Abstract
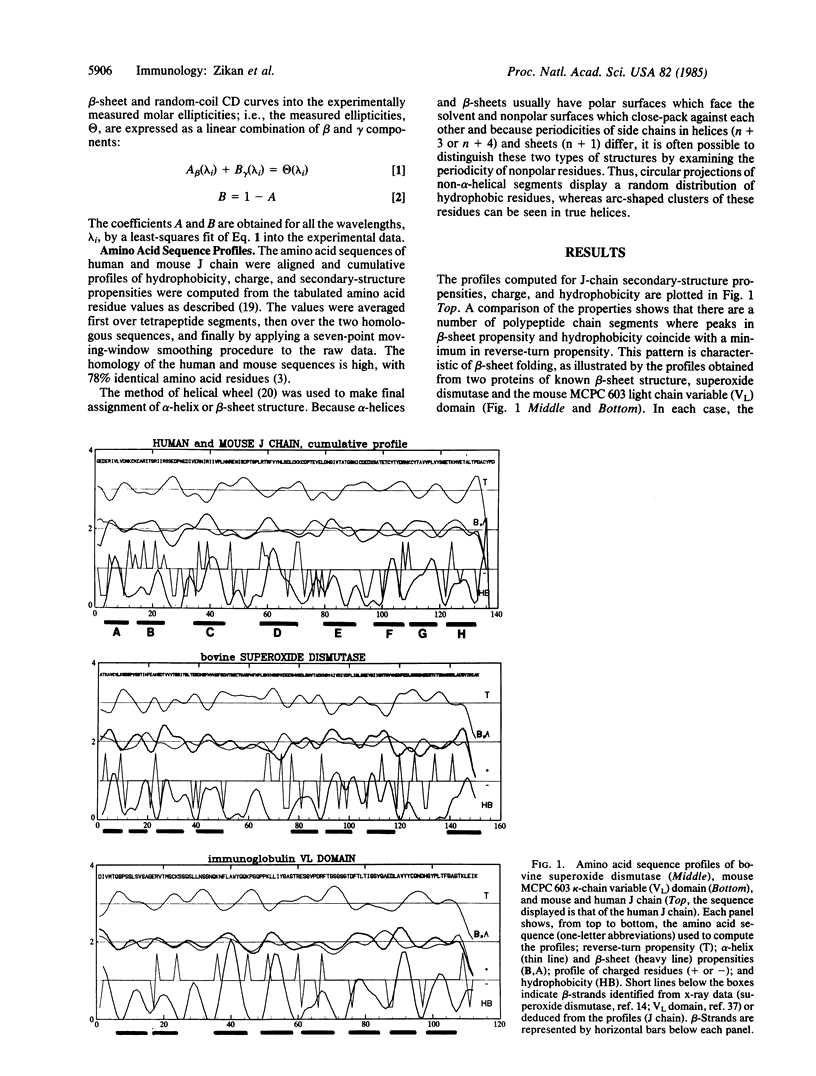
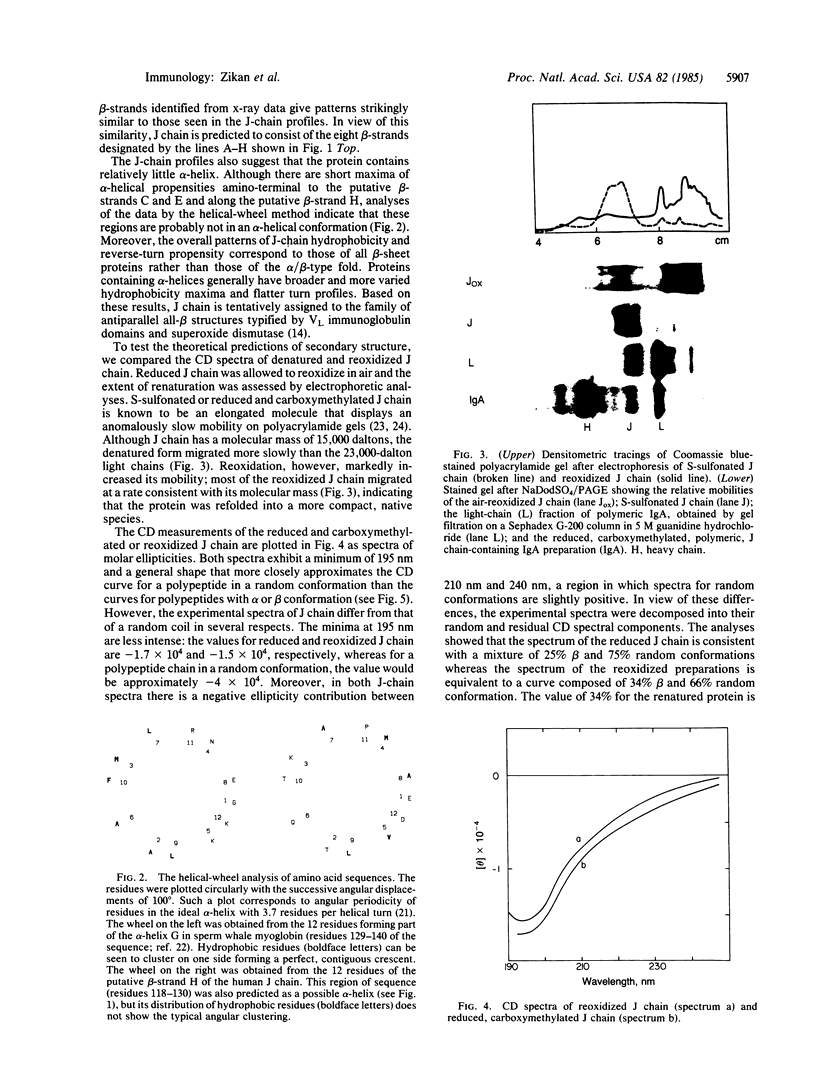
J chain is a 137-residue polypeptide that is covalently linked to polymeric immunoglobulins and participates in their synthesis and transport to external secretions. To clarify these roles, the secondary structure of J chain was characterized by computer-assisted analyses of human and mouse sequences and by circular dichroism measurements of the isolated J chain. The secondary-structure profiles obtained were very similar to those of superoxide dismutase or immunoglobulin light chain variable domains, suggesting that the J chain folds into an eight-stranded antiparallel beta-barrel and should contain approximately 37% beta-sheet conformation, with the rest of the structure existing as reverse turns (random coil). The circular dichroism measurements indicated that the conformation of denatured, S-carboxymethylated or S-sulfonated J chain consists of 75% random coil and 25% beta-structure. Upon reformation of disulfide bonds the percentage of beta-structure in the air-oxidized J chain increased to 34%, a value that is in good agreement with the secondary-structure analysis. Two alternative models of J-chain structure, a two-domain model [Cann, G., Zaritsky, A. & Koshland, M.E. (1982) Proc. Natl. Acad. Sci. USA 79, 6656-6660] and a single-domain antiparallel beta-sheet bilayer model (proposed in this paper), are compared.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ANFINSEN C. B., HABER E. Studies on the reduction and re-formation of protein disulfide bonds. J Biol Chem. 1961 May;236:1361–1363. [PubMed] [Google Scholar]
- Brandtzaeg P., Prydz H. Direct evidence for an integrated function of J chain and secretory component in epithelial transport of immunoglobulins. Nature. 1984 Sep 6;311(5981):71–73. doi: 10.1038/311071a0. [DOI] [PubMed] [Google Scholar]
- Brandtzaeg P. Transport models for secretory IgA and secretory IgM. Clin Exp Immunol. 1981 May;44(2):221–232. [PMC free article] [PubMed] [Google Scholar]
- Cann G. M., Zaritsky A., Koshland M. E. Primary structure of the immunoglobulin J chain from the mouse. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6656–6660. doi: 10.1073/pnas.79.21.6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. Principles that determine the structure of proteins. Annu Rev Biochem. 1984;53:537–572. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- Chothia C. Structural invariants in protein folding. Nature. 1975 Mar 27;254(5498):304–308. doi: 10.1038/254304a0. [DOI] [PubMed] [Google Scholar]
- Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry. 1981 Apr 28;20(9):2361–2370. [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Elliott B. W., Jr, Steiner L. A. Amino- and carboxy-terminal sequence of mouse J chain and analysis of tryptic peptides. J Immunol. 1984 Jun;132(6):2968–2974. [PubMed] [Google Scholar]
- Halpern M. S., Koshland M. E. Noval subunit in secretory IgA. Nature. 1970 Dec 26;228(5278):1276–1278. doi: 10.1038/2281276a0. [DOI] [PubMed] [Google Scholar]
- KENDREW J. C., WATSON H. C., STRANDBERG B. E., DICKERSON R. E., PHILLIPS D. C., SHORE V. C. The amino-acid sequence x-ray methods, and its correlation with chemical data. Nature. 1961 May 20;190:666–670. doi: 10.1038/190666a0. [DOI] [PubMed] [Google Scholar]
- Koshland M. E., Chapuis R. M., Recht B., Brown J. C. Selective proteolysis of the J chain component in human polymeric immunoglobulin. J Immunol. 1977 Mar;118(3):775–781. [PubMed] [Google Scholar]
- Koshland M. E. The coming of age of the immunoglobulin J chain. Annu Rev Immunol. 1985;3:425–453. doi: 10.1146/annurev.iy.03.040185.002233. [DOI] [PubMed] [Google Scholar]
- Kutteh W. H., Moldoveanu Z., Prince S. J., Kulhavy R., Alonso F., Mestecky J. Biosynthesis of J-chain in human lymphoid cells producing immunoglobulins of various isotypes. Mol Immunol. 1983 Sep;20(9):967–976. doi: 10.1016/0161-5890(83)90037-8. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levitt M., Chothia C. Structural patterns in globular proteins. Nature. 1976 Jun 17;261(5561):552–558. doi: 10.1038/261552a0. [DOI] [PubMed] [Google Scholar]
- Max E. E., Korsmeyer S. J. Human J chain gene. Structure and expression in B lymphoid cells. J Exp Med. 1985 Apr 1;161(4):832–849. doi: 10.1084/jem.161.4.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestecky J., Schrohenloher R. E., Kulhavy R., Wright G. P., Tomana M. Site of J chain attachment to human polymeric IgA. Proc Natl Acad Sci U S A. 1974 Feb;71(2):544–548. doi: 10.1073/pnas.71.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestecky J., Schrohenloher R. E. Site of attachment of J chain to human immunoglobulin M. Nature. 1974 Jun 14;249(458):650–652. doi: 10.1038/249650a0. [DOI] [PubMed] [Google Scholar]
- Mestecky J., Zikan J., Butler W. T. Immunoglobulin M and secretory immunoglobulin A: presence of a common polypeptide chain different from light chains. Science. 1971 Mar 19;171(3976):1163–1165. doi: 10.1126/science.171.3976.1163. [DOI] [PubMed] [Google Scholar]
- Mole J. E., Bhown A. D., Bennett J. Sequence analysis of human J chain. Amino terminal location of a disulfide bond linking the immunoglobulin heavy chain. Biochem Biophys Res Commun. 1976 Nov 8;73(1):92–97. doi: 10.1016/0006-291x(76)90501-5. [DOI] [PubMed] [Google Scholar]
- Mole J. E., Bhown A. S., Bennett J. C. Primary structure of human J chain: alignment of peptides from chemical and enzymatic hydrolyses. Biochemistry. 1977 Aug 9;16(16):3507–3513. doi: 10.1021/bi00635a002. [DOI] [PubMed] [Google Scholar]
- Mostov K. E., Friedlander M., Blobel G. The receptor for transepithelial transport of IgA and IgM contains multiple immunoglobulin-like domains. Nature. 1984 Mar 1;308(5954):37–43. doi: 10.1038/308037a0. [DOI] [PubMed] [Google Scholar]
- Novotný J., Auffray C. A program for prediction of protein secondary structure from nucleotide sequence data: application to histocompatibility antigens. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):243–255. doi: 10.1093/nar/12.1part1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAULING L., COREY R. B., BRANSON H. R. The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain. Proc Natl Acad Sci U S A. 1951 Apr;37(4):205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisfeld R. A., Small P. A., Jr Electrophoretic heterogeneity of polypeptide chains of specific antibodies. Science. 1966 May 27;152(3726):1253–1255. doi: 10.1126/science.152.3726.1253. [DOI] [PubMed] [Google Scholar]
- Richardson J. S., Richardson D. C., Thomas K. A., Silverton E. W., Davies D. R. Similarity of three-dimensional structure between the immunoglobulin domain and the copper, zinc superoxide dismutase subunit. J Mol Biol. 1976 Apr 5;102(2):221–235. doi: 10.1016/s0022-2836(76)80050-2. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Schiffer M., Edmundson A. B. Correlation of amino acid sequence and conformation in tobacco mosaic virus. Biophys J. 1968 Jan;8(1):29–39. doi: 10.1016/S0006-3495(68)86472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer M., Girling R. L., Ely K. R., Edmundson A. B. Structure of a lambda-type Bence-Jones protein at 3.5-A resolution. Biochemistry. 1973 Nov 6;12(23):4620–4631. doi: 10.1021/bi00747a013. [DOI] [PubMed] [Google Scholar]
- Schrohenloher R. E., Mestecky J., Stanton T. H. Molecular weight of a human J chain. Biochim Biophys Acta. 1973 Feb 21;295(2):576–581. doi: 10.1016/0005-2795(73)90055-x. [DOI] [PubMed] [Google Scholar]
- Segal D. M., Padlan E. A., Cohen G. H., Rudikoff S., Potter M., Davies D. R. The three-dimensional structure of a phosphorylcholine-binding mouse immunoglobulin Fab and the nature of the antigen binding site. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4298–4302. doi: 10.1073/pnas.71.11.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde C. E., 3rd, Koshland M. E. Molecular size and shape of the J chain from polymeric immunoglobulins. Biochemistry. 1973 Aug 14;12(17):3218–3224. doi: 10.1021/bi00741a012. [DOI] [PubMed] [Google Scholar]
- Word C. J., Mushinski J. F., Tucker P. W. The murine immunoglobulin alpha gene expresses multiple transcripts from a unique membrane exon. EMBO J. 1983;2(6):887–898. doi: 10.1002/j.1460-2075.1983.tb01518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]