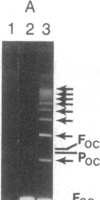
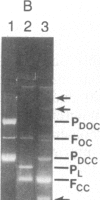
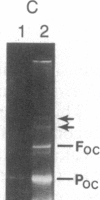
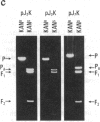
Abstract
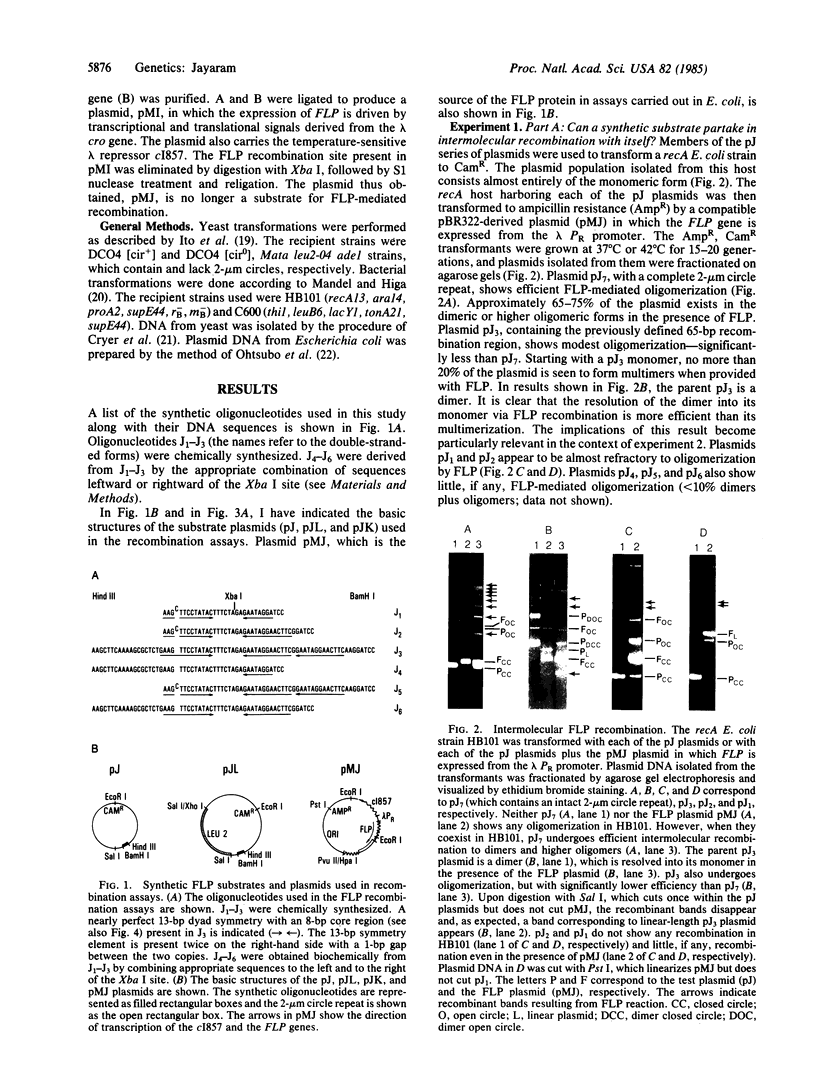
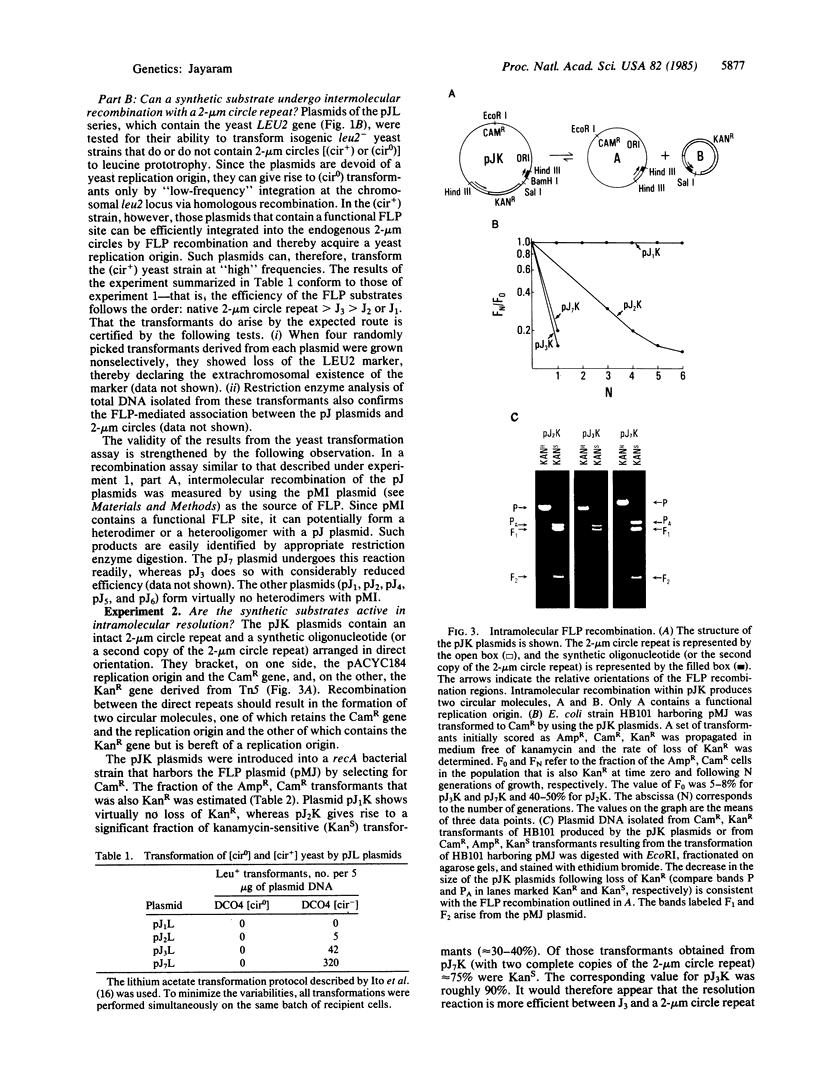
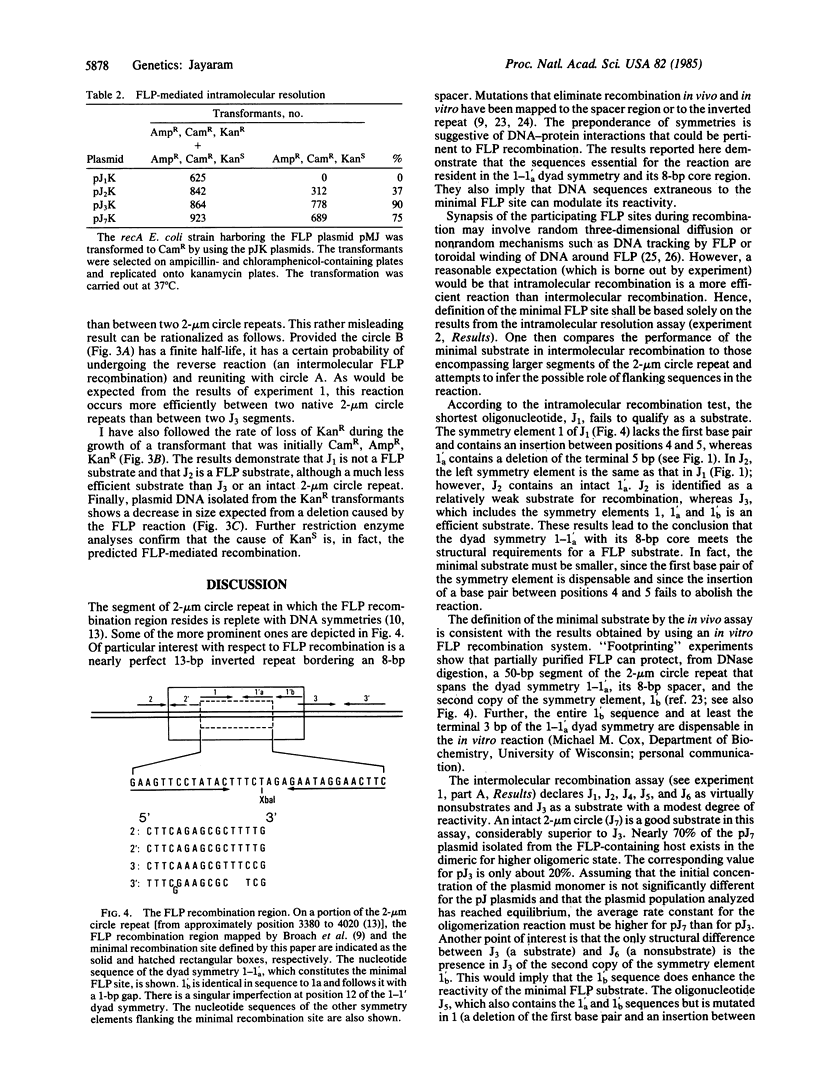
The 2-mum circle DNA of yeast encodes a site-specific recombination system (FLP recombination). The recombination region had been mapped earlier to a 65-base-pair (bp) segment within the 599-bp-long inverted repeats of the molecule. I have shown that the "minimal" FLP substrate resides in a 13-bp dyad symmetry plus an 8-bp core located within the 65-bp recombination region. Further, as determined by different in vivo assays, sequences extraneous to the minimal FLP site and the 65-bp recombination region can affect the efficiency of the recombination reaction.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abremski K., Hoess R., Sternberg N. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell. 1983 Apr;32(4):1301–1311. doi: 10.1016/0092-8674(83)90311-2. [DOI] [PubMed] [Google Scholar]
- Andrews B. J., Proteau G. A., Beatty L. G., Sadowski P. D. The FLP recombinase of the 2 micron circle DNA of yeast: interaction with its target sequences. Cell. 1985 Apr;40(4):795–803. doi: 10.1016/0092-8674(85)90339-3. [DOI] [PubMed] [Google Scholar]
- Benjamin H. W., Matzuk M. M., Krasnow M. A., Cozzarelli N. R. Recombination site selection by Tn3 resolvase: topological tests of a tracking mechanism. Cell. 1985 Jan;40(1):147–158. doi: 10.1016/0092-8674(85)90318-6. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Guarascio V. R., Jayaram M. Recombination within the yeast plasmid 2mu circle is site-specific. Cell. 1982 May;29(1):227–234. doi: 10.1016/0092-8674(82)90107-6. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Hicks J. B. Replication and recombination functions associated with the yeast plasmid, 2 mu circle. Cell. 1980 Sep;21(2):501–508. doi: 10.1016/0092-8674(80)90487-0. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Cox M. M. The FLP protein of the yeast 2-microns plasmid: expression of a eukaryotic genetic recombination system in Escherichia coli. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4223–4227. doi: 10.1073/pnas.80.14.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. The mechanism of phage lambda site-specific recombination: site-specific breakage of DNA by Int topoisomerase. Cell. 1983 Dec;35(3 Pt 2):795–803. doi: 10.1016/0092-8674(83)90112-5. [DOI] [PubMed] [Google Scholar]
- Hartley J. L., Donelson J. E. Nucleotide sequence of the yeast plasmid. Nature. 1980 Aug 28;286(5776):860–865. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]
- Hoess R. H., Abremski K. Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1026–1029. doi: 10.1073/pnas.81.4.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess R. H., Abremski K. Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol. 1985 Feb 5;181(3):351–362. doi: 10.1016/0022-2836(85)90224-4. [DOI] [PubMed] [Google Scholar]
- Hoess R., Abremski K., Sternberg N. The nature of the interaction of the P1 recombinase Cre with the recombining site loxP. Cold Spring Harb Symp Quant Biol. 1984;49:761–768. doi: 10.1101/sqb.1984.049.01.086. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Meyer B. J., Ptashne M. Interactions between DNA-bound repressors govern regulation by the lambda phage repressor. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5061–5065. doi: 10.1073/pnas.76.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A., Meyer B. J., Ptashne M. Mechanism of action of the cro protein of bacteriophage lambda. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1783–1787. doi: 10.1073/pnas.75.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Simon M. I. Hin-mediated site-specific recombination requires two 26 bp recombination sites and a 60 bp recombinational enhancer. Cell. 1985 Jul;41(3):781–791. doi: 10.1016/s0092-8674(85)80059-3. [DOI] [PubMed] [Google Scholar]
- Kahmann R., Rudt F., Mertens G. Substrate and enzyme requirements for in vitro site-specific recombination in bacteriophage mu. Cold Spring Harb Symp Quant Biol. 1984;49:285–294. doi: 10.1101/sqb.1984.049.01.034. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod M., Volkert F., Broach J. Components of the site-specific recombination system encoded by the yeast plasmid 2-micron circle. Cold Spring Harb Symp Quant Biol. 1984;49:779–787. doi: 10.1101/sqb.1984.049.01.088. [DOI] [PubMed] [Google Scholar]
- Nash H. A. Integration and excision of bacteriophage lambda. Curr Top Microbiol Immunol. 1977;78:171–199. doi: 10.1007/978-3-642-66800-5_6. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Rosenbloom M., Schrempf H., Goebel W., Rosen J. Site specific recombination involved in the generation of small plasmids. Mol Gen Genet. 1978 Feb 16;159(2):131–141. doi: 10.1007/BF00270886. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Van de Putte P. Genetic switches by DNA inversions in prokaryotes. Biochim Biophys Acta. 1984 Jun 16;782(2):111–119. doi: 10.1016/0167-4781(84)90013-7. [DOI] [PubMed] [Google Scholar]
- Queen C. A vector that uses phage signals for efficient synthesis of proteins in Escherichia coli. J Mol Appl Genet. 1983;2(1):1–10. [PubMed] [Google Scholar]
- Reed R. R. Transposon-mediated site-specific recombination: a defined in vitro system. Cell. 1981 Sep;25(3):713–719. doi: 10.1016/0092-8674(81)90178-1. [DOI] [PubMed] [Google Scholar]
- Simon M., Zieg J., Silverman M., Mandel G., Doolittle R. Phase variation: evolution of a controlling element. Science. 1980 Sep 19;209(4463):1370–1374. doi: 10.1126/science.6251543. [DOI] [PubMed] [Google Scholar]
- Tonegawa S., Sakano H., Make R., Traunecker A., Heinrich G., Roeder W., Kurosawa Y. Somatic reorganization of immunoglobulin genes during lymphocyte differentiation. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):839–858. doi: 10.1101/sqb.1981.045.01.102. [DOI] [PubMed] [Google Scholar]
- Vetter D., Andrews B. J., Roberts-Beatty L., Sadowski P. D. Site-specific recombination of yeast 2-micron DNA in vitro. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7284–7288. doi: 10.1073/pnas.80.23.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]