Abstract
Recent studies of the low abundant signaling lipid, phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2), reveal an intriguingly diverse list of downstream pathways, the intertwined relationship between PI(3,5)P2 and PI5P, as well as links to neurodegenerative diseases. Derived from the structural lipid phosphatidylinositol, PI(3,5)P2 is dynamically generated on multiple cellular compartments where interactions with an increasing list effectors regulate many cellular pathways. A complex of proteins that includes Fab1/PIKfyve, Vac14 and Fig4/Sac3 mediates the biosynthesis of PI(3,5)P2, and mutations that disrupt complex function and/or formation cause profound consequences in cells. Surprisingly, mutations in this pathway are linked with neurological diseases, including Charcot-Marie-Tooth Syndrome and Amyotrophic Lateral Sclerosis. Future studies of PI(3,5)P2 and PI5P are likely to expand the roles of these lipids in regulation of cellular functions, as well as provide new approaches for treatment of some neurological diseases.
INTRODUCTION
Phosphorylated phosphatidylinositol (PIP) signaling lipids play regulatory roles. These low-abundance lipids are produced from phosphatidylinositol (PI), an abundant structural component of membranes, which can be phosphorylated in any combination on positions three, four or five. Highly regulated PIP kinases and phosphatases generate and turn over the resultant seven PIP lipids (Fig. 1).
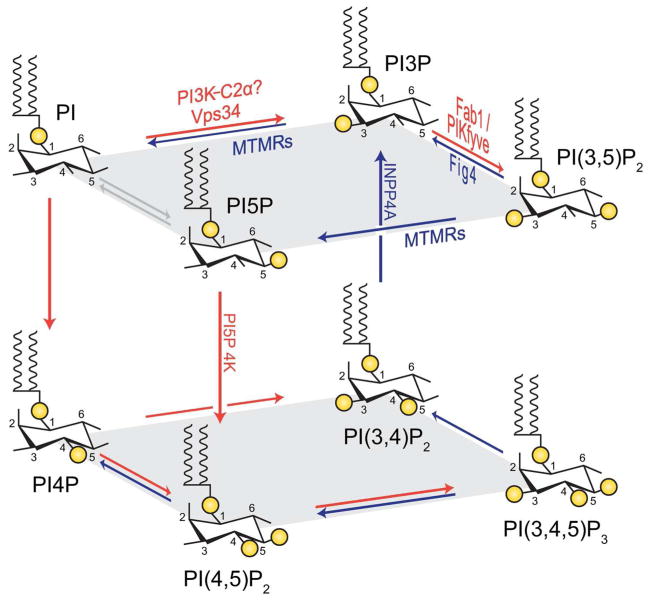
Figure 1.
Interconversion among the seven known phosphoinositide lipids occurs via action of specific lipid kinases (red arrows) and phosphatases (blue arrows). Selected kinases and phosphatases are shown. While controversial, direct conversion of PI to PI5P via PIKfyve activity may contribute to the PI5P pool (gray arrows). INPP4A phosphatase, which causes neurodegeneration in mice [118], and the type II PI5P 4-kinase [119,120], which has a role in the regulation of PI5P levels, were not discussed in this review.
PIP lipids provide spatial and temporal regulation of complex protein machines. The interconvertibility of PIPs enables rapid changes in the identity of the signaling lipid to dynamically recruit effector proteins to specific membranes at the right time. For example, synthesis of phosphatidylinositol 3-phosphate (PI3P) [1] at a confined region is predicted to assemble a large complex of multiple PI3P binding proteins and their associated binding partners. Notably, the lipid kinase, Fab1, binds PI3P [2] (Fig. 2) and catalyzes the conversion of PI3P to PI(3,5)P2 [1]. Recruitment of Fab1 causes local depletion of PI3P and an increase in the levels of PI(3,5)P2, which releases PI3P binding proteins and recruits a distinct set of PI(3,5)P2 binding proteins.
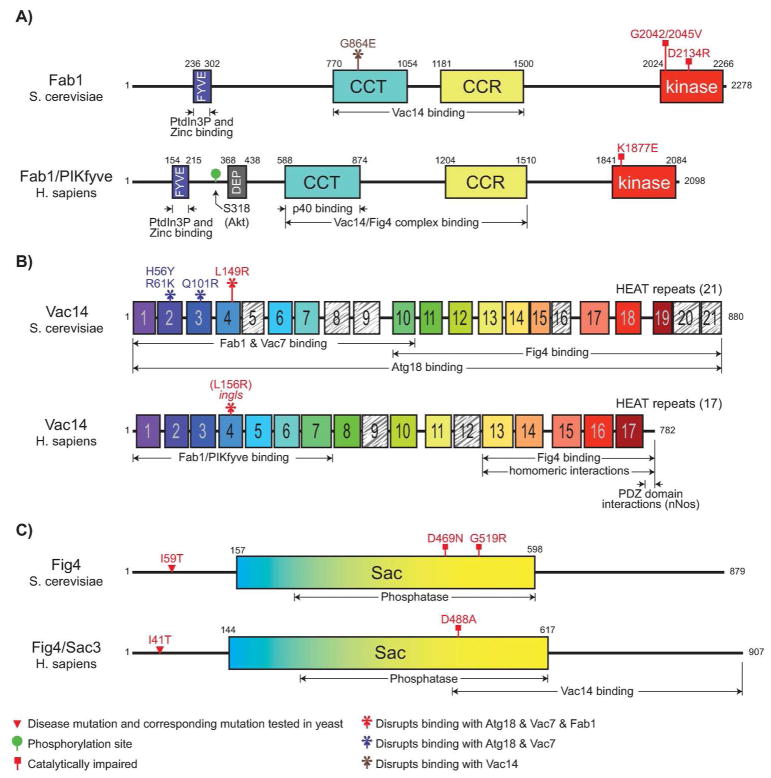
Figure 2.
Fab1/PIKfyve, Vac14 and Fig4 are conserved in most eukaryotes. Domains of S. cerevisiae and human Fab1/PIKfyve, Vac14 and Fig4/Sac3 are shown. A: Fab1 domains include FYVE (binds PI3P), DEP (unknown function; present in chordate and insect Fab1), CCT (homologous to the chaperone Cpn60/TCP-1 family; mediates interactions with Vac14), CCR (a conserved cysteine rich domain found only in Fab1/PIKfyve; part of the Vac14 binding region), kinase (catalytic site for conversion of PI3P to PI(3,5)P2). B: Vac14 is composed of tandem HEAT repeats, which are rod-like helical structures that mediate protein-protein interactions. C: Fig4 contains a Sac domain, which is a module found in several lipid phosphatases. Note, the number of amino acids in mouse PIKfyve and human PIKfyve are not identical. The catalytically impaired mutation in mouse PIKfyve, K1831E, is indicated on the schematic of human PIKfyve, K1877E. The boundaries for FYVE, CCT, CCR, kinase, and Sac domains were identified as follows: 1) conserved in multiple sequence alignments and 2) contained unbroken secondary structure elements predicted by the program Jpred. Sequences for Fab1/PIKfyve were from the following species: Saccharomyces cerevisiae (budding yeast, NP_116674), Schizosaccharomyces pombe (fission yeast, NP_596090), Candida albicans (human pathogen, CAC42810), Ashbya gossypii (cotton pathogen, NP_985045), Arabidopsis thaliana (plant, NP_001078484), Drosophila melanogaster (fly, NP_611269), Apis mellifera (honey bee, XP_393666), Anopheles gambiae (mosquito, XP_314118), Caenorhabditis elegans (worm, CAA19436), and Homo sapiens (human, NP_055855). The Sac domain in Fig4 was defined through alignment of the following Sac domain proteins in S. cerevisiae: Inp51, Inp52, Inp53, Sac1 and Fig4.
Since the discovery of PI(3,5)P2 in 1997 [3,4], the number of known PI(3,5)P2 regulated pathways has expanded greatly. Identification of a comprehensive list of these pathways and downstream effector proteins will be required to fully understand PI(3,5)P2 signaling. Similarly, stimuli that regulate PI(3,5)P2 levels remain to be identified. Here we assess current knowledge and suggest future directions for the study of this very low abundance lipid.
PI(3,5)P2 is much less abundant than most PIPs, including PI4P and PI(4,5)P2. PI(3,5)P2 is present at about 0.1% and 0.04% of total phosphatidylinositol in yeast and mammalian fibroblasts, respectively. The amount of PI(3,5)P2 is 17-fold and 125-fold less abundant than PI(4,5)P2 in yeast [5] and mammalian fibroblasts [6], respectively. The scarcity of PI(3,5)P2 likely contributed to the twenty-five year delay in its discovery [3,4] relative to PI4P and PI(4,5)P2 [7]. Utilizing dilute perchloric acid to precipitate cells followed by deacylation of lipids significantly improved the yield of glycerol-inositol head-groups and the identification of PI(3,5)P2 over the traditional Folch extraction [4,5].
SYNTHESIS AND TURNOVER OF PI(3,5)P2 IS TIGHTLY CONTROLLED BY A LARGE PROTEIN COMPLEX
In yeast, Fab1 [8] is the sole PI3P 5-kinase [1,9] and Vps34 is the sole PI 3-kinase [10]. Both PI(3,5)P2 and PI3P levels dynamically and transiently change in response to specific stimuli. Prolonged introduction of yeast into hyperosmotic media causes a 20-fold transient elevation of PI(3,5)P2 [3] that lasts for about ten minutes before a precipitous drop to basal levels [11]. Concomitant with the rise in PI(3,5)P2, synthesis of PI3P increases. These data suggest that PI(3,5)P2 and PI3P play early roles in adaptation of yeast to hyperosmotic stress. Similarly, these lipids may regulate adaptation in plants and animals, such as transient responses to hormonal or sensory stimuli.
Fab1, commonly called PIKfyve in mammals, is present in most eukaryotes [12]. In this review, “Fab1” refers to Fab1 in all non-mammalian species and “PIKfyve” to mammals. “Fab1/PIKfyve” refers to the mammalian and non-mammalian enzyme. In yeast and mouse embryonic fibroblasts (MEF), Fab1/PIKfyve provides all of the PI(3,5)P2 [1,6,9,13–18]. Across species, the domain structure is similar (Fig. 2).
The PI(3,5)P2 Synthesis Complex
The dynamic and rapid changes in PI(3,5)P2 observed in yeast suggests that Fab1 is tightly regulated. Moreover, overexpression of Fab1 does not increase PI(3,5)P2 levels [1]. Indeed, Fab1 activity requires formation of a complex of regulatory proteins, including Fig4, Vac14, Vac7 and Atg18.
Fig4, a PI(3,5)P2 5-phosphatase, catalyzes the turnover of PI(3,5)P2 in yeast [11,19–21]. Unexpectedly, Fig4 is also required for the activation of Fab1/PIKfyve [6,11,19,22]. Mutations in the catalytic site of Fig4 negatively affect both the turnover of PI(3,5)P2 and the elevation in PI(3,5)P2 in response to hyperosmotic stress [19]. In addition to Fig4 catalytic activity, other regions in Fig4 may play a role. Several disease mutations in Fig4 reside in a non-catalytic, N-terminal domain [23]. Analysis of a corresponding mutation (Fig4-I59T) in yeast revealed a defect in hyperosmotic shock induced activation of Fab1 [22]. Analysis of this N-terminal domain may provide insight into how Fig4 activates Fab1/PIKfyve.
Vac14 regulates both Fab1 and Fig4 [5,24] and is required for the synthesis and turnover of PI(3,5)P2 [11]. Vac14, composed of virtually all HEAT repeats (Fig. 2), functions as a scaffold protein that nucleates the formation of a complex including Fab1, Fig4 and other regulators [25,26] (Fig. 3). Vac14 forms dimers or oligomers [24–27]. In the cytoplasm, Fig4 and Vac14 interact without Fab1 [25]. There may be additional proteins required to form the complex.
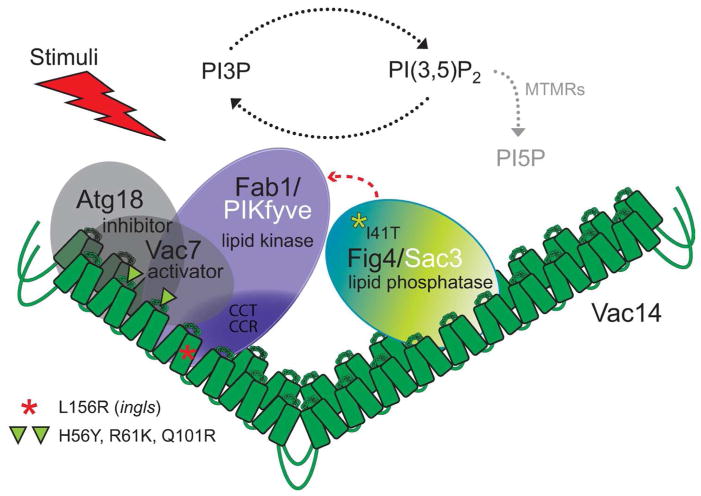
Figure 3.
Schematic of the Fab1/PIKfyve, Vac14, Fig4/Sac3 complex. Vac14 oligomerizes with itself and nucleates the complex through direct interactions with Fab1/PIKfyve and Fig4/Sac3. In yeast, Vac14 also directly interacts with Atg18 and Vac7. The yeast Vac14 point mutants, H56Y (HEAT repeat loop 2), R61K (HEAT repeat loop 2) and Q101R (HEAT repeats loop 3), each disrupt binding of Atg18 and Vac7. Thus, Atg18 and Vac7 may bind overlapping or identical sites of Vac14 [26]. The Vac14-L156R mutation, found in ingls mice, and corresponding mutation Vac14-L149R in yeast, disrupts Vac14 interaction with Atg18, Vac7 and Fab1. This suggests that all three proteins bind overlapping sites on Vac14. The point mutation, Fig4-I41T found in patients with CMT4J, disrupts the interaction between Fig4 and Vac14, although the major portion of human Fig4 that interacts with Vac14 resides within residues 478–907 [28]. In mammalian cells, myotubularin related proteins (MTMRs) can convert PI(3,5)P2 to PI5P and may provide the majority of cellular PI5P.
Similarly, mammalian Vac14 (ArPIKfyve) forms a complex with PIKfyve, and Fig4 (Sac3) [26,28–31]. The interaction sites between the yeast and mammalian complexes are likely conserved. The binding site for Fab1 on Vac14 is conserved in the mammalian complex [26,28]. Fig4 binds to Vac14 through the conserved C-terminal region [28]. Additionally, the N-terminal pathogenic point mutation, Fig4-I41T, disrupts the interaction of Fig4 with Vac14 and destabilizes Fig4 [29,32]. These observations raise the possibility that both the N- and C-termini of Fig4 interact with Vac14.
Yeast Vac7, a critical activator of Fab1, has no recognizable motifs; its mode of action is unknown [1,5,11,19,20,33]. Vac7 resides within the Fab1 complex, but is not required for formation or localization of the complex [1,21,26]. This is surprising because Vac7 is the only protein in the complex with a transmembrane domain [5]. Vac7 function is likely conserved in metazoans. However, based on sequence similarity, Vac7 is only present in some fungi. Either, alternative mechanisms activate Fab1/PIKfyve in metazoans, or proteins with functions analogous to Vac7 cannot be identified by BLAST search.
Yeast Atg18, a negative regulator of PI(3,5)P2 levels, resides within the Fab1 complex. Through two adjacent binding sites [34–36], Atg18 binds to PI(3,5)P2 and PI3P [37]. These sites are essential for Atg18 to negatively regulate PI(3,5)P2 levels and for localization of Atg18 on the vacuole [38]. Relief of Fab1 inhibition in an atg18Δ strain requires Fab1 activators. Thus, Atg18 likely inhibits the activators rather than acting on Fab1 directly. Metazoans may have unidentified proteins that function similarly to yeast Atg18. The mammalian genes, WIPI1, WIPI2, WIPI3 and WIPI4, encode proteins with greater than 20% identity to yeast Atg18 [15,39]. WIPI1 and WIPI2, like Atg18, function in autophagy. However, whether they function as negative regulators of PI(3,5)P2 levels has not been tested.
Orchestrating Fab1/PIKfyve activity
At least three mechanisms within the Fab1 complex contribute to the dynamic regulation of PI(3,5)P2. First, the lipid kinase and phosphatase reside within the complex. Second, the Fab1 activator, Vac7, and inhibitor, Atg18, bind overlapping sites on Vac14 and likely compete for access to Fab1. Third, catalytic activity of Fig4 is required for the activation of Fab1. Tight coordination between synthesis and turnover of PI(3,5)P2 likely explains how a sustained stimulus of hyperosmotic shock causes a steep transient increase in PI(3,5)P2 levels.
Other opposing lipid kinases and phosphatases reside in the same complex or have coordinated regulation (reviewed in [40]). MTM1, a lipid 3-phosphatase, resides in a complex with the PI3-kinase, Vps34. Inositol polyphosphate 4-phosphatase is in a complex with a class I PI3-kinase. The added complexity in the Fab1 complex, that the opposing lipid phosphatase has a second role as activator of the lipid kinase, underscores the importance of directly measuring phosphoinositide lipid levels to determine cellular functions of predicted lipid phosphatases.
Comparison of PI(3,5)P2 synthesis in yeast and metazoans
In metazoans, several PI 3-kinases, in addition to Vps34, may produce the PIKfyve substrate, PI3P. Indeed, knockdown of either PIKfyve or PI3K-C2α, but not Vps34, affects TORC1 activity in adipocytes [41]. Thus, in some cases PI3K-C2α may provide the pool of PI3P utilized to generate PI(3,5)P2.
A major difference between the yeast and mammalian Fab1/PIKfyve complex are the lipid pools that they control. Surprisingly, in MEF cells the PIKfyve complex is required for most of the PI5P and all of the PI(3,5)P2 pool [6,14,17,42]. An independent study concluded that PIKfyve does not contribute to PI5P levels [43]; however, that study assumed that PIKfyve inhibition did not impact the lipids used to standardize the samples.
The relative importance of Vac14 for Fab1/PIKfyve activity differs between the yeast and mammalian complexes. In vac14Δ yeast, PI(3,5)P2 levels are reduced at least 10-fold [11,19], while Vac14−/− and Fig4−/− MEF cells reveal a more modest 2-fold reduction in PI(3,5)P2 and PI5P [6,22,42]. Since Vac14 or Fig4 are required for only half of the PI(3,5)P2 pool and PIKfyve is required for the entire pool, PIKfyve either retains partial function in the absence of Vac14 or Fig4 and/or PIKfyve has additional regulators.
Localization of the Vac14, Fab1/PIKfyve and Fig4
In yeast, Fab1, Vac14, Fig4 are found on the limiting membrane of vacuoles and adjacent foci, which are likely endosomes [21,25,26]. In metazoan cells, Fab1/PIKfyve and Vac14 are found on early and late endosomes, lysosomes and in the cytosol [44–50]. Questions remain about how the complex is associated with membranes. Is the FYVE domain of Fab1/PIKfyve sufficient for localization of the complex? Are there other lipid-binding or transmembrane containing subunit(s)?
PI(3,5)P2 is a precursor for PI5P synthesis
PI(3,5)P2 likely serves as a precursor for most of the cellular PI5P pool. The strongest evidence for this hypothesis comes from heterologous expression of PIKfyve in yeast, which greatly increases PI(3,5)P2, decreases its precursor, PI3P. Importantly, the combined total of PI3P, PI5P and PI(3,5)P2 remains constant in the presence or absence of heterologous PIKfyve [6]. If PI5P were generated directly by PIKfyve, then new direct conversion of PI to PI5P would raise the combined total of PI3P, PI5P and PI(3,5)P2. Additionally, transient activation or inhibition of endogenous PIKfyve in fibroblasts, causes PI(3,5)P2 levels to reach a new steady-state faster than PI5P, an outcome consistent with a precursor-product relationship [6].
Generation of PI5P from PI(3,5)P2 requires proteins with 3-phosphatase activity [6], which may be provided by myotubularins (MTMRs) [51]. Indeed, mouse MTMR2 and Drosophila MTMR3 function with Fab1/PIKfyve to control PI5P and PI(3,5)P2 [52,53]. That both PI(3,5)P2 and PI5P are embedded in membranes, and cannot freely diffuse, raises the possibility that MTMRs reside within the PIKfyve complex. This would provide rapid access of MTMRs to the newly synthesized PI(3,5)P2.
An alternative hypothesis, that PIKfyve directly generates most of the cellular PI5P, has been recently reviewed [54]. Briefly, there is controversy between independent in vitro studies about whether PIKfyve directly generates PI5P [12]. In some studies PIKfyve was immunoprecipitated from cells that express many lipid 3-phosphatases. Tight association of PIKfyve with 3-phosphatases during immunoprecipitation may explain some discrepancies. Strong in vitro evidence that PIKfyve can directly generate PI5P comes from studies of PIKfyve expressed from insect Sf9 cells [55]. Development of a general inhibitor of myotubularin function may help resolve whether most of the cellular PI5P pools are generated directly or indirectly by PIKfyve.
That PIKfyve is either directly or indirectly responsible for most of the PI5P in fibroblasts raises questions about whether PI(3,5)P2 and PI5P reside on the same membrane. Localization of Vac14, Fab1/PIKfyve, and Fig4 provide insights into the subcellular locations of PI(3,5)P2 in yeast and metazoans. If MTMRs are associated with this complex, then MTMR localization would provide information for the subcellular distribution of PI5P as well (Fig. 4A). However, the presence of Vac14, Fab1/PIKfyve, and Fig4 does not a priori indicate enzymatic activity. Thus, development of probes will be critical to determine the spatial and temporal dynamics of PI(3,5)P2 and PI5P.
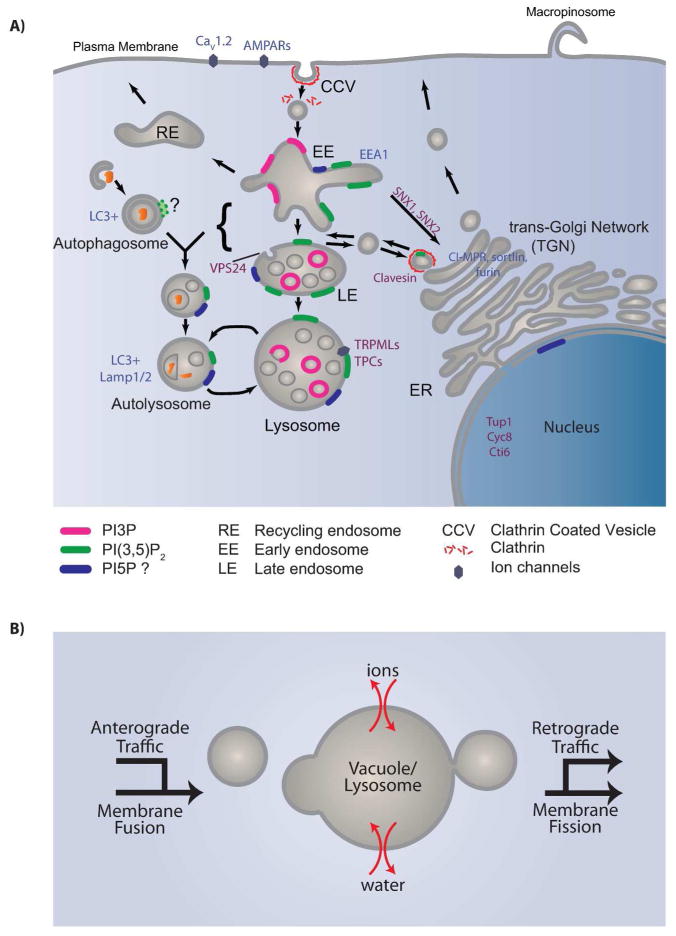
Figure 4.
A: Localization of PI(3,5)P2 and PI5P inferred from the localization of PIKfyve and Vac14. PI(3,5)P2 localizes on early endosomes, late endosomes and lysosomes. Localization of PI(3,5)P2 on autophagosomes is less clear. PI5P may also be present at all or some of these locations. To further establish the locations of these lipids, suitable lipid probes need to be developed. PI(3,5)P2 effectors and trafficking pathways affected in PIKfyve/Vac14/Fig4 deficient cells are also indicated. Purple: known PI(3,5)P2 effectors. Blue: proteins affected by PI(3,5)P2 and/or PI5P. B: The size of a yeast vacuole or mammalian lysosome is dependent on ion and water homeostasis, as well as the net sum of anterograde traffic, retrograde traffic, membrane fusion and membrane fission.
PI(3,5)P2 BINDING PROTEINS
Based on the pleiotropic defects observed in cells and organisms with defects in PIKfyve activity, most PI(3,5)P2 binding proteins are likely not yet identified. To date, multiple types of motifs as well as novel sequences have been shown to interact with PI(3,5)P2. PI(3,5)P2 binds directly to some WD40 domain containing proteins, including Atg18, Atg21, Hsv2, Tup1 (in yeast) and Raptor (in adipocytes), and regulates their functions in vivo [37,41,56]. Additionally, sorting nexin proteins, SNX1 and SNX2 (PX domain) [57,58], Cti6 (PHD domain) [56], clavesin (Sec14 domain) [59] and class II formins (PTEN domain) [60] interact with PI(3,5)P2. TRPML1 and RyR1 also bind PI(3,5)P2 [61,62]. In these latter examples, no lipid binding motif is apparent. Thus, bioinformatic approaches are not sufficient to determine which proteins bind PI(3,5)P2.
Atg18 binds PI(3,5)P2 with a high affinity, in the nanomolar range [37], likely due to tandem lipid binding sites. Lower affinity interactions may be of equal biological significance but are more difficult to detect. Moreover, some effectors require simultaneous interactions with other proteins (reviewed in [63]). For example, the FYVE domain containing protein, EEA1, associates with membranes via simultaneous interaction with PI3P and Rab5 GTPase. The development of strategies to detect relatively low affinity binding will be necessary to identify the full set of PI(3,5)P2 and PI5P effectors.
PATHWAYS REGULATED BY Vac14, Fab1/PIKfyve and Fig4
Mutants deficient in Fab1 or its regulators provide information on its cellular (Table 1) and physiological roles (Table 2). The PIKfyve inhibitors, YM201636 and MF4, have also facilitated studies of cellular functions of PIKfyve [13,64]. However, off-target effects need to be considered [65].
Table 1.
Pathways regulated by Fab1/PIKfyve, Vac14, and Fig4 complex
| Pathways affected | Known PI(3,5)P2 Effectors | Species and reference |
|---|---|---|
| PI5P biogenesis | MTMRs† [6,111] | Cell culture [6,111] |
| Vacuole fission | Atg18 [37] | S. cerevisiae [33,37] |
| Retrograde traffic from the vacuole | Atg18 [37] | S. cerevisiae [37,79] |
| Acidification of vacuoles or endolysosomes | S. cerevisiae [8],[33]; C. elegans [75]; D. melanogaster [49]; A. thaliana [66]; | |
| Ion channel function | TRPML1, TRPML2, TRPML3 [61]; TPC1, TPC2, TPC3 [72]; RyR1, RyR2 [62,73,112] | Cell culture and mouse [61,62,72,73,112] |
| Autophagy in metazoans | C. elegans [75]; D. melanogaster [88]; Cell culture [13,64]; Mouse [32,53,69,90] | |
| Fluid phase endocytosis | Cell culture [13,42,113]; D. melanogaster [49] | |
| Sorting of cargoes at the MVB | Vps24† [81] | S. cerevisiae (Reviewed in [82]) |
| Traffic of cell surface receptors to lysosomes | Cell culture [13]; D. melanogaster [49] | |
| Endosome-to-TGN traffic | SNX1, SNX2 [58] | Cell culture [13,42,48,58,85] |
| Glut4 translocation in response to insulin | 3T3 adipocytes (Reviewed in [54]) | |
| mTORC1 translocation to the plasma membrane in response to insulin | Raptor [41] | 3T3 adipocytes [41]; |
| Cortical actin array dynamics | Formins [60] | P. patens [60] |
| GAL1 induction in the absence of GAL4 pathway | Tup1, Cti6 [56] | S. cerevisiae [56] |
| AMPA receptor and Cav1.2 trafficking | Neuron culture [44,100,101] | |
| Exocytosis | Neuron culture [44,102] |
Table 2.
Phenotypes of PI(3,5)P2 deficiency in model organisms.
| Phenotypes | Species and reference |
|---|---|
| Enlarged endosomes and lysosomes | S. cerevisiae [1,8,9,33]; S. pombe [16]; C. albicans [74]; Cell culture [6,13,22,42,48,64,68]; C. elegans [75]; D. melanogaster [49]; A. thaliana [67] |
| Sensitivity to heat and calcium | S. cerevisiae [8]; S. pombe [16] |
| Secretion and response to mating hormones | S. pombe [16] |
| Hyphae formation on solid medium | C. albicans [74] |
| Inviable pollen, slow root growth, curled leaves, dwarfed plants, abnormal flowers | A. thaliana [66,67] |
| Stunted growth | P. patens [60] |
| Early lethality | C. elegans [75]; D. melanogaster [49]; mouse [6,14,22,26,42] |
| Dilute coat color | Mouse [22,26] |
| Neurodegeneration | Mouse [6,22,26,42,53,90,97,116,117] |
| Increased pre- and post-synaptic strength | Neuron culture [44] |
| Myelination defects | Mouse [53,97,116,117] |
| Abnormalities in heart, lung, kidney, thymus, and spleen | Mouse [6,22] |
In mammals, loss of PIKfyve function decreases both PI(3,5)P2 and PI5P [6,14,42], thus phenotypes linked to mutations in this pathway may be due to loss of PI(3,5)P2, PI5P or both lipids. In addition, disagreement among studies about whether a specific pathway requires PIKfyve may be due to differences in the extent of decrease of PIKfyve activity. In most cell-based studies some PIKfyve function remains, including mutant cells and RNAi experiments. For example, in PIKfyveβgeo/βgeo MEF cells, 5% of the normal levels of PIKfyve provide half of the normal levels of PI(3,5)P2 and PI5P [6]. Here, we present pathways that require PIKfyve. Those known to be directly regulated by PI(3,5)P2, because PI(3,5)P2 protein effectors have been identified, will be indicated. In other cases, the regulatory lipid may either be PI(3,5)P2 or PI5P.
Formation of large vacuoles
A striking feature in PI(3,5)P2 deficient organisms are enlarged vacuoles [8,18,22,33,42,48,64,66–68]. In mutant yeast, the vacuole/lysosome is enlarged. In Vac14−/− and Fig4−/− MEF cells, the vacuoles are heterogeneous, arising from both late endosomes and lysosomes, as well as enlarged autophagosomes [22,42,44,69]. Complete inhibition of PIKfyve causes vacuoles to form from early endosomes as well [64].
The enlarged vacuoles in PI(3,5)P2-defective yeast mutants cannot release water even when exposed to hyperosmotic shock [5], which suggests an inability to regulate the water content of the vacuole. Similarly, vacuoles in mammalian cells are likely due to defects in the regulation of osmolarity within the endomembrane system. The Vac14Ingls/Ingls and Fig4−/− mouse mutants exhibit extreme hydrocephalus [26,32], and the vacuoles that form in Vac14−/− or Fig4−/− MEF cells are not filled with lipid [22,42].
PI(3,5)P2 regulates some ion channels
The defects in water homeostasis may be linked to defects in ion homeostasis. Indeed, overexpression of the TRPML1 calcium channel in Vac14−/− MEF cells suppresses the formation of vacuoles [61]. In S. pombe, mutations in a calcium permease (SPAC521.04c) rescue the enlarged vacuoles in the fab1Δ mutant [70]. Suppression in both cases may be due to regulation of calcium flux.
A role for phosphoinositide regulation of ion channels is better understood on the plasma membrane where multiple ions channels are activated by PI(4,5)P2. In some cases, PI(4,5)P2 directly interacts with the channel; in other cases, PI(4,5)P2 recruits regulators [71]. Similarly, PI(3,5)P2 activates ion channels on endosomes and lysosomes, including mucolipin transient receptor potential channels (TRPML1, TRPML2, TRPML3) and yeast homolog, yeast vacuolar conductance (Yvc1), and two-pore channels (TPC1, TPC2, TPC3) [61,72]. While the mechanism of PI(3,5)P2 regulation of TPCs is not known, PI(3,5)P2 interacts directly with the cytoplasmic N-terminus of TRPML1. PI(3,5)P2 is also important for calcium dynamics in muscles. PI(3,5)P2 directly activates the ryanodine receptors (RyR1, RyR2), which release calcium from the sarcoplasmic reticulum in skeletal and cardiac muscles, respectively [62,73].
PI(3,5)P2 plays a role in the acidification of the vacuole
Vacuoles in fab1Δ, vac7Δ, and vac14Δ yeast are less acidified than wild-type vacuoles [8,33]. Similar phenotypes occur in Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster and Arabidopsis thaliana [49,66,74–76]. These acidification defects may contribute to the formation of large vacuoles; vacuolar ATPase function is required for both vacuole fission and fusion [77]. However, PI(3,5)P2 plays additional roles in vacuole morphology. A limited increase in PI(3,5)P2 corrects acidification of the vacuole without correcting vacuole size [1,5]. PI(3,5)P2 effectors involved in acidification have not been identified, although the vacuolar ATPase is a likely candidate. While assembly of the vacuolar ATPase does not require PI(3,5)P2 [5], PI(3,5)P2 may regulate vacuolar ATPase activity.
Fab1/PIKfyve is required for multiple pathways in the endomembrane system
Membrane trafficking defects also contribute to the formation of enlarged vacuoles. In yeast, PI(3,5)P2 and Atg18 are required for fission of the vacuole [38,78] and retrograde traffic from the vacuole to the Golgi [37,79]. These defects contribute to but do not fully account for the large vacuoles in the fab1Δ mutant. The vacuoles in atg18Δ are not as enlarged as observed in fab1Δ yeast [38]. Thus, defects in water and ion homeostasis, vacuole acidification, as well as defects in membrane trafficking and vacuole fission, each contribute to the enlarged vacuoles caused by low levels of PI(3,5)P2 (Fig. 4B).
Roles for Fab1/PIKfyve in the multivesicular body pathway
Fab1/PIKfyve function has been linked to multivesicular body (MVB) formation, a protein degradation pathway. MVB formation involves the ubiquitination and capture of cargoes on the limiting membrane of late endosomes, which are then internalized via invagination and formation of intraluminal vesicles (ILV). Degradation of protein cargoes occurs when the MVB fuses with the lysosome. In fab1Δ yeast, fewer ILVs are formed [1]. Formation of ILVs requires several ESCRT proteins, including Vps24. While controversial [80], one study suggested that Vps24 binds PI(3,5)P2 [81]. Thus, the partial defect in ILV formation may be due to a requirement for PI(3,5)P2 in Vps24 function. In addition to a possible role in forming ILV vesicles, PI(3,5)P2 may be required for sorting some protein cargoes (reviewed in [82]). How PI(3,5)P2 regulates cargo sorting remains to be determined.
Loss of PIKfyve activity has also been linked to events that occur after cargo sorting. Drosophila Notch, Wingless and Dpp (a fly homologue of TGFβ) accumulate in the MVB and are not degraded in a Drosophila fab1 mutant. Thus, Fab1 may also function downstream of cargo internalization [49]. Delayed epidermal growth factor receptor (EGFR) degradation due to inhibition of PIKfyve [64,83] may also be due to similar defects: either trafficking problems after sorting ligands into the MVB or loss of protease activity in lysosomes that are not properly acidified.
PIKfyve is required for protein trafficking from endosomes to the TGN
Similar to yeast [37,79], PIKfyve is required for retrograde traffic of proteins from endosomes to the trans-Golgi network (TGN). Knockdown of PIKfyve inhibits retrograde traffic of the cation-independent mannose-6-phosphate receptor (CI-MPR), sortillin (a related receptor) and furin [48]. In Vac14−/− fibroblasts [42], or following inhibition of PIKfyve [13], CI-MPR localizes to endosomes and cathepsin D, one of its ligands, is missorted [42]. Inhibition of PIKfyve also delays trafficking of the Shiga Toxin B subunit from endosomes to the TGN [13]. These defects may be due in part to misregulation of SNX1 and SNX2, retromer proteins that bind directly to PI(3,5)P2. In addition, PIKfyve binds to two proteins required for retrograde trafficking: p40, a Rab9 effector, and JLP, a kinesin adaptor required for microtubule based transport from endosomes to the TGN [84,85].
Regulation of PIKfyve in response to insulin
PIKfyve is required for insulin-mediated Glut4 translocation. Insulin stimulates the glucose transporter (Glut4) to transiently translocate to the plasma membrane, which facilitates glucose uptake. Regulated Glut4 trafficking occurs in both adipocytes and muscle. Suppression of PIKfyve activity reduces insulin induced Glut4 translocation in cultured adipocytes (reviewed in [54]). PIKfyve is also required for Glut4 trafficking in animals; a muscle-specific knock-out of PIKfyve in mice causes a defects in Glut4 translocation and glucose uptake [86]. The precise role(s) of PIKfyve in Glut4 translocation are not known.
Insulin stimulation regulates PIKfyve activity [41,46,87], in part by Akt, which phosphorylates PIKfyve on serine 318 [87]. Moreover, EGF stimulation, which promotes EGFR internalization and degradation, also induces Akt phosphorylation of PIKfyve on serine 318 [83]. However, in cells the degree of activation of PIKfyve due to phosphorylation of serine 318 is relatively modest. Thus, there are may be additional Akt phosphorylation sites on PIKfyve, as well as additional PIKfyve activators.
One outcome of insulin activation of PIKfyve in adipocytes is an effect on mTOR, a major regulator of cell metabolism. Insulin-induced translocation of mTOR to the plasma membrane, as well as mTOR activity, requires PIKfyve [41]. The recruitment of mTOR to the plasma membrane in response to insulin may occur through direct interactions with PI(3,5)P2. However, whether PI(3,5)P2 is found on the plasma membrane of adipocytes is not known.
Fab1/PIKfyve is required for autophagy in metazoans
Autophagy requires Fab1/PIKfyve. Autophagy delivers cargoes to the lysosome for degradation. Suppression of Fab1/PIKfyve results in impaired clearance of autophagic organelles. In C. elegans, mutations in PPK-3 (Fab1) cause an increase in autophagosomes [75]. Similarly, Drosophila fab1 mutant larvae accumulate autophagosomes and amphisomes [88]. In NIH3T3 or HEK293 cells, and in primary cultured hippocampal neurons, inhibition of PIKfyve with YM201636 or MF4 causes an accumulation of autophagosomes and the autophagic marker, LC3-II [13,64,89]. Similarly, the brains of mice with mutations in Fig4 have elevated levels of LC3-II and p62, another marker of autophagy [32,53,69,90]. Together, the above studies indicate that PIKfyve has multiple roles in the endomembrane system.
Roles for lysosomal PI(3,5)P2 in the regulation of transcription
Endosomal PI(3,5)P2 may also regulate some transcriptional pathways. Expression of pheromone responsive genes in S. pombe is defective in a fab1Δ mutant [16]. Similarly, in S. cerevisiae PI(3,5)P2 modulates transcription via interaction with Tup1 and Cti6 [56]. PI(3,5)P2 provides a site on the yeast vacuole for assembly of the Tup1/Cyc8/Cti6 transcription complex. These findings predict that PI(3,5)P2 on lysosomes may regulate additional transcription pathways.
Fab1/PIKfyve may function at the plasma membrane
In addition to multiple functions on endosomes, a small pool of Fab1/PIKfyve may function at or near the plasma membrane. In adipocytes, PIKfyve activity may contribute to localization of mTORC1 to the plasma membrane. Furthermore, PIKfyve has been implicated in phagocytosis and pinocytosis [91,92]. Further evidence for a potential role for Fab1/PIKfyve at the plasma membrane comes from the Physcomitrella patens class II formins, which bind PI(3,5)P2 [60] and require Fab1 activity for their localization at the cell cortex. PIKfyve has also been implicated in actin remodeling in mammalian cells (reviewed in [54]). In addition, when expressed heterologously in Xenopus oocytes, several plasma membrane localized ion channels and carrier proteins require PIKfyve activity (reviewed in [54,82,93]). Thus, while most PIKfyve is associated with endosomal membranes, PIKfyve may also have roles at the plasma membrane.
Fab1/PIKfyve, Vac14, and Fig4 IN PLANT AND ANIMAL PHYSIOLOGY
PIKfyve plays critical roles in development. Knockout of PIKfyve in mice results in very early lethality: PIKfyve−/− embryos did not survive past E3.5 [14] and, in an independent knock-out, embryos did not survive past E8.5 [18]. Similarly, Drosophila fab1 and C. elegans (ppk-3) mutants display early lethality [49,75]. In Arabidopsis thaliana the two Fab1 genes, FAB1A and FAB1B, play critical roles in development [66,67]}, perhaps due in part to hyposensitivity to auxin signaling [94].
Analysis of a PIKfyveβgeo/βgeo hypomorphic mutant mouse with partial PIKfyve activity, which dies perinatally, has revealed post-development roles of PI(3,5)P2 and PI5P in animal physiology [6]. Similarly, Vac14−/− mutant mice, which also have less Fig4 protein [6,32], die perinatally [42]. Fig4−/− mice can live up to 6 weeks [22]. Vac14Ingls/Ingls, a missense mutation that disrupts binding of Vac14 with PIKfyve, survives up to 3 weeks [26]. Differences in lethality may be largely due to differences in strain background. Early lethality is rescued in Fig4−/− mice by neuronal-specific, but not astrocyte-specific, expression of Fig4 [95]. Thus, loss of PI(3,5)P2 and PI5P in neurons likely contributes to early lethality of the Fig4−/− mice and other PIKfyve-related mouse models.
Multiple tissues require PIKfyve
Vac14, Fig4 and PIKfyve are expressed globally. Accordingly, defects in the corresponding mouse mutants occur in multiple tissues. Hearts of the Vac14−/−, Fig4−/− and PIKfyve hypomorph mutants have vacuoles [6] and in the two latter mutants, there is a spongiform-like phenotype in the spleen as well. Moreover, the lungs and kidneys of the PIKfyve hypomorph have a spongiform-like appearance. Conditional knock-out of PIKfyve (PIPKIII), in intestinal cells, causes vacuole formation and defects in membrane trafficking in the gut epithelia, which ultimately lead to early lethality [18].
Vac14, PIKfyve and Fig4 proteins are most abundant in the nervous system, which fits with findings that the nervous system is profoundly affected in the corresponding mutant animals [6,44]. Fig4−/−, Vac14−/− and Vac14Ingls/Ingls mice display degeneration of the brain, including enlarged ventricles, increased apoptosis and severe spongiform encephalopathy; large vacuoles in the cell bodies of neurons are also observed in the peripheral nervous system [22,26,42]. The PIKfyve hypomorph has similar defects [6]. Consistent with the importance of PI(3,5)P2 and PI5P in the nervous system, a mouse with a neuron-specific knock-out of Vps34, displays juvenile lethality and neurodegeneration, and has reduced PI3P and PI(3,5)P2. PI5P was not measured [96].
Myelination is reduced in the central and peripheral nerves of Fig4−/− mice [22,95,97]. Fig4 may be particularly abundant during development of myelinating cells and dorsal root ganglia sensory neurons [98], although Fig4−/− controls, which would indicate whether the antigen detected by the anti-Fig4 antibody was bona fide Fig4, were missing. Interestingly, hypomyelination in Fig4−/− mice is rescued by neuron-specific expression of Fig4 [97]. Heterozygous Fig4+/− mice show no signs of neurodegeneration or increased susceptibility to trauma induced degeneration [99]. Mtmr2−/− Fig4−/− double mutant mice have more severe hypomyelination and neurodegeneration, which suggest that loss of PI5P contributes to these phenotypes [53].
PIKfyve in neurons
Vac14, PIKfyve and Fig4 have specialized roles at the synapse. AMPA-type glutamate receptors, which mediate fast neurotransmission in the brain, cycle between endosomes and the plasma membrane. Notably, trafficking of the AMPA receptor subunits, GluA1 and GluA2, are modulated by the PIKfyve complex. shRNA silencing of PIKfyve impairs trafficking of GFP-HA-GluA2 [100], and addition of PI(3,5)P2 promotes trafficking of heterologously expressed GluA1 [101]. In Vac14−/− cultured hippocampal neurons, GluA1 and GluA2 are increased on the plasma membrane with a concomitant increase in postsynaptic strength [44]. Similarly, in cultured cortical neurons, internalization and degradation of the L-type voltage-gated calcium channel subunit, CaV1.2, requires PIKfyve [100].
In addition to postsynaptic defects, Vac14−/− neurons also displayed an increased probability of presynaptic vesicle fusion [44]. Similarly, PIKfyve is a negative regulator of calcium-dependent exocytosis in neurosecretory cells [102]. Together, PIKfyve and potentially PI(3,5)P2, PI5P or both negatively regulate the excitatory response of neurons, which may explain why defects in the PIKfyve complex are linked to excitotoxic neuronal death.
Further determination of roles for PI(3,5)P2 and PI5P signaling at the synapse will likely come from identification of proteins that binds these lipids and/or interact with the Vac14, PIKfyve, or Fig4. Potential candidates include clavesin and nitric oxide synthase (nNOS). Clavesin (clathrin vesicle-associated Sec14 protein), is expressed solely in the brain and binds PI(3,5)P2. Knockdown of clavesin causes enlarged late-endosomes/lysosomes similar to those seen with suppression of PIKfyve activity [59]. nNOS, which functions at the synapse in the regulation of neurotransmission, binds Vac14 through a PDZ domain in vitro [103]; a functional interaction between Vac14 and nNOS at the synapse has not been tested.
MUTATIONS IN GENES THAT ENCODE THE VAC14, PIKFYVE, AND FIG4 COMPLEX ASSOCIATED WITH HUMAN DISEASES
Mutations in FIG4 underlie a severe form Charcot Marie-Tooth (CMT) type 4J [22]. In CMT, progressive deterioration of nerves and/or demyelination throughout the peripheral nervous system results in reduced nerve conduction velocity and sensory sensation. These defects overlap with those observed in the Fig4−/− mouse. The most common genotype in CMT4J patients is FIG4 compound heterozygosity: one null allele and the other encoding the missense mutation, isoleucine 41 to threonine (I41T) [22]. The mutation retains partial function. In Fig4−/− mice, overexpression of a Fig4-I41T transgene significantly suppresses the early lethality [22,32]. That Fig4-I14T has a modest functional defect, yet causes peripheral neuropathy, underscores the importance of precise modulation of PI(3,5)P2 and/or PI5P levels in the nervous system.
CMT4B1 and CMT4B2 are caused by loss-of-function mutations in MTMR2 and MTMR13 respectively (reviewed in [104]) and have clinical symptoms that overlap with those observed in CMT4J. The clinical symptoms in common between CMT4B and CMT4J may be due to either less PI5P or elevated PI3P.
A range of mutations in Fig4 were found in 7 out of 473 patients with ALS and 2 patients with PLS [105]. Mutations in Fig4 may be causative in other neurological diseases as well. Moreover, mutations in Fig4 can cause defects in additional tissues. Homozygous null mutations in Fig4 cause Yunis-Varón syndrome, a severe autosomal-recessive congenital disorder, which affects multiple tissues, including the heart, skeletal muscle, skeleton and brain [106]. The diversity of affected tissues underscores the importance of the Vac14/PIKfyve/Fig4 complex in human physiology.
To date, neither PIKfyve nor Vac14 have been linked to neurological disease. Heterozygous null mutations in PIKfyve are associated with Francois-Mouchetee Fleck Corneal Dystrophy (CFD) [107], which results in white flecks throughout the corneal stroma of the eye that do not affect vision. Corneal flecks are thought to be enlarged vacuoles in swollen keratocytes [108]. Interestingly, Vac14 mRNA is down-regulated in a large subset of patients with chronic fatigue syndrome [109]. Based on the common molecular functions of PIKfyve, Vac14 and Fig4, it is tempting to speculate that mutations in PIKfyve and Vac14 will be discovered that are linked to neurological disorders.
Conclusions
The roles and regulation of PI(3,5)P2 parallel those of other PIP species. Notably, PI3P is a precursor for PI(3,5)P2, which in turn is a precursor for PI5P. The interconversion between these lipids predicts that there are pathways where these lipids spatially and temporally control multi-step pathways.
Compared with PI3P, PI4P and PI(4,5)P2, the levels of PI(3,5)P2 are exceedingly low. The difficulty of measuring the low levels of PI(3,5)P2 in cells, and the lack of a fluorescent probe to monitor its spatial and temporal dynamics have provided major hurdles towards elucidating the roles and regulation of PI(3,5)P2. A more complete picture of the pathways that rely on PI(3,5)P2 and PI5P will likely provide insights into how minor defects in the regulation of these lipids leads to profound human diseases. Recent observations that mutations in Fig4 cause defects with striking similarities to lysosomal storage disorders may also provide insight into the links between these lipids and disease [110]. Moreover, as whole exome sequencing of patients becomes feasible, more diseases linked to this pathway will likely be discovered. The severity of CMT4J, ALS and Yunis-Varon syndrome underscores the importance of uncovering the molecular mechanisms that regulate the Vac14/PIKfyve/Fig4 complex, as well as the discovery of new cellular pathways that are regulated by PI(3,5)P2 and PI5P.
Table 3.
Human Disease
| Disease | Affected Gene | Reference |
|---|---|---|
| Charcot Marie-Tooth type 4J (CMT4J) | Fig4 I41T* Fig4 L17P* |
[22,105,116] |
| Amyotrophic lateral sclerosis (ALS) | Mutations in Fig4 | [105] |
| Primary lateral sclerosis (PLS) | Mutations in Fig4 | [105] |
| Yunis–Varon syndrome | Mutations in Fig4 | [106] |
| Charcot Marie-Tooth type 4B1 | Mutations in MTMR2 | Reviewed in [104] |
| Charcot Marie-Tooth type 4B2 | Mutations in MTMR13 | Reviewed in [104] |
| Francois-Mouchetee Fleck Corneal Dystrophy | Mutations in PIKfyve | [107,108] |
| Chronic Fatigue Syndrome | Down regulation of Vac14 | [109] |
Patients are compound heterozygotes with a null allele of Fig4 and Fig4-I41T, or Fig4-L17P.
Acknowledgments
Due to space limitations, we apologize to our friends and colleagues for omission of some critical citations. We thank Drs. Miriam Meisler and Michael Sutton for discussions of this manuscript. The yeast and metazoan portions of this review were supported by R01-GM50403 and R01 NS064015, respectively. AJM was supported in part by NRSA F31NS074740 and Rackham Predoctoral Fellowship.
Footnotes
The authors certify that they have no conflict of interest.
References
- 1.Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, et al. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. The Journal of cell biology. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Molecular cell. 1998;2:157–62. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 3.Dove SK, Cooke FT, Douglas MR, Sayers LG, et al. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–92. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 4.Whiteford CC, Brearley CA, Ulug ET. Phosphatidylinositol 3,5-bisphosphate defines a novel PI 3-kinase pathway in resting mouse fibroblasts. Biochem J. 1997;323 (Pt 3):597–601. doi: 10.1042/bj3230597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonangelino CJ, Nau JJ, Duex JE, Brinkman M, et al. Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. The Journal of cell biology. 2002;156:1015–28. doi: 10.1083/jcb.200201002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zolov SN, Bridges D, Zhang Y, Lee W-W, et al. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proceedings of the National Academy of Sciences U S A. 2012;109(43):17472–7. doi: 10.1073/pnas.1203106109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brockerhoff H, Ballou CE. Phosphate incorporation in brain phosphionositides. J Biol Chem. 1962;237:49–52. [PubMed] [Google Scholar]
- 8.Yamamoto A, Dewald DB, Boronenkov IV, Anderson RA, et al. Novel Pi(4)P 5-Kinase Homolog, Fab1p, Essential for Normal Vacuole Function and Morphology in Yeast. Mol Biol Cell. 1995;6:525–39. doi: 10.1091/mbc.6.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooke FT, Dove SK, McEwen RK, Painter G, et al. The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S-cerevisiae. Current Biology. 1998;8:1219–22. doi: 10.1016/s0960-9822(07)00513-1. [DOI] [PubMed] [Google Scholar]
- 10.Schu PV, Takegawa K, Fry MJ, Stack JH, et al. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 11.Duex JE, Nau JJ, Kauffman EJ, Weisman LS. Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryotic cell. 2006;5:723–31. doi: 10.1128/EC.5.4.723-731.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McEwen RK, Dove SK, Cooke FT, Painter GF, et al. Complementation analysis in PtdInsP kinase-deficient yeast mutants demonstrates that Schizosaccharomyces pombe and murine Fab1p homologues are phosphatidylinositol 3-phosphate 5-kinases. Journal of Biological Chemistry. 1999;274:33905–12. doi: 10.1074/jbc.274.48.33905. [DOI] [PubMed] [Google Scholar]
- 13.de Lartigue J, Polson H, Feldman M, Shokat K, et al. PIKfyve regulation of endosome-linked pathways. Traffic. 2009;10:883–93. doi: 10.1111/j.1600-0854.2009.00915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikonomov OC, Sbrissa D, Delvecchio K, Xie YF, et al. The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve(−/−) embryos but normality of PIKfyve(+/−) mice. Journal of Biological Chemistry. 2011;286:13404–13. doi: 10.1074/jbc.M111.222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeffries TR, Dove SK, Michell RH, Parker PJ. PtdIns-specific MPR pathway association of a novel WD40 repeat protein, WIPI49. Mol Biol Cell. 2004;15:2652–63. doi: 10.1091/mbc.E03-10-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morishita M, Morimoto F, Kitamura K, Koga T, et al. Phosphatidylinositol 3-phosphate 5-kinase is required for the cellular response to nutritional starvation and mating pheromone signals in Schizosaccharomyces pombe. Genes Cells. 2002;7:199–215. doi: 10.1046/j.1356-9597.2001.00510.x. [DOI] [PubMed] [Google Scholar]
- 17.Sbrissa D, Ikonomov OC, Filios C, Delvecchio K, et al. Functional dissociation between PIKfyve-synthesized PtdIns5P and PtdIns(3,5)P-2 by means of the PIKfyve inhibitor YM201636. American Journal of Physiology-Cell Physiology. 2012;303:C436–C46. doi: 10.1152/ajpcell.00105.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takasuga S, Horie Y, Sasaki J, Ge-Hong Sun-Wada G, et al. Critical roles of type III phosphatidylinositol phosphate kinase in murine embryonic visceral endoderm and adult intestine. Proc Natl Acad Sci U S A. 2013;110(5):1726–31. doi: 10.1073/pnas.1213212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duex JE, Tang F, Weisman LS. The Vac14p-Fig4p complex acts independently of Vac7p and couples PI3,5P2 synthesis and turnover. The Journal of cell biology. 2006;172:693–704. doi: 10.1083/jcb.200512105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gary JD, Sato TK, Stefan CJ, Bonangelino CJ, et al. Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase pathway by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. Mol Biol Cell. 2002;13:1238–51. doi: 10.1091/mbc.01-10-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudge SA, Anderson DM, Emr SD. Vacuole size control: Regulation of PtdIns(3,5)P-2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3.5)P-2-specific phosphatase. Mol Biol Cell. 2004;15:24–36. doi: 10.1091/mbc.E03-05-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chow CY, Zhang Y, Dowling JJ, Jin N, et al. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manford A, Xia TA, Saxena AK, Stefan C, et al. Crystal structure of the yeast Sac1: implications for its phosphoinositide phosphatase function. Embo Journal. 2010;29:1489–98. doi: 10.1038/emboj.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dove SK, McEwen RK, Mayes A, Hughes DC, et al. Vac14 controls PtdIns(3,5)P-2 synthesis and Fab1-dependent protein trafficking to the multivesicular body. Current Biology. 2002;12:885–93. doi: 10.1016/s0960-9822(02)00891-6. [DOI] [PubMed] [Google Scholar]
- 25.Botelho RJ, Efe JA, Teis D, Emr SD. Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol Biol Cell. 2008;19:4273–86. doi: 10.1091/mbc.E08-04-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin N, Chow CY, Liu L, Zolov SN, et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. The EMBO journal. 2008;27:3221–34. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alghamdi TA, Ho CY, Mrakovic A, Taylor D, et al. Vac14 protein multimerization is a prerequisite step for Fab1 protein complex assembly and function. J Biol Chem. 2013;288:9363–72. doi: 10.1074/jbc.M113.453712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikonomov OC, Sbrissa D, Fenner H, Shisheva A. PIKfyve-ArPIKfyve-Sac3 core complex: contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis. J Biol Chem. 2009;284:35794–806. doi: 10.1074/jbc.M109.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikonomov OC, Sbrissa D, Fligger J, Delvecchio K, et al. ArPIKfyve regulates Sac3 protein abundance and turnover: disruption of the mechanism by Sac3I41T mutation causing Charcot-Marie-Tooth 4J disorder. J Biol Chem. 2010;285:26760–4. doi: 10.1074/jbc.C110.154658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sbrissa D, Ikonomov OC, Fenner H, Shisheva A. ArPIKfyve homomeric and heteromeric interactions scaffold PIKfyve and Sac3 in a complex to promote PIKfyve activity and functionality. Journal of molecular biology. 2008;384:766–79. doi: 10.1016/j.jmb.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sbrissa D, Ikonomov OC, Fu ZY, Ijuin T, et al. Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport - Novel sac phosphatase joins the arpikfyve-pikfyve complex. Journal of Biological Chemistry. 2007;282:23878–91. doi: 10.1074/jbc.M611678200. [DOI] [PubMed] [Google Scholar]
- 32.Lenk GM, Ferguson CJ, Chow CY, Jin N, et al. Pathogenic mechanism of the FIG4 mutation responsible for Charcot-Marie-Tooth disease CMT4J. PLoS genetics. 2011;7(6):e1002104. doi: 10.1371/journal.pgen.1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonangelino CJ, Catlett NL, Weisman LS. Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Molecular and cellular biology. 1997;17:6847–58. doi: 10.1128/mcb.17.12.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baskaran S, Ragusa MJ, Boura E, Hurley JH. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Molecular cell. 2012;47:339–48. doi: 10.1016/j.molcel.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe Y, Kobayashi T, Yamamoto H, Hoshida H, et al. Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J Biol Chem. 2012;287:31681–90. doi: 10.1074/jbc.M112.397570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krick R, Busse RA, Scacioc A, Stephan M, et al. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a beta-propeller protein family. Proc Natl Acad Sci U S A. 2012;109:E2042–9. doi: 10.1073/pnas.1205128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dove SK, Piper RC, McEwen RK, Yu JW, et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. Embo Journal. 2004;23:1922–33. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Efe JA, Botelho RJ, Emr SD. Atg18 regulates organelle morphology and Fab1 kinase activity independent of its membrane recruitment by phosphatidylinositol 3,5-bisphosphate. Mol Biol Cell. 2007;18:4232–44. doi: 10.1091/mbc.E07-04-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Proikas-Cezanne T, Ruckerbauer S, Stierhof YD, Berg C, et al. Human WIPI-1 puncta-formation: a novel assay to assess mammalian autophagy. FEBS Lett. 2007;581:3396–404. doi: 10.1016/j.febslet.2007.06.040. [DOI] [PubMed] [Google Scholar]
- 40.Botelho RJ. Changing phosphoinositides “on the fly”: how trafficking vesicles avoid an identity crisis. Bioessays. 2009;31:1127–36. doi: 10.1002/bies.200900060. [DOI] [PubMed] [Google Scholar]
- 41.Bridges D, Ma JT, Park S, Inoki K, et al. Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol Biol Cell. 2012;23:2955–62. doi: 10.1091/mbc.E11-12-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Zolov SN, Chow CY, Slutsky SG, et al. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc Natl Acad Sci U S A. 2007;104:17518–23. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones DR, Foulger R, Keune WJ, Bultsma Y, et al. PtdIns5P is an oxidative stress-induced second messenger that regulates PKB activation. FASEB J. 2012;27(4):1644–56. doi: 10.1096/fj.12-218842. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, McCartney AJ, Zolov SN, Ferguson CJ, et al. Modulation of synaptic function by VAC14, a protein that regulates the phosphoinositides PI(3,5)P(2) and PI(5)P. EMBO J. 2012;31:3442–56. doi: 10.1038/emboj.2012.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ikonomov OC, Sbrissa D, Shisheva A. Localized PtdIns 3,5-P-2 synthesis to regulate early endosome dynamics and. American Journal of Physiology-Cell Physiology. 2006;291:C393–C404. doi: 10.1152/ajpcell.00019.2006. [DOI] [PubMed] [Google Scholar]
- 46.Shisheva A, Rusin B, Ikonomov OC, DeMarco C, et al. Localization and insulin-regulated relocation of phosphoinositide 5-kinase PIKfyve in 3T3-L1 adipocytes. Journal of Biological Chemistry. 2001;276:11859–69. doi: 10.1074/jbc.M008437200. [DOI] [PubMed] [Google Scholar]
- 47.Cabezas A, Pattni K, Stenmark H. Cloning and subcellular localization of a human phosphatidylinositol 3-phosphate 5-kinase, PIKfyve/Fab1. Gene. 2006;371:34–41. doi: 10.1016/j.gene.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 48.Rutherford AC, Traer C, Wassmer T, Pattni K, et al. The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J Cell Sci. 2006;119:3944–57. doi: 10.1242/jcs.03153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rusten TE, Rodahl LMW, Pattni K, Englund C, et al. Fab1 phosphatidylinositol 3-phosphate 5-kinase controls trafficking but not silencing of endocytosed receptors. Mol Biol Cell. 2006;17:3989–4001. doi: 10.1091/mbc.E06-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sbrissa D, Ikonomov OC, Strakova J, Dondapati R, et al. A mammalian ortholog of Saccharomyces cerevisiae Vac14 that associates with and up-regulates PIKfyve phosphoinositide 5-kinase activity. Mol Cell Biol. 2004;24:10437–47. doi: 10.1128/MCB.24.23.10437-10447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tronchere H, Laporte J, Pendaries C, Chaussade C, et al. Production of phosphatidylinositol 5-phosphate by the phosphoinositide 3-phosphatase myotubularin in mammalian cells. J Biol Chem. 2004;279:7304–12. doi: 10.1074/jbc.M311071200. [DOI] [PubMed] [Google Scholar]
- 52.Oppelt A, Lobert VH, Haglund K, Mackey AM, et al. Production of phosphatidylinositol 5-phosphate via PIKfyve and MTMR3 regulates cell migration. EMBO Rep. 2013;14:149–59. doi: 10.1038/embor.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaccari I, Dina G, Tronchere H, Kaufman E, et al. Genetic interaction between MTMR2 and FIG4 phospholipid phosphatases involved in Charcot-Marie-Tooth neuropathies. PLoS genetics. 2011;7(10):e1002319. doi: 10.1371/journal.pgen.1002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shisheva A. PIKfyve and its Lipid products in health and in sickness. Curr Top Microbiol Immunol. 2012;362:127–62. doi: 10.1007/978-94-007-5025-8_7. [DOI] [PubMed] [Google Scholar]
- 55.Sbrissa D, Ikonomov OC, Shisheva A. PIKfyve lipid kinase is a protein kinase: Downregulation of 5′-phosphoinositide product formation by autophosphorylation. Biochemistry. 2000;39:15980–9. doi: 10.1021/bi001897f. [DOI] [PubMed] [Google Scholar]
- 56.Han BK, Emr SD. Phosphoinositide [PI(3,5)P2] lipid-dependent regulation of the general transcriptional regulator Tup1. Genes Dev. 2011;25:984–95. doi: 10.1101/gad.1998611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carlton J, Bujny M, Peter BJ, Oorschot VM, et al. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high- curvature membranes and 3-phosphoinositides. Curr Biol. 2004;14:1791–800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- 58.Carlton JG, Bujny MV, Peter BJ, Oorschot VM, et al. Sorting nexin-2 is associated with tubular elements of the early endosome, but is not essential for retromer-mediated endosome-to-TGN transport. J Cell Sci. 2005;118:4527–39. doi: 10.1242/jcs.02568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katoh Y, Ritter B, Gaffry T, Blondeau F, et al. The clavesin family, neuron-specific lipid- and clathrin-binding Sec14 proteins regulating lysosomal morphology. J Biol Chem. 2009;284:27646–54. doi: 10.1074/jbc.M109.034884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Gisbergen PA, Li M, Wu SZ, Bezanilla M. Class II formin targeting to the cell cortex by binding PI(3,5)P(2) is essential for polarized growth. J Cell Biol. 2012;198:235–50. doi: 10.1083/jcb.201112085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong XP, Shen D, Wang X, Dawson T, et al. PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nature communications. 2010;1:38. doi: 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen J, Yu W-M, Brotto M, Scherman JA, et al. Deficiency of MIP/MTMR14 phosphatase induces a muscle disorder by disrupting Ca2+ homeostasis. Nat Cell Biol. 2009;11:769–76. doi: 10.1038/ncb1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 64.Jefferies HB, Cooke FT, Jat P, Boucheron C, et al. A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding. EMBO Rep. 2008;9:164–70. doi: 10.1038/sj.embor.7401155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ikonomov OC, Sbrissa D, Shisheva A. YM201636, an inhibitor of retroviral budding and PIKfyve-catalyzed PtdIns(3,5)P-2 synthesis, halts glucose entry by insulin in adipocytes. Biochemical and Biophysical Research Communications. 2009;382:566–70. doi: 10.1016/j.bbrc.2009.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hirano T, Matsuzawa T, Takegawa K, Sato MH. Loss-of-Function and Gain-of-Function Mutations in FAB1A/B Impair Endomembrane Homeostasis, Conferring Pleiotropic Developmental Abnormalities in Arabidopsis. Plant Physiology. 2011;155:797–807. doi: 10.1104/pp.110.167981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whitley P, Hinz S, Doughty J. Arabidopsis FAB1/PIKfyve Proteins Are Essential for Development of Viable Pollen. Plant Physiology. 2009;151:1812–22. doi: 10.1104/pp.109.146159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ikonomov OC, Sbrissa D, Shisheva A. Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 6-kinase PIKfyve. Journal of Biological Chemistry. 2001;276:26141–7. doi: 10.1074/jbc.M101722200. [DOI] [PubMed] [Google Scholar]
- 69.Ferguson CJ, Lenk GM, Meisler MH. Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum Mol Genet. 2009;18:4868–78. doi: 10.1093/hmg/ddp460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Onishi M, Nakamura Y, Koga T, Takegawa K, et al. Isolation of suppressor mutants of phosphatidylinositol 3-phosphate 5-kinase deficient cells in Schizosaccharomyces pombe. Bioscience, biotechnology, and biochemistry. 2003;67:1772–9. doi: 10.1271/bbb.67.1772. [DOI] [PubMed] [Google Scholar]
- 71.Falkenburger BH, Jensen JB, Dickson EJ, Suh BC, et al. Phosphoinositides: lipid regulators of membrane proteins. J Physiol. 2010;588:3179–85. doi: 10.1113/jphysiol.2010.192153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, Zhang X, Dong XP, Samie M, et al. TPC Proteins Are Phosphoinositide- Activated Sodium-Selective Ion Channels in Endosomes and Lysosomes. Cell. 2012;151:372–83. doi: 10.1016/j.cell.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Touchberry CD, Bales IK, Stone JK, Rohrberg TJ, et al. Phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) potentiates cardiac contractility via activation of the ryanodine receptor. J Biol Chem. 2010;285:40312–21. doi: 10.1074/jbc.M110.179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Augsten M, Hubner C, Nguyen M, Kunkel W, et al. Defective Hyphal induction of a Candida albicans phosphatidylinositol 3-phosphate 5-kinase null mutant on solid media does not lead to decreased virulence. Infect Immun. 2002;70:4462–70. doi: 10.1128/IAI.70.8.4462-4470.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nicot AS, Fares H, Payrastre B, Chisholm AD, et al. The phosphoinositide kinase PIKfyve/Fab1p regulates terminal lysosome maturation in Caenorhabditis elegans. Mol Biol Cell. 2006;17:3062–74. doi: 10.1091/mbc.E05-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bak G, Lee EJ, Lee Y, Kato M, et al. Rapid Structural Changes and Acidification of Guard Cell Vacuoles during Stomatal Closure Require Phosphatidylinositol 3,5-Bisphosphate. Plant Cell. 2013;25:2202–16. doi: 10.1105/tpc.113.110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baars TL, Petri S, Peters C, Mayer A. Role of the V-ATPase in regulation of the vacuolar fission-fusion equilibrium. Mol Biol Cell. 2007;18:3873–82. doi: 10.1091/mbc.E07-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zieger M, Mayer A. Yeast vacuoles fragment in an asymmetrical two-phase process with distinct protein requirements. Mol Biol Cell. 2012;23:3438–49. doi: 10.1091/mbc.E12-05-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bryant NJ, Piper RC, Weisman LS, Stevens TH. Retrograde traffic out of the yeast vacuole to the TGN occurs via the prevacuolar/endosomal compartment. J Cell Biol. 1998;142:651–63. doi: 10.1083/jcb.142.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Michell RH, Heath VL, Lemmon MA, Dove SK. Phosphatidylinositol 3,5-bisphosphate: metabolism and cellular functions. Trends Biochem Sci. 2006;31:52–63. doi: 10.1016/j.tibs.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 81.Whitley P, Reaves BJ, Hashimoto M, Riley AM, et al. Identification of mammalian Vps24p as an effector of phosphatidylinositol 3,5-bisphosphate-dependent endosome compartmentalization. J Biol Chem. 2003;278:38786–95. doi: 10.1074/jbc.M306864200. [DOI] [PubMed] [Google Scholar]
- 82.Ho CY, Alghamdi TA, Botelho RJ. Phosphatidylinositol-3,5-bisphosphate: no longer the poor PIP2. Traffic. 2012;13:1–8. doi: 10.1111/j.1600-0854.2011.01246.x. [DOI] [PubMed] [Google Scholar]
- 83.Er EE, Mendoza MC, Mackey AM, Rameh LE, et al. AKT Facilitates EGFR Trafficking and Degradation by Phosphorylating and Activating PIKfyve. Sci Signal. 2013;6:ra45. doi: 10.1126/scisignal.2004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ikonomov OC, Sbrissa D, Mlak K, Deeb R, et al. Active PIKfyve associates with and promotes the membrane attachment of the late endosome-to-trans-Golgi network transport factor Rab9 effector p40. Journal of Biological Chemistry. 2003;278:50863–71. doi: 10.1074/jbc.M307260200. [DOI] [PubMed] [Google Scholar]
- 85.Ikonomov OC, Fligger J, Sbrissa D, Dondapati R, et al. Kinesin Adapter JLP Links PIKfyve to Microtubule-based Endosome-to-Trans-Golgi Network Traffic of Furin. Journal of Biological Chemistry. 2009;284:3750–61. doi: 10.1074/jbc.M806539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ikonomov OC, Sbrissa D, Delvecchio K, Feng HZ, et al. Muscle-specific Pikfyve gene disruption causes glucose intolerance, insulin resistance, adiposity, and hyperinsulinemia but not muscle fiber-type switching. Am J Physiol Endocrinol Metab. 2013;305:E119–31. doi: 10.1152/ajpendo.00030.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Berwick DC, Dell GC, Welsh GI, Heesom KJ, et al. Protein kinase B phosphorylation of PIKfyve regulates the trafficking of GLUT4 vesicles. J Cell Sci. 2004;117:5985–93. doi: 10.1242/jcs.01517. [DOI] [PubMed] [Google Scholar]
- 88.Rusten TE, Vaccari T, Lindmo K, Rodahl LMW, et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Current Biology. 2007;17:1817–25. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 89.Martin S, Harper CB, May LM, Coulson EJ, et al. Inhibition of PIKfyve by YM-201636 dysregulates autophagy and leads to apoptosis-independent neuronal cell death. PloS one. 2013;8:e60152. doi: 10.1371/journal.pone.0060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Katona I, Zhang X, Bai Y, Shy ME, et al. Distinct pathogenic processes between Fig4-deficient motor and sensory neurons. Eur J Neurosci. 2011;33:1401–10. doi: 10.1111/j.1460-9568.2011.07651.x. [DOI] [PubMed] [Google Scholar]
- 91.Hazeki K, Nigorikawa K, Takaba Y, Segawa T, et al. Essential roles of PIKfyve and PTEN on phagosomal phosphatidylinositol 3-phosphate dynamics. FEBS Lett. 2012;586:4010–5. doi: 10.1016/j.febslet.2012.09.043. [DOI] [PubMed] [Google Scholar]
- 92.Kerr MC, Wang JTH, Castro NA, Hamilton NA, et al. Inhibition of the PtdIns(5) kinase PIKfyve disrupts intracellular replication of Salmonella. Embo Journal. 2010;29:1331–47. doi: 10.1038/emboj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dove SK, Dong K, Kobayashi T, Williams FK, et al. Phosphatidylinositol 3,5-bisphosphate and Fab1p/PlKfyve underPPIn endo-lysosome function. Biochemical Journal. 2009;419:1–13. doi: 10.1042/BJ20081950. [DOI] [PubMed] [Google Scholar]
- 94.Hirano T, Sato MH. Arabidopsis FAB1A/B is possibly involved in the recycling of auxin transporters. Plant Signal Behav. 2011;6:583–5. doi: 10.4161/psb.6.4.15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ferguson CJ, Lenk GM, Jones JM, Grant AE, et al. Neuronal expression of Fig4 is both necessary and sufficient to prevent spongiform neurodegeneration. Human Molecular Genetics. 2012;21:3525–34. doi: 10.1093/hmg/dds179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou X, Wang L, Hasegawa H, Amin P, et al. Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proc Natl Acad Sci U S A. 2010;107:9424–9. doi: 10.1073/pnas.0914725107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Winters JJ, Ferguson CJ, Lenk GM, Giger-Mateeva VI, et al. Congenital CNS hypomyelination in the Fig4 null mouse is rescued by neuronal expression of the PI(3,5)P(2) phosphatase Fig4. J Neurosci. 2011;31:17736–51. doi: 10.1523/JNEUROSCI.1482-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guo JS, Ma YH, Yan Q, Wang L, et al. Fig4 Expression in the Rodent Nervous System and Its Potential Role in Preventing Abnormal Lysosomal Accumulation. Journal of Neuropathology and Experimental Neurology. 2012;71:28–39. doi: 10.1097/NEN.0b013e31823deda8. [DOI] [PubMed] [Google Scholar]
- 99.Yan Q, Guo J, Zhang X, Bai Y, et al. Trauma does not accelerate neuronal degeneration in Fig4 insufficient mice. J Neurol Sci. 2012;312:102–7. doi: 10.1016/j.jns.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 100.Tsuruta F, Green EM, Rousset M, Dolmetsch RE. PIKfyve regulates CaV1.2 degradation and prevents excitotoxic cell death. J Cell Biol. 2009;187:279–94. doi: 10.1083/jcb.200903028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Seebohm G, Neumann S, Theiss C, Novkovic T, et al. Identification of a Novel Signaling Pathway and Its Relevance for GluA1 Recycling. PloS one. 2012;7:e33889. doi: 10.1371/journal.pone.0033889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Osborne SL, Wen PJ, Boucheron C, Nguyen HN, et al. PIKfyve negatively regulates exocytosis in neurosecretory cells. J Biol Chem. 2008;283:2804–13. doi: 10.1074/jbc.M704856200. [DOI] [PubMed] [Google Scholar]
- 103.Lemaire JF, McPherson PS. Binding of Vac14 to neuronal nitric oxide synthase: Characterisation of a new internal PDZ-recognition motif. FEBS Lett. 2006;580:6948–54. doi: 10.1016/j.febslet.2006.11.061. [DOI] [PubMed] [Google Scholar]
- 104.Bolis A, Zordan P, Coviello S, Bolino A. Myotubularin-related (MTMR) phospholipid phosphatase proteins in the peripheral nervous system. Mol Neurobiol. 2007;35:308–16. doi: 10.1007/s12035-007-0031-0. [DOI] [PubMed] [Google Scholar]
- 105.Chow CY, Landers JE, Bergren SK, Sapp PC, et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. American journal of human genetics. 2009;84:85–8. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Campeau PM, Lenk GM, Lu JT, Bae Y, et al. Yunis-Varón syndrome is caused by mutations in FIG4 encoding a phosphoinositide phosphatase. American Journal of Human Genetics. 2013;92:781–91. doi: 10.1016/j.ajhg.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li SL, Tiab L, Jiao XD, Munier FL, et al. Mutations in PIP5K3 are associated with Francois-Neetens Mouchetee fleck corneal dystrophy. American Journal of Human Genetics. 2005;77:54–63. doi: 10.1086/431346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kotoulas A, Kokotas H, Kopsidas K, Droutsas K, et al. novel PIKFYVE mutation in fleck corneal dystrophy. Molecular Vision. 2011;17:2776–81. [PMC free article] [PubMed] [Google Scholar]
- 109.Carmel L, Efroni S, White PD, Aslakson E, et al. Gene expression profile of empirically delineated classes of unexplained chronic fatigue. Pharmacogenomics. 2006;7:375–86. doi: 10.2217/14622416.7.3.375. [DOI] [PubMed] [Google Scholar]
- 110.Martyn C, Li J. Fig4 deficiency: a newly emerged lysosomal storage disorder? Prog Neurobiol. 2013;101–102:35–45. doi: 10.1016/j.pneurobio.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Berger P, Schaffitzel C, Berger I, Ban N, et al. Membrane association of myotubularin-related protein 2 is mediated by a pleckstrin homology-GRAM domain and a coiled-coil dimerization module. Proc Natl Acad Sci U S A. 2003;100:12177–82. doi: 10.1073/pnas.2132732100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Silswal N, Parelkar NK, Wacker MJ, Brotto M, et al. Phosphatidylinositol 3,5-bisphosphate increases intracellular free Ca2+ in arterial smooth muscle cells and elicits vasocontraction. Am J Physiol Heart Circ Physiol. 2011;300:H2016–26. doi: 10.1152/ajpheart.01011.2010. [DOI] [PubMed] [Google Scholar]
- 113.Ikonomov OC, Sbrissa D, Foti M, Carpentier JL, et al. PIKfyve controls fluid phase endocytosis but not recycling/degradation of endocytosed receptors or sorting of procathepsin D by regulating multivesicular body morphogenesis. Mol Biol Cell. 2003;14:4581–91. doi: 10.1091/mbc.E03-04-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Narayan K, Lemmon MA. Determining selectivity of phosphoinositide-binding domains. Methods. 2006;39:122–33. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Choudhury P, Srivastava S, Li Z, Ko K, et al. Specificity of the myotubularin family of phosphatidylinositol-3-phosphatase is determined by the PH/GRAM domain. J Biol Chem. 2006;281:31762–9. doi: 10.1074/jbc.M606344200. [DOI] [PubMed] [Google Scholar]
- 116.Zhang X, Chow CY, Sahenk Z, Shy ME, et al. Mutation of FIG4 causes a rapidly progressive, asymmetric neuronal degeneration. Brain. 2008;131:1990–2001. doi: 10.1093/brain/awn114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bolino A, Bolis A, Previtali SC, Dina G, et al. Disruption of Mtmr2 produces CMT4B1-like neuropathy with myelin outfolding and impaired spermatogenesis. J Cell Biol. 2004;167:711–21. doi: 10.1083/jcb.200407010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Norris FA, Auethavekiat V, Majerus PW. The isolation and characterization of cDNA encoding human and rat brain inositol polyphosphate 4-phosphatase. J Biol Chem. 1995;270:16128–33. doi: 10.1074/jbc.270.27.16128. [DOI] [PubMed] [Google Scholar]
- 119.Carricaburu V, Lamia KA, Lo E, Favereaux L, et al. The phosphatidylinositol (PI)-5-phosphate 4-kinase type II enzyme controls insulin signaling by regulating PI-3,4,5-trisphosphate degradation. Proc Natl Acad Sci U S A. 2003;100:9867–72. doi: 10.1073/pnas.1734038100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rameh LE, Tolias KF, Duckworth BC, Cantley LC. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature. 1997;390:192–6. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]