in mouse blastocysts. Mol Cells. 2008;25(2):211–215. [PubMed] [Google Scholar]