Abstract
The nucleus is the defining intracellular organelle of eukaryotic cells and represents a major structural innovation that differentiates the eukaryotic and prokaryotic cellular form. The presence of a nuclear envelope (NE) encapsulating the nucleus necessitates a mechanism for interchange between the contents of the nuclear interior and the cytoplasm, which is mediated via the nuclear pore complex (NPC), a large protein assembly residing in nuclear pores in the NE. Recent advances have begun to map the structure and functions of the NPC in multiple organisms, and to allow reconstruction of some of the evolutionary events that underpin the modern NPC form, highlighting common and differential NPC features across the eukaryotes. Here we discuss some of these advances and the questions being pursued, consider how the evolution of the NPC has been constrained, and finally propose a model for how the nuclear pore complex evolved.
Keywords: nuclear pore complex, nucleus, molecular evolution, comparative genomics, protocoatomer, eukaryogenesis
Introduction
The nucleus is generally acknowledged as a fundamental evolutionary innovation, and is bounded by a double membrane to form the container for almost all of the genetic material of eukaryotic cells. We can presume that the original proto-nucleus was freely permeable, but this is no longer the case for the modern nucleus and the double-membraned nuclear envelope (NE) is perforated throughout by membrane-lined nuclear pores containing proteinaceous assembles termed nuclear pore complexes (NPCs) (1,2). These NPCs serve as gatekeepers to actively regulate and mediate all trafficking between the internal nucleoplasm and the surrounding cytoplasm, allowing only specific macromolecules to enter and exit. Conceptually, this presents a rather significant conundrum: How did evolutionary processes retro-fit such a trafficking system into cells where previously all molecules had facile access to the DNA, and why were cells under pressure to evolve such as system?
Nucleocytoplasmic transport is an essential process, and describes the movement of macromolecules and solutes between the cytoplasmic and nuclear compartments of eukaryotic cells (3). As the nucleus is encapsulated within the NE, the distinct functions residing within the nucleus or cytoplasm rely on the discrete protein and nucleic acid compositions of the respective compartments. Exchange of molecules between these compartments must therefore be selective, both an evolutionary consequence of the presence of the nucleus, and a necessity arising from the presence of the NE barrier itself (4) The translocation of RNA species from the nucleus to the cytoplasm and the import of proteins required for nuclear functions after translation on the cytoplasmic ribosome are clearly core processes and essential for viability, whilst the transduction of signals from plasma membrane or cytoplasmic receptors must also be transmitted across the NE to effect transcriptional changes and/or RNA processing (5).
Both the outer and inner NE are contiguous with the endoplasmic reticulum, due to invagination of NE membrane at nuclear pores, and consequently the NE also shares some components and functionality with the ER. The presence of two membranes in the NE likely is a remnant of this ER origin. The nuclear pores are, as far as we know, the sole sites for movement of molecules between the nucleus and cytoplasm, and occupied by NPCs or, in a few cases, a spindle pole body (6). Those pores containing NPCs are the sites for selective bidirectional transport across the NE.
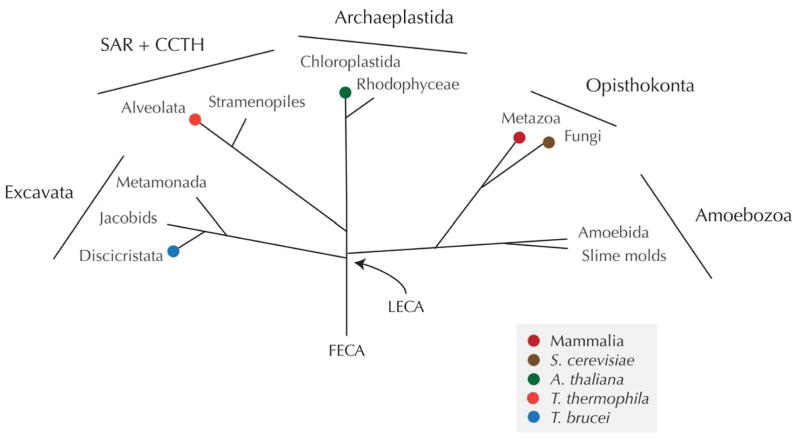
Remarkably, through a combination of in silico and direct experimental approaches, it has emerged that there is a considerable level of conservation of the NE/NPC/KAP/Ran system. Since some of the earliest ultrastructural studies of diverse eukaryotes, and particularly amoebae, it has been clear that the basic organisation of the NE was probably highly conserved, with both NE membranes closely opposed in nearly all organisms. The presence of nuclear pores was also obvious from these studies, as well as the presence of diffuse material embedded within the pores, the NPC, together with a lamina (7,8,9). Ultrastructure alone, however, is insufficient to determine if there is structural or mechanistic conservation between NPCs from different taxa, but sequencing of diverse eukaryotic genomes in the last decade or so, together with pioneering proteomic studies in metazoan and fungal model systems provided a parts list for the NPC in opisthokont taxa (Saccharomyces cerevisiae and Rattus rattus) and facilitated comparative genomics and proteomics to extend this understanding into additional supergroups (10,11,12,13) (Figure 1).
Figure 1. Schematic phylogenetic tree of the eukaryotes.
The major taxonomic groupings are shown, and the overall topology corresponds to the most likely based on present data and analytical methods. Supergroups are indicated by bars, and orders are labelled at the various nodes. Colored blobs indicate the positions of specific taxa that are discussed in the article and specified in the key. LECA and FECA are the last and first eukaryotic common ancestors respectively.
Here we will discuss the basic architecture of the NPC, as we know it from work (mainly) in S. cerevisiae, describe proteomic studies in several non-classical model systems that have yielded partial or near complete lists of NPC components, together with in silico approaches and functional interrogation. Together these analyses have indicated that there is considerable conservation of architecture, but that this appears under relaxed constraints for sequence conservation, and also indicates that lineage-specific evolutionary processes are indeed at work. Finally, we will consider what these findings may tell us concerning the evolutionary mechanisms that underpin NPC diversity, together with possible insights into how the NPC and nuclear pore originated.
Ancient origins – the coat at the heart of the machine
The NPC is made of a set of proteins termed nucleoporins or Nups. A protein is operationally considered as a Nup if the majority of its cellular pool spends most of the cell’s life cycle associated with the immediate vicinity of the nuclear pore (10). The NPC is immediately recognizable in electron micrographs by its distinctive morphology, well conserved among all eukaryotes examined to date. It is a disc of greater than 100 nm in diameter, embedded within the nuclear pore (above) and displaying a clear octagonal symmetry around its cylindrical axis. Eight spokes surround a central tube through which the bulk of nucleocytoplasmic macromolecular trafficking occurs (3) (Figure 2). These spokes are connected by three coaxial rings: the inner rings, at the NPC’s equator facing the central tube, composed of: the Nup170 complex in yeast and homologous Nup155 complex in metazoa; the outer rings, sandwiching the inner rings and composed of the Nup84 complex (yeast) / Nup107-160 complex (vertebrates); and the membrane rings on the lumenal side of the pore membrane containing Pom152 in yeast and gp210 in metazoa (12)(Figure 2).
Figure 2. The NPC; conserved and non-conserved regions.
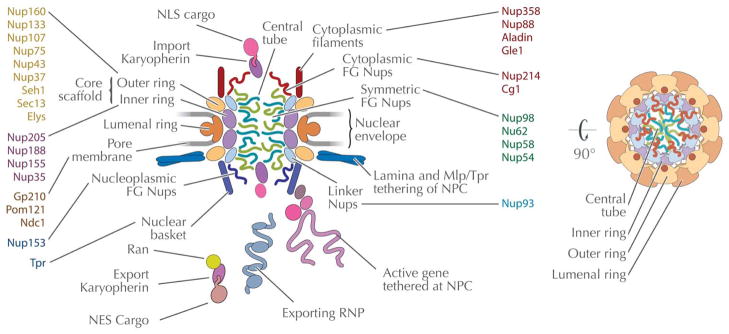
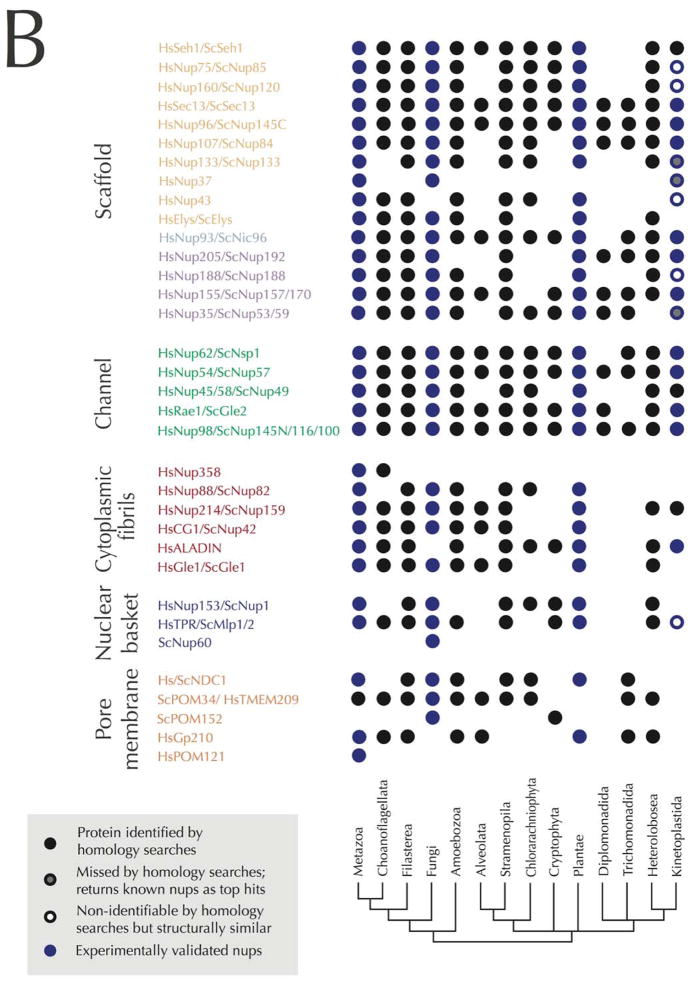
Panel A: Schematic of the nuclear pore complex color-coded to emphasis distinct structural modules within the structure. The central channel is shown with FG-repeat Nups as noodle-like structures, while the scaffold is shown as globular lozenges. Connections between the NPC core and the lamina are also shown. Several transport cargo systems are also shown. The structure is rotated through 90° at right to shown the organelle from the cytoplasmic face. Panel B: Schematics of the nuclear pore complex color-coded to match Panel A and C with individual Nups listed inside the major structural modules. The nomenclature is according to the human Nups with the exception of the names starting with Sc for Nups restricted to yeasts. The names of the subunits that were likely present in LECA based on comparative genomics (Panel C) are in black. The presence of the Nups with names in gray in LECA is more speculative. Lineage specific Nups are in white. Panel C: Coulson plot representation of nucleoporins across the eukaryotes as revealed by genomics. Well characterized Nups from vertebrates (Hs), yeast (Sc) and their homologs as identified in other eukaryotic lineages are shown as a Coulson plot. The data are based on the analysis of Neumann et al. (36) and our own additional homology searches using the HMMER software and protein alignments of clearly homologous protein sequences of the known Nups of metazoa and/or fungi as queries. Positive hits in other eukaryotic lineages were verified by reverse BLAST or HMMER homology searches. The composition of the NPC of Cryptophytes and Chlorarachniophytes is based on annotation of the NPC subunits in these algae in Curtis et al. (80). The color scheme of the nucleoporin modules is the same as in Panel A and B. Some of the subunits in the kinetoplastid lineage were found by protein-interaction analyses in T. brucei and are assigned as Nups based on similar secondary structure even when low amino-acid sequence similarity with validated Nups from vertebrates and fungi was found (empty circles). Blue symbols indicate experimentally validated Nups.
Remarkably, the NPC has at its heart a set of Nups that in a sense represent molecular fossils, revealing its evolutionary origin. This heart, comprising the structural core of the NPC and termed appropriately the “core scaffold”, serve both to stabilize the reflexed curvature of the pore membrane (14,15) and to anchor all other Nups, as well as many accessory NPC-associated proteins, at the NPC. A few years ago, concerted biochemical and informatics analyses of the NPC revealed that the core scaffold was composed of a set of cage-like structures containing Nups composed entirely of proteins with strong structural similarities to the proteins that coat transport vesicles. It was already known that the COPI and clathrin/adaptin vesicle coating complexes were related (16); both complexes contain proteins carrying iterations of repeat motif-containing folds - either a β-propeller fold, an α-solenoid-like fold, or both folds in tandem in the order β-α (17,18). However, it was surprising to discover how wide-ranging the re-use of this leitmotif has been in cells. The core scaffold’s inner and outer rings and the linker Nup Nic96/Nup93, together comprising over half the NPC’s mass, all consist entirely of such proteins - even going as far as incorporating the protein Sec13, shared by both the COPII vesicle coats and the NPC (10,11) (Figure 2). Based on these similarities, the protocoatomer hypothesis proposed that NPCs and clathrin, COPI, and COPII vesicle coats share a common evolutionary origin in an early membrane-curving module, the ‘protocoatomer’ (17). Recent discoveries have shown that this leitmotif is inscribed in even more complexes with even more diverse roles, including the intraflagellar transport complex involved in trafficking of materials along a cilia’s or flagella’s microtubules and the SEA and HOPS/CORVET complexes, whose functionalities in signaling and trafficking are still being determined (19,20,21). The remarkably adaptable nature of this leitmotif, both in terms of form and function, seems to have led to it becoming a predominant mechanism for membrane bending and tethering, and hence a mainstay of the cell’s trafficking repertoire (22). Further structural studies strongly support the protocoatomer hypothesis, and lessening further the chances that these complexes arose through convergent evolution.
The NPC as a macromolecular transporter
The central tube of the NPC is not empty – rather, it is occluded by a class of Nups termed “FG Nups”, doing so via their eponymous FG repeat domains (Figure 2). Each such domain consists of multiple Phe-Gly (i.e., FG) repeats spaced by very hydrophilic, and sometimes charged, spacer sequences of up to ~20 amino acids in length. These domains are characteristically unstructured and so highly flexible (23). FG repeat domains come in different “flavors”, distinguished by the exact nature of their Phe-containing repeats and the composition and size of their spacers. Transport is mediated by the interactions between soluble transport factors and FG-Nups, thus FG-Nups are at the heart of the NPC’s transport mechanism and are key to understanding transport; approximately one-third of all Nups contain FG repeat domains.
For a protein cargo to be transported across the NPC, it must carry a nuclear localisation signal (NLSs) or a nuclear export signal (NESs). This signal is recognized by a cognate transport factor, variously termed importins, exportins or transportins, but virtually all belonging to a related superfamily of proteins termed karyopherins (Kaps). The overall directionality of transport is driven by a gradient of Ran•GTP/Ran•GDP, a small Ras superfamily GTPase. Smaller RNA classes, (for example tRNAs), are also trafficked by karyopherins, but ribosomal subunits also require other ancilliary transport factors, and mRNA export requires a dedicated set of export factors unrelated to karyopherins (24,25). Nevertheless, all such cargoes still require a transport factor to mediate their transient association with FG Nups in order for them to transit the NPC’s central tube. While several models for the selective mechanism have been suggested, all are arguably based around a “virtual gating” mechanism, in that: “virtual gating implies that transport is based on local stochastic molecular interactions within the NPC while the gross structure of the NPC is maintained” (26,27). However, the precise molecular details of the selective transport mechanism remain unclear.
The NPC as a platform for nuclear organization
Projecting from the nuclear side of the NPC is a structure termed the nuclear basket, comprising eight filaments conjoining at a distal ring (Figure 2). Evidence increasingly supports the idea that the basket plays major roles in functionally linking the NPC to processes elsewhere in the nucleus. These include interactions with the SAGA complex, responsible for transcriptional control in mammalian cells and yeasts (28,29,30), plus binding the TREX-2 complex, important for mRNA quality control and export (31). Both these connections are now well established for animals, fungi and plants, providing evidence for broad conservation of these pathways (32). In more divergent protists, such as trypanosomes and chromalveolates, an association of these complexes with the NPC has not been established; this may reflect sequence or functional divergence, for example the rather different mechanisms for control of mRNA copy number in these taxa, but the details remain to be established. Interactions between the NPC and the nuclear lamina are also vital for the control of gene expression via the creation of heterochromatin, this being well established in metazoa and appearing conserved across the eukaryotes, for example in plants and trypanosomes.
Evolution of the NPC across the eukarytotes; evidence from genome sequences
The evidence above suggests that control of heterochromatin and participation in transcriptional regulation are likely near-universal aspects of Nups, and hence, together with nucleocytoplasmic transport, ancient functions for this group of proteins. In silico reconstruction of the NPC has been difficult due to low sequence conservation (33,34,35,36), but does serve to provide substantial evidence for retention of the Nups across eukaryotes, as well as to provide insights into the likely configuration in the last eukaryotic common ancestor (LECA), a hypothetical lineage that re-dates the differentiation of the modern eukaryotic lineages (Figure 1).
From the distribution of the NPC homologs among eukaryotes, it is clear that an NPC very similar to that in humans was already present in LECA (Figure 2C). The vast majority of Nups are shared by several unrelated eukaryotic lineages and their presence in LECA is thus the most parsimonious explanation; this also suggests that the scaffold and much of the FG repeat family are well conserved, while there is evidence for greater divergence in the nuclear basket and trans-membrane Nups. Examples of lineage-restricted Nups include POM152 of S. cerevisiae, only found in fungi and a cryptophyte alga Guillardia theta, which may suggest a horizontal gene transfer event and homologs of human Nup37 only found in metazoa and some fungi (36) suggesting an origin in the Opisthokont lineage (although HMMER identifies Nup37 in trypanosoma, which makes the presence of Nup37 in LECA possible). Only three Nups appear to be phylogenetically restricted to either fungi or metazoans: POM121 in vertebrates, Nup60 in fungi and metazoan Nup358. Overall, the predicted composition of the NPC of LECA (Figure 2C) is strikingly similar to that of H. sapiens and is even more elaborate than S. cerevisiae, which lacks Nup43, Aladin and Gp210.
Another interesting outcome of homology searches is the detection of relationships between proteins that have been characterized but were not considered related. For instance Boruc et al. (37) reported that Arabidosis lacking an ortholog of human Nup153, although HMMER searches identify plant Nup136, which was suggested to play analogous functions to the Nup153 but was considered to be plant-specific (13,37,38). We also identified human trans-membrane protein 209 (TMEM209) as a putative ortholog of S. cerevisiae POM34; both proteins share the same domain architecture and human TMEM209 was recently found to interact with Nup205 (39). It is therefore possible that TMEM209 is a novel trans-membrane nucleoporin of the human NPC.
In general, some eukaryotic taxa seem to retain a more conserved NPC, while in other lineages many of the components have either been lost or diverged to the level that they are not identifiable. For example Oomycetes seem to have near complete NPC while many of the NPC proteins were not found in other stramenopiles (e.g. diatoms) and even more identifiable homologs are missing from Alveolates, that are the stramenopiles’ sister lineage (Figure 2C). Similarly, orthologs of most of the known Nups are present in a heterolobosean protist Naegleria gruberi, while two other excavates Giardia and Trichomonas have the least conserved NPC among all eukaryotes. It is likely that many of the NPC subunits that were not found by sequence searches are in fact present, and with similar secondary structure as exemplified by studies in Trypanosoma (see below).
While in silico analysis of the NPC across eukaryotes is possible, functional and structural studies of the NPC are most advanced in yeast, with metazoan cells a close second. However, the unique biology of many taxa makes them valuable organisms from which to investigate the evolution of NPC function and how this relates to the selective pressures arising from individual adaptations and lifestyle. We restrict our discussion below to three lineages where considerable work has been performed recently: Tetrahymena thermophila (Alveolata); Arabidopsis thaliana (Plantae); and Trypanosoma brucei (Excavata), and where there is a level of characterisation of the NPC proteome and some direct experimental information (Figure 1).
The NPC as a transporter in different eukaryotes
In T. thermophila the major unique feature is nuclear dimorphism. The macronucleus (MAC) is a somatic structure that contains gene-sized fragments in multiple copies, alongside a more conventional micronucleus (MIC) that is involved in meiotic cell division but is transcriptionally silent during vegetative growth (40). All transcription is from the MAC during the non-sexual cell cycle. Whilst the NPC composition is not fully characterised in T. thermophila, about half of the Nups have been identified (assuming a similar total number to yeast), and the vast majority localise to both the MAC and MIC, suggesting very similar architectures for the NPCs of these organelles. The sole exception at present is the ortholog of the FG repeat nucleoporin, Nup98. In T. thermophila Nup98 is present as four paralogs, two of which localize to the MIC and the other two to the MAC (41). The the MAC paralogs possess conventional FG repeats, but uniquely the MIC paralogs possess poly-N tracts and NIFN repeats. Domain swap experiments indicate that the NIFN repeats contribute to selective import into the MIC. Significantly there is more selectivity in nuclear transport into the MIC, as cytoplasmic GFP (~28kDa) cannot diffuse into the MIC in the absence of an NLS but can diffuse into the MAC (42). Hence MIC TtNup98 NIFN repeats appear capable of excluding cargo efficiently, even when mistargeted to the MAC. This also suggests that the size exclusion for the MIC is rather lower than reported for NPCs in most organisms, so that the NIFN repeats may restrict the selective channel somewhat.
Further, up to thirteen KAP-α paralogs have been identified in T. thermophila, all of which contain an IBB domain. Specific targeting to the MAC or MIC for the individual KAP-α paralogs is apparent, but by contrast all KAP-β proteins analysed appear to target to both nuclei (42). These studies indicate rather robustly that transport pathways into the MAC and MIC are non-equivalent, and that apparent increased selectivity in import to the MIC may be associated with a novel variant repeat Nup98, as well as the use of specific KAP-α paralogs. Presumably, as the MIC is transcriptionally silent (although presumably retaining a requirement for maintenance of the nuclear proteome and repair of DNA damage), there is a significant need to facilitate efficient export of mRNA from the MAC, while ensuring protection of the MIC from aberrant transcriptional activity. Significantly, Nup98 is important in control of NPC disassembly in mammalian cells, and, assuming the function is conserved, the presence of distinct Nup98 paralogs in the T. thermophila MIC and MAC may also facilitate differential NPC turnover for each nucleus. This therefore represents a significant example of recent evolution of the NPC/KAP system to support the division of labour between the MAC and MIC, contributing to nuclear dimorphism.
In higher plants NPC morphology has been described based on ultrastructure and is highly similar to vertebrates (43). Interestingly, in some stages of the Arabidopsis development the NPCs are organised into rows, suggesting an intimate connection with a plant lamina (43). The composition of the Arabidopsis NPC has also been defined, using a novel proteomics strategy based around sequential immunoisolation (13). This approach identified the vast majority of Arabidopsis Nups (again assuming similar numbers of subunits to yeast), and these data suggest a remarkable level of conservation between plant, yeast and metazoan NPCs. This is the case both in terms of total Nup repertoire as well as sequence similarity between individual Nup orthologs. Importantly, several functional connections appear conserved as well, including interactions between AtNup50 and transport factors, and a FG Nup, AtNup136, which appears equivalent to Nup153 and associates with membranes surrounding chromatin during mitosis. Analogs for both the trans-membrane Nups gp210 and NDC1 (but not Pom121, which is key to NPC reassembly processes in mammalian cells), have also been identified, which stands in quite sharp contrast to T. brucei where no membrane Nups have been identified as yet (see below). Furthermore, the connections between the metazoan and yeast NPCs and additional complexes mediating control of mRNA export (e.g. TREX-2) or transcription, e.g. SAGA, also have counterparts in plants. The connections between TREX-2 and the NPC basket are apparently conserved and mediated via AtNup1, an ortholog of a yeast nuclear basket protein (44).
A prominent absence from the A. thaliana NPC is an analog of Nup358, a component of the cytoplasmic fibrils, also missing from yeast (13,36). This is functionally significant as in metazoa Nup358 constitutes a major binding site for RanGAP (45), but in Arabidopsis an alternate anchor for RanGAP is present in the nuclear envelope and specifically proteins of the WIT and WIP families (46,47,48). These data indicate that even though the plant NPC is driven by a Rab GTP/GDP gradient, the precise mechanistic details can vary between lineages; what impact this has on cellular functions remains unclear and may simply reflect alternate strategies towards the same end, but such differences can often manifest in constraints leading to further differentiation of otherwise similar systems.
Attempts to understand the functions of specific Nups in Arabidopsis have met with issues of complexity and likely redundancy (see ref. 49 for an excellent recent discussion of this area in some detail, 44,50,51). However, several phenotypes map to Arabidopsis Nups, and specifically resistance to infection and the control of autoimmunity is connected with specific alleles of the AtNup107/160 complex, the equivalent of the ScNup84 complex, and also KAP-α3 (52,53). Interestingly the evidence suggests that the mechanism underpinning this process may involve control of mRNA export, although it remains unclear if this reflects a specific requirement for this complex in the export of a subset of mRNAs or simply that the affected factors are more susceptible to disruption of post-transcriptional processes than others.
The third organism under consideration, Trypanosoma brucei, belongs to a eukaryotic supergroup Excavata, and may represent a very early branching taxon that separated from the remaining eukaryotes shortly after the LECA (Figure 1) (54). Trypanosomes are in general quite unusual, and the dominance of polycistronic transcription together with trans-splicing governing mRNA production for the vast majority of the protein-coding genes, coupled to a near total absence of cis-spicing (and hence conventional introns) indicates unique mechanisms for control of mRNA copy number are present (55). Signals for mRNA stability are based mainly within 3′-end elements of mature mRNAs, but the difficulty in fully mapping these signals suggests that additional factors for controlling mRNA copy number are also important. Further, for the vast majority of genes these features preclude promoter-based control of gene expression. As mRNA export and control of gene expression (both via promoters and heterochromatinization) are both intimately connected to the NPC, these considerations suggest that the trypanosome NPC may hold novel features and insights into these functions.
Trypanosome nucleoporins have been identified by a combination of subcellular proteomics, structure prediction and localisation (35,56 and Obado, S., et al., in preparation). The level of sequence divergence of trypanosome Nups is very significant and there is low conservation compared with yeast, plants and animals. Even with validated assignment as NPC components, determination of a precise orthologous relationship remains tentative in some cases. However, analysis does demonstrate a significant retention of scaffold Nups and their α/β protocoatomer domain architectures, and conservation of FG-repeat number although their FG repeat sequences are highly divergent. Trypanosomes are also the first non-metazoan NPC to be shown to contain ALADIN, previously thought to be animal-specific; ALADIN was subsequently found in A. thaliana, confirming that absence from yeast is a secondary loss (13).
Therefore, significant parts of the higher order architecture appears well conserved with the other characterised NPCs. Significant divergence is, however, manifest within the nucleoporin composition; for example there are probable additional components within the trypanosome Nup84 complex, several likely absences from the mRNA recognition/export system and a rather divergent nuclear basket. Additionally, other Nups found in opisthokonts appear absent (Obado, S., et al., in preparation). Further, based on knockdown experiments most nucleoporins are essential in at least one life stage (57).
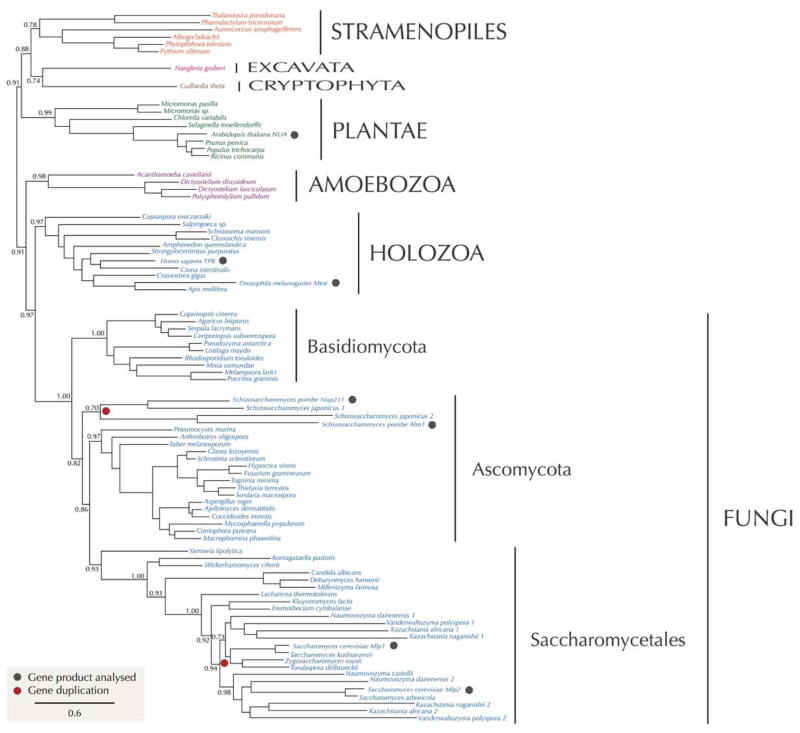
The NPC nuclear basket has been observed morphologically in a wide range of eukaryotes including plants, amoebae, metazoa, fungi and trypanosomes, strongly suggesting that this is a highly conserved feature. However, the level of conservation becomes less clear when we attempt to identify homologous components. The vertebrate basket’s major component, Tpr, has two yeast orthologs, Mlp1 and Mlp2, the latter of which is also associated with the spindle organizer. Indeed, both Tpr and the Mlp proteins appear to extend beyond the immediate vicinity of the basket, connecting between NPCs. Tpr and the Mlps are large (~200 kDa) coiled-coil proteins, each with their NPC attachment site some ⅓ along their length. Phylogenetic analyses of Tpr/Mlp indcate that these proteins are quite widely represented, with likely orthologs detected in Amoebae, plants, stramenopiles and Naegleria gruberi, an excavate (Figure 3).
Figure 3. Phylogenetic reconstruction of Mlp nucleoporin evolution.
Maximum likelihood phylogenetic tree of the TPR protein family. Broad distribution of these proteins in distinct eukaryotic lineages indicates that it was present in LECA. Vast majority of taxa possess a single gene in their genomes; the gene duplications that led to Mlp1/Mlp2 in S. cerevisiae and Nup211/Alm1 of S. pombe (indicated by red symbols) are two independent events that occurred later in the evolution of fungi. Numbers at nodes are SH-like aLRT values calculated in PhyML 3.1.
Such a phylogenetic distribution is strongly indicative that the LECA possessed a Tpr/Mlp ortholog. However, in the trypanosomes, which are also excavates, the situation appears to be different, and no clear Tpr/Mlp homolog is detected, which likely suggests compositional divergence for the basket in these organisms. Two nucleoporins, TbNup92 and TbNup110, both predominantly coiled-coil proteins that appear to be on the nuclear side of the NPC, have been suggested to be trypanosome basket proteins, with one - similar to Mlp2 - being associated at the spindle poles. However, TbNup92 also has a BRCT domain at the C-terminus, and both are only ½ the length of Tpr or the Mlps. Thus, their identity as basket proteins, and Tpr homologs, remains uncertain. Furthermore, other basket components (if any) remain to be unequivocally identified in any eukaryote, although some FG Nups have been implicated as such. Overall, these data suggest that, despite morphologicial conservation, the basket’s composition can be quite variable or divergent between lineages.
In conclusion, we can infer that the NPC was present in a form that we would recognise in the LECA, and its participation in chromatin organization as well as nucleocytoplasmic transport was likely also a major role of the LECA NPC. However, the apparent lineage-specific nature of several factors, as identified in trypanosomes and other organisms, presents a challenge to reconstruction of the molecular players that operated in concert with the NPC at early points of eukaryotic evolution.
Assembly, mitosis and the NPC
LIttle is known about how different eukaryotes manage their NPC numbers during their life cycle. For all cells, there is a requirement for assembly of NPCs during interphase, as a doubling of NPC number is required to maintain a constant NPC copy number in daughter nuclei. However, different eukaryotic lineages display a bewildering variety of strategies to manage their NEs - and NPCs - during mitosis, with distinct strategies being found even between relatively closely-related organisms. These mitotic strategies broadly fall into two catagories, “open” or “closed”. In the former, the NPCs and the NE are disassembled at the onset of mitosis to allow the spindle apparatus access to the cell’s chromosomes, and are reassembled at its completion; in the latter, the NPCs and the NE remain assembled throughout mitosis, with the spindle being assembled within the nucleoplasm (58). Additional controlled turnover pathways may also operate to regulate changes to NPC numbers under differing growth conditions for example, and the counting mechanism for maintaining a constant copy number is presently unclear (59).
How NPC assembly or disassembly is controlled is becoming somewhat clearer, and there are suggestions for some conservation of these pathways, at least between animals and fungi. In mammalian cells the interphase and mitotic assembly pathways have a distinct mechanism, with the former dependent on the trans-membrane nucleoporin Pom121 and the latter requiring ELYS, a peripheral Nup that contains membrane-deforming ALPS motifs (60). ELYS itself associates with chromatin, and is important for recruitment of the Nup107/160 core scaffold complex, a process which is likely modulated by KAP-β; however some of these factors are also part of the more general interphase NPC assembly system, and genetic screens in yeast also pinpoint a similar cohort of gene products, where mitosis is closed (reviewed in 14). Significantly, in mammalian cells NPC disassembly is one of the earliest mitotic events and which is initiated by the disengagement of Nup98 from the NPC, itself a result of hyperphosphorylation by NIMA, Cdk1 and probably additional kinases (61). Therefore, while there are distinct requirements for the mitotic and interphase pathways it is likely that the former is simply a specialisation of the universal need to insert new NPCs into the NE. In mammalian cells at least, the creation of new NPCs is a nonconservative mechanism, i.e. it does not relay on nucleoporins from pre-existing NPCs, is non-template encoded, and takes place from both faces of the NE, where cohorts of nucleoporins accumulate (62).
Interestingly, in Aspergillus nigerans, which formally has closed mitosis, many Nups become dispersed during mitosis (63,64). These are predominantly the FG Nups as well as a few other peripheral Nups, but not the core scaffold, and this partial disassembly results in the NPCs becoming highly permeable. This dispersal is again controlled by NIMA and Cdk1 kinase, indicating a conserved mechanism for control of NPC dynamics. Interestingly, the Nup107/160 complex core scaffold can be induced to disperse in these cells by deletion of ELYS and Nup37, providing additional evidence for similar requirements between mitotic and interphase NPC assembly (65). In these organisms, ELYS appears critical for recruiting the Nup107/160 complex as well as other Nups, essentially merging the two mechanisms as described for mammalian cells (58). Additional cryptic remodeling of the NPC has been observed in Schizosaccharomyces pombe, where during meiosis exclusion of RanGAP from the nucleus is lost, resulting in complete breakdown of nucleocytoplasmic transport, and which may hint at rather more extensive NPC remodeling processes accompanying mitosis than so far uncovered (66). Overall, what these examples are probably indicating is that the underlying mechanisms for control of the NPC are intimately entwined with the mitotic apparatus, even in cells where the NE does not break down, and that at their core these are probably also a highly conserved pathways. Once again, there are hints of some likely lineage-specific features, but these remain to be fully investigated.
The karyopherin transport system
A great many transport systems operate across the NPC, and many of the cargo receptors mediating this transport do not share obvious common origins, perhaps suggesting independent and possible stepwise evolution of these pathways. One system, that powered by the karyopherins, is mediated by a large paralagous family, and has been studied in an evolutionary context, and therefore can be discussed in the present context.
Most known KAP proteins belong to the KAP-β family, ~100kDa polypeptides predominantly folded into α-helical HEAT repeats with an acidic pI. A minority belong to the much smaller KAP-α family, which are ~60kDa proteins and consist of three domains, an N-terminal IBB domain, a central armadillo (ARM) repeat region and a small hydrophobic C-terminus (67). In opisthokonts KAP-α operates in complex with a member of the KAP-β family, KAP-β1/Kap60, but it is unknown if this is the case in other supergroups. The clear similarity in the ARM and HEAT α-helical repeat architectures indicates that KAP-α and KAP-β are closely related at the secondary structural level and hence possibly share a common ancestor. As the HEAT repeat is common to prokaryotes and the ARM domain is eukaryotic restricted, this suggests that the KAP-α family probably evolved after the KAP-β system, and maybe from them (33,68,69).
All members of the KAP family appear to be able to recognize either NLSs or NESs. Cargo recognition within the KAP family is an interesting combination of sequence- and conformer-specific associations, and while the directional transport of specific classes of cargoes (such as ribosomal protein import or tRNA export) has been attributed to individual KAPs in model organisms, it is presently unknown how well these functions are conserved between orthologs from different taxa. The HEAT and ARM repeat architectures likely also represent a highly flexible and adaptable platform for the recognition of the diverse NLS and NES, a clear necessity when only a score of transport receptors are responsible for the import and export of up to thousands of polypeptides and RNAs. In fact, it may be the case that the use of the flexible KAP system, together with the physicochemical recognition mechanisms provided by FG Nups, is important for evolvability, as too specific a recognition system would necessitate co-evolution of cargo, KAPs and the NPC, likely providing a lock against rapid evolution and response to alterations in the proteome. Interestingly, recent data indicates that evolution of NLSs within the H. sapiens mRNA export factor NXF1 is somewhat flexible. NXF1 can interact with several KAPs and contains at least two NLSs; the promiscuity of NXF1 KAP interactions appears to correlate with organismal complexity within a sampling of opistokhonts, providing a likely example of the great adaptability that is facilitated by the KAP system (70).
Both KAP-α and KAP-β were present in LECA. KAP-α is mainly present as a single gene in many taxa, but is expanded to multiple paralogs in metazoa, plants and some fungi, and likely parallels increased complexity within the substrates requiring transport, although exactly what this means in terms of KAP-cargo specificity, number of different substrates, tissue-specific expression and flux for individual cargos is completely unexplored (71). However, this also indicates that the KAP-α/KAP-β1 system was present in the LECA. In Arabidopsis, which has nine KAP-α family members, most of the members are ubiquitously expressed and the functional discrimination between them is unclear. However, KAP-α3 has been implicated as having a specific role in autoimmunity and is a suppressor of snc1, an autoimmunity gene (52), while KAP-α4 knockouts are fully resistant to infection by Agrobacterium tumefaciens due to defective nuclear import of VirD2 and VirE2, two Agrobacterium products required for transformation (72). Interestingly this phenotype can be rescued by overexpression of other AtKAP-α family members, suggesting a similar level of overlapping specificity/affinity as seen in S. cerevisiae. An additional example of such redundancy/flexibility is also provided from A. nidulans, where only four of 14 possible transport pathways appear essential (73).
Phylogenetic reconstruction of the large KAP-β family demonstrated that the LECA contained an extensive KAP-β repertoire, of likely similar complexity to extant eukaryotes (74). Therefore it can be predicted that the LECA possessed multiple KAP-β pathways as well. Due to the nature of phylogenetic reconstruction, close paralogs cannot be simply resolved, so it remains possible that both the KAP-α and KAP-β families were larger in the LECA than reconstruction suggests.
There is little evidence for substantial innovation of new KAP-β subfamilies within individual lineages (74). Both expansions of individual KAP-β members and secondary losses are frequent, with only KAP-β1 and KAP-β3 being universally retained; the formation of a complex between KAP-β and KAP-α likely explains KAP-β1 retention. The roles of more recent, lineage-specific expansions are unknown, but while this analysis found almost no evidence for massive KAP-β innovations, birth and death within the KAP-β family has been frequent and occurs across the eukaryotes, and it is possible that some of these paralogous expansions may represent the evolution of novel heterodimeric systems similar to the KAP-α/KAP-β system. We speculate that the flickering of KAP-β paralog loss and production indicates coevolution of the KAP-β transport receptors and the proteomes within their respective taxa. This would suggest that the flexibility of KAP-β interactions with transport cargo has been sufficient to accommodate proteomic diversity.
Splendid complexity from humble origins
At a very basic level, there are two rather different blueprints for cellular architecture. Prokaryotes embody an apparently simple approach, where in general a single, albeit elaborate, membranous structure suffices to contain all of the macromolecular assemblies that drive living processes. If prokaryotes do have internal membranes, they tend to be both simple and structurally homogenous. However, some one and a half billion years ago, over two billion years after cellular life evolved, the ancestral eukaryotes acquired the ability to invaginate their membranes. While the precise reason for this is unknown, it possibly arose as a predatory adaptation, allowing these early eukaryotic cells to engulf and digest their less fortunate neighbors. Ultimately, eukaryotes exploited this ability to compartmentalize their protoplasm into diverse, specialized membrane-bound organelles. The protocoatomer hypothesis implies mechanisms by which new coat systems and compartments can arise with comparative ease from pre-exisiting systems (22) (Figure 4). How can these observations and speculations contribute to an understanding of the origins of the NPC?
Figure 4. Evolution of the NPC, nucleus and the ER.
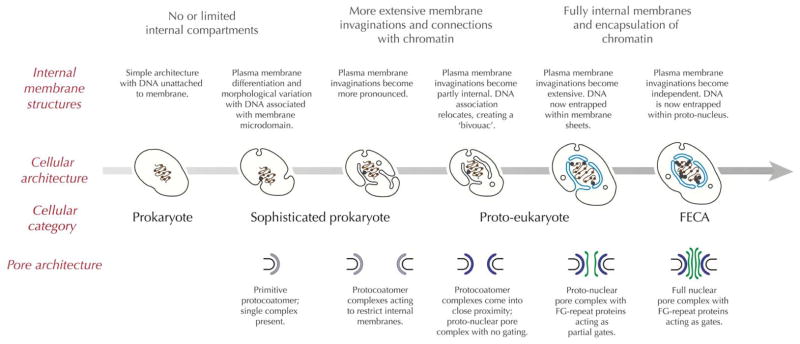
A simple model is proposed for how the nucleus could have evolved from a prokaryotic ancestor that lacked a differentiated cytoplasm. Initially the DNA is untethered to membrane (left), but in some bacterial lineages both the presence of membrane microdomains and DNA tethering is known (second left). Invaginations of membrane, which may have been accompanied by the presence of a primitive protocoatomer complex are envisaged. As these structures become more complex, stable internal membrane have arisen, which may maintain a connection with the plasma membrane (third left). At this point the internal membranes are in essence evolving into a proto-ER, as they may share protein export machinery and other factors with the bacterial plasma membrane. Maintaining these structures within the cytosol requires a coating system at the rim/apex of the membrane tubules or sheets for stability, control of proliferation and possible membrane fission. Once the DNA relocates to these membranes, these membrane act as a bivouac for the DNA, and are essentially transformed into a proto-nucleus (third right). Close proximity of membranes, which would have been fenestrated to facilitate exchange of molecules between the DNA and cytoplasm, requires a coat complex to bring stability to these membrane structures, and which also serves as the beginning of a nuclear pore and NPC; at this stage we suggest that the system would be a non-specific pore lacking a gating function. Later the system becomes more differentiated, and the appearance of FG-repeat nucleoporins serve to introduce gating, specificity and to increase targeting efficiency (second right), until a fully differentiated nucleus has arisen, which finally parts company with the plasma membrane (right). Note that the model ignores many critical aspects of eukaryogenesis, including endosymbiosis and membrane trafficking and many other process, as well as evidence that such states constitute true intermediates. It also assumes that the nucleus arose by a fully autogenous mechanism, which is generally, if not universally, accepted. Schematics of idealised cells are shown at center; with the true nuclear envelope and DNA in blue. The implied configuration of the protocoatomer ancestor leading up to the NPC is shown below; FG-repeat NUPs are shown in green, the nuclear envelope in gray and the NPC core scaffold in blue. The large gray arrow indicates a timebase.
The secondary structures of the NPC scaffold and other protocoatomer components are well conserved, and similarities between the NPC and COPII may provide a clue to NPC origins. Let us assume that organisms in the evolutionary transition from sophisticated prokaryotes to simple eukaryotes need to control the proliferation of their internal membranes as well as the budding of membrane for transport processes. Let us also assume that these internal membranes functionally represented a primitive endoplasmic reticulum (Figure 4). With these assumptions in place, the construction of a simple bivouac of membrane around the cell’s DNA/chromatin could have arisen simply by the extension of plasma membrane associations to the ER, itself not a massive assumption given the presence of other prokaryotic plasma membrane descendants, i.e. Sec61, at the ER membrane. If, as would seem likely, that budding from the ER was mediated by a primitive protocoatomer complex, then a primitive system for control of fenestrations around the chromatin would arise as a consequence. It is then a rather small step for such fenestrations to evolve into primitive nuclear pores, where the boundary of the pore is defined by a newly differentiated protocoatomer complex. Such a system could diversify from the original ER-associated protocoatomer system by paralogous expansion, and the gradual replacement of proteins originally common between the resulting protocoatomer systems by specialised subunits that eventually become targeted to only one or the other system; this is in essence identical to the ratchet like mechanism of the organellar paralogy hypothesis originally proposed for the evolution of new systems defining endomembrane compartments (75,76).
The protocoatomer hypothesis suggests that the coating leitmotif developed to help early eukaryotes invaginate their membranes and were so successful an adaptation that the genes encoding their proteins duplicated, diverged, and specialized to originate the multiple coatomer-related complexes we see today. Evidence for such duplication events remains fossilized in the core scaffold; each NPC spoke seems to be made of two similar parallel columns, in which every Nup in one column contains a similarly-positioned homolog in the adjacent column, a pattern that can be explained if ancient duplication events gave rise to the two columns comprising each spoke. Clear structural similarities also exist between the inner and outer rings, and between components within each of these rings, likely indicating the even earlier duplication events very early in eukaryogenesis leading eventually to the NPC (12,17). Once the primitive cell arrived at this point, then the NPC had evolved, but in a form that lacked much in the way of either selectivity or chromatin organizational functions. What has been gained however, is some compartmentalisation of the nuclear material, together with mechanical protection of chromatin and with the potential to begin subdivision of chromatin into peripheral and non-peripheral domains - the beginnings of hetero- and euchromatin. Elaboration of the basic scaffold, by further gene duplications and protein associations, could have then led to an ever more sophisticated platform for chromatin organization and regulation. The arrival of selective transport, with the evolution of FG-repeat Nup precursors (perhaps from the repeat motifs of the β-propeller folds or α-solenoid-like folds already in the core scaffold proteins), would then provide further refinement, and facilitate the beginnings of nucleocytoplasmic transport. The structure of the KAP superfamily also closely resembles that of the apparently paralogous inner ring Nups, Nup192 and Nup188 (77), suggesting a possible common evolutionary origin, in further agreement with the protocoatomer hypothesis. Significantly, this model also suggests a very simple ratchet-like mechanism for the evolution of increasing complexity, by the simple expansion of genes that subsequently gain new function. Such evolutionary trajectories have been argued as potentially being the products of non-adaptive evolution, at least in their initial stages (78,79), and hence may not necessarily arise from selective pressure. This perhaps removes the need to seek for a specific functional rational for acquisition of each component of the NPC, as well as KAP superfamily.
Of course, our scenario above remains very speculative and has yet to address a number of important questions. Understanding the structure and functions of the NPC in more diverse taxa has become a reality in the last ten years, and we are now confident of a complex NPC and nucleocytoplasmic transport system in the LECA. However, our data lack great depth in terms of detailed functional dissection, and in the number of organisms that have been analysed. It is clear that there are a great many evolutionary insights that can be gleaned from such studies, and as distinct aspects of NPC biology begin to emerge. Like Leifur Eiríksson in 986, we are standing at the edge of an Old World, with a New World of evolutionary and comparative biology potential to explore; these journeys have the power to contribute to our understanding of eukaryotic origins, as well as offer possible mechanisms for the manipulation of crop species, pathogens and the environment.
Synopsis.
The nuclear pore complex (NPC) is responsible for nucleocytoplasmic traffic, chromatin organisation and regulation of gene expression. The NPC is an ancient feature of the eukaryotic cell, with origins over a billion years ago, predating the radiation of modern eukaryotic lineages. Here we discuss emerging views of NPC evolution and the evidence for specialisations in many organisms that may reflect specific adaptations. We consider the selective pressures that have shaped NPC functions and propose a simple model for how the NPC arose during eukaryogenesis.
Acknowledgments
Work in the Field laboratory is supported by the Wellcome Trust and the MRC (090007/Z/09/Z and MR/K008749/1 respectively). LK is supported by a Marie Curie fellowship. The Rout laboratory acknowledges the support of the National Institutes of Health (R21 AI096069, U54 GM103511, U01 GM098256). We are also grateful to the following for discussions of various aspects of the topics covered in this article; Aaron Turkewitz (University of Chicago), Samson Obado (Rockefeller University) and Jennifer Holden (Univeristy of Cambridge).
References
- 1.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4. New York: Garland Science; 2002. [Google Scholar]
- 2.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 3.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 4.Tetenbaum-Novatt J, Rout MP. The mechanism of nucleocytoplasmic transport through the nuclear pore complex. Cold Spring Harb Symp Quant Biol. 2010;75:567–584. doi: 10.1101/sqb.2010.75.033. [DOI] [PubMed] [Google Scholar]
- 5.Heym RG, Niessing D. Principles of mRNA transport in yeast. Cell Mol Life Sci. 2012;69:1843–1853. doi: 10.1007/s00018-011-0902-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldherr CM. Nucleocytoplasmic exchanges during early interphase. J Cell Biol. 1968;39:49–54. doi: 10.1083/jcb.39.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leeson TS, Bhatnagar R. Microfibrillar structures in the nucleus and cytoplasm of amoeba proteus. J Exp Zool. 1975;192:265–270. doi: 10.1002/jez.1401920216. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt M, Grossmann U, Krohne G. The nuclear membrane-associated honeycomb structure of the unicellular organism Amoeba proteus: on the search for homologies with the nuclear lamina of metazoa. Eur J Cell Biol. 1995;67:199–208. [PubMed] [Google Scholar]
- 10.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 13.Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell. 2010;22:4084–4097. doi: 10.1105/tpc.110.079947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez-Martinez J, Rout MP. Nuclear pore complex biogenesis. Curr Opin Cell Biol. 2009;21:603–612. doi: 10.1016/j.ceb.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez-Martinez J, Phillips J, Sekedat MD, Diaz-Avalos R, Velazquez-Muriel J, Franke JD, Williams R, Stokes DL, Chait BT, Sali A, Rout MP. Structure-function mapping of a heptameric module in the nuclear pore complex. J Cell Biol. 2012;196:419–434. doi: 10.1083/jcb.201109008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boehm M, Bonifacino JS. Adaptins: the final recount. Mol Biol Cell. 2001;12:2907–2920. doi: 10.1091/mbc.12.10.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci U S A. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dokudovskaya S, Waharte F, Schlessinger A, Pieper U, Devos DP, Cristea IM, Williams R, Salamero J, Chait BT, Sali A, Field MC, Rout MP, Dargemont C. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol Cell Proteomics. 2011;10:M110.006478. doi: 10.1074/mcp.M110.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Dam TJ, Townsend MJ, Turk M, Schlessinger A, Sali A, Field MC, Huynen MA. Evolution of modular intraflagellar transport from a coatomer-like progenitor. Proc Natl Acad Sci U S A. 2013;110:6943–6948. doi: 10.1073/pnas.1221011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balderhaar HJ, Ungermann C. CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J Cell Sci. 2013;126:1307–1316. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- 22.Field MC, Sali A, Rout MP. Evolution: On a bender--BARs, ESCRTs, COPs, and finally getting your coat. J Cell Biol. 2011;193:963–972. doi: 10.1083/jcb.201102042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang W. ‘Natively unfolded’ nucleoporins in nucleocytoplasmic transport: clustered or evenly distributed? Nucleus. 2011;2:10–16. doi: 10.4161/nucl.2.1.13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faza MB, Chang Y, Occhipinti L, Kemmler S, Panse VG. Role of Mex67-Mtr2 in the nuclear export of 40S pre-ribosomes. PLoS Genet. 2012;8:e1002915. doi: 10.1371/journal.pgen.1002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aitchison JD, Rout MP. The yeast nuclear pore complex and transport through it. Genetics. 2012;190:855–883. doi: 10.1534/genetics.111.127803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters R. Functionalization of a nanopore: the nuclear pore complex paradigm. Biochim Biophys Acta. 2009;1793:1533–1539. doi: 10.1016/j.bbamcr.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters R. Translocation through the nuclear pore: Kaps pave the way. Bioessays. 2009;31:466–477. doi: 10.1002/bies.200800159. [DOI] [PubMed] [Google Scholar]
- 28.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 29.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, Chaurasia S, Valentini SR, Corbett AH. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem. 2007;282:3042–3049. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- 31.Thakurta AG, Gopal G, Yoon JH, Kozak L, Dhar R. Homolog of BRCA2-interacting Dss1p and Uap56p link Mlo3p and Rae1p for mRNA export in fission yeast. EMBO J. 2005;24:2512–2523. doi: 10.1038/sj.emboj.7600713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.García-Oliver E, García-Molinero V, Rodríguez-Navarro S. mRNA export and gene expression: the SAGA-TREX-2 connection. Biochim Biophys Acta. 2012;1819:555–565. doi: 10.1016/j.bbagrm.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 33.Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- 34.Bapteste E, Charlebois RL, MacLeod D, Brochier C. The two tempos of nuclear pore complex evolution: highly adapting proteins in an ancient frozen structure. Genome Biol. 2005;6:R85. doi: 10.1186/gb-2005-6-10-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics. 2009;8:2119–2130. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neumann N, Lundin D, Poole AM. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS One. 2010;5:e13241. doi: 10.1371/journal.pone.0013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boruc J, Zhou X, Meier I. Dynamics of the plant nuclear envelope and nuclear pore. Plant Physiol. 2012;158:78–86. doi: 10.1104/pp.111.185256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K, Hara-Nishimura I. Involvement of the nuclear pore complex in morphology of the plant nucleus. Nucleus. 2011;2:168–172. doi: 10.4161/nucl.2.3.16175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujitomo T, Daigo Y, Matsuda K, Ueda K, Nakamura Y. Critical function for nuclear envelope protein TMEM209 in human pulmonary carcinogenesis. Cancer Res. 2012;72:4110–4118. doi: 10.1158/0008-5472.CAN-12-0159. [DOI] [PubMed] [Google Scholar]
- 40.Miao W, Xiong J, Bowen J, Wang W, Liu Y, Braguinets O, Grigull J, Pearlman RE, Orias E, Gorovsky MA. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One. 2009;4:e4429. doi: 10.1371/journal.pone.0004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwamoto M, Mori C, Kojidani T, Bunai F, Hori T, Fukagawa T, Hiraoka Y, Haraguchi T. Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate tetrahymena. Curr Biol. 2009;19:843–847. doi: 10.1016/j.cub.2009.03.055. [DOI] [PubMed] [Google Scholar]
- 42.Malone CD, Falkowska KA, Li AY, Galanti SE, Kanuru RC, LaMont EG, Mazzarella KC, Micev AJ, Osman MM, Piotrowski NK, Suszko JW, Timm AC, Xu MM, Liu L, Chalker DL. Nucleus-specific importin alpha proteins and nucleoporins regulate protein import and nuclear division in the binucleate Tetrahymena thermophila. Eukaryot Cell. 2008;7:1487–1499. doi: 10.1128/EC.00193-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fiserova J, Kiseleva E, Goldberg MW. Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J. 2009;59:243–255. doi: 10.1111/j.1365-313X.2009.03865.x. [DOI] [PubMed] [Google Scholar]
- 44.Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, Kohalmi SE, Keller WA, Tsang EW, Harada JJ, Rothstein SJ, Cui Y. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 2010;61:259–270. doi: 10.1111/j.1365-313X.2009.04048.x. [DOI] [PubMed] [Google Scholar]
- 45.Hutten S, Flotho A, Melchior F, Kehlenbach RH. The Nup358-RanGAP complex is required for efficient importin alpha/beta-dependent nuclear import. Mol Biol Cell. 2008;19:2300–2310. doi: 10.1091/mbc.E07-12-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu XM, Meulia T, Meier I. Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr Biol. 2007;17:1157–1163. doi: 10.1016/j.cub.2007.05.076. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Q, Brkljacic J, Meier I. Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell. 2008;20:1639–1651. doi: 10.1105/tpc.108.059220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brkljacic J, Zhao Q, Meier I. WPP-domain proteins mimic the activity of the HSC70-1 chaperone in preventing mistargeting of RanGAP1-anchoring protein WIT1. Plant Physiol. 2009;151:142–154. doi: 10.1104/pp.109.143404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parry G. Assessing the function of the plant nuclear pore complex and the search for specificity. J Exp Bot. 2013;64:833–845. doi: 10.1093/jxb/ers289. [DOI] [PubMed] [Google Scholar]
- 50.Parry G, Ward S, Cernac A, Dharmasiri S, Estelle M. The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell. 2006;18:1590–1603. doi: 10.1105/tpc.106.041566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng YT, Germain H, Wiermer M, Bi D, Xu F, García AV, Wirthmueller L, Després C, Parker JE, Zhang Y, Li X. Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell. 2009;21:2503–2516. doi: 10.1105/tpc.108.064519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiermer M, Cheng YT, Imkampe J, Li M, Wang D, Lipka V, Li X. Putative members of the Arabidopsis Nup107-160 nuclear pore sub-complex contribute to pathogen defense. Plant J. 2012;70:796–808. doi: 10.1111/j.1365-313X.2012.04928.x. [DOI] [PubMed] [Google Scholar]
- 53.Roth C, Wiermer M. Nucleoporins Nup160 and Seh1 are required for disease resistance in Arabidopsis. Plant Signal Behav. 2012;7:1212–1214. doi: 10.4161/psb.21426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett. 2010;6:342–345. doi: 10.1098/rsbl.2009.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelly S, Kramer S, Schwede A, Maini PK, Gull K, Carrington M. Genome organization is a major component of gene expression control in response to stress and during the cell division cycle in trypanosomes. Open Biol. 2012;2:120033. doi: 10.1098/rsob.120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rout MP, Field MC. Isolation and characterization of subnuclear compartments from Trypanosoma brucei. Identification of a major repetitive nuclear lamina component. J Biol Chem. 2001;276:38261–38271. doi: 10.1074/jbc.M104024200. [DOI] [PubMed] [Google Scholar]
- 57.Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21:915–924. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imamoto N, Funakoshi T. Nuclear pore dynamics during the cell cycle. Curr Opin Cell Biol. 2012;24:453–459. doi: 10.1016/j.ceb.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 59.Jaspersen SL, Ghosh S. Nuclear envelope insertion of spindle pole bodies and nuclear pore complexes. Nucleus. 2012;3:226–236. doi: 10.4161/nucl.20148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doucet CM, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 2010;141:1030–1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, Kutay U. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 2011;144:539–550. doi: 10.1016/j.cell.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 62.D’Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW. A change in nuclear pore complex composition regulates cell differentiation. Dev Cell. 2012;22:446–458. doi: 10.1016/j.devcel.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Souza CP, Osmani AH, Hashmi SB, Osmani SA. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr Biol. 2004;14:1973–1984. doi: 10.1016/j.cub.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 64.Osmani AH, Davies J, Liu HL, Nile A, Osmani SA. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol Biol Cell. 2006;17:4946–4961. doi: 10.1091/mbc.E06-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu HL, De Souza CP, Osmani AH, Osmani SA. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol Biol Cell. 2009;20:616–630. doi: 10.1091/mbc.E08-06-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Asakawa H, Hiraoka Y, Haraguchi T. Nuclear translocation of RanGAP1 coincides with virtual nuclear envelope breakdown in fission yeast meiosis. Commun Integr Biol. 2011;4:312–314. doi: 10.4161/cib.4.3.14808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 68.Andrade MA, Bork P. HEAT repeats in the Huntington’s disease protein. Nat Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- 69.Malik HS, Eickbush TH, Goldfarb DS. Evolutionary specialization of the nuclear targeting apparatus. Proc Natl Acad Sci U S A. 1997;94:13738–13742. doi: 10.1073/pnas.94.25.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang ZC, Satterly N, Fontoura BM, Chook YM. Evolutionary development of redundant nuclear localization signals in the mRNA export factor NXF1. Mol Biol Cell. 2011;22:4657–4668. doi: 10.1091/mbc.E11-03-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mason DA, Stage DE, Goldfarb DS. Evolution of the metazoan-specific importin alpha gene family. J Mol Evol. 2009;68:351–365. doi: 10.1007/s00239-009-9215-8. [DOI] [PubMed] [Google Scholar]
- 72.Gelvin SB. Finding a way to the nucleus. Curr Opin Microbiol. 2010;13:53–58. doi: 10.1016/j.mib.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 73.Markina-Iñarrairaegui A, Etxebeste O, Herrero-García E, Araújo-Bazán L, Fernández-Martínez J, Flores JA, Osmani SA, Espeso EA. Nuclear transporters in a multinucleated organism: functional and localization analyses in Aspergillus nidulans. Mol Biol Cell. 2011;22:3874–3886. doi: 10.1091/mbc.E11-03-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O’Reilly AJ, Dacks JB, Field MC. Evolution of the karyopherin-β family of nucleocytoplasmic transport factors; ancient origins and continued specialization. PLoS One. 2011;6:e19308. doi: 10.1371/journal.pone.0019308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dacks JB, Field MC. Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J Cell Sci. 2007;120:2977–2985. doi: 10.1242/jcs.013250. [DOI] [PubMed] [Google Scholar]
- 76.Dacks JB, Peden AA, Field MC. Evolution of specificity in the eukaryotic endomembrane system. Int J Biochem Cell Biol. 2009;41:330–340. doi: 10.1016/j.biocel.2008.08.041. [DOI] [PubMed] [Google Scholar]
- 77.Sampathkumar P, Kim SJ, Upla P, Rice WJ, Phillips J, Timney BL, Pieper U, Bonanno JB, Fernandez-Martinez J, Hakhverdyan Z, Ketaren NE, Matsui T, Weiss TM, Stokes DL, Sauder JM, et al. Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Structure. 2013;21:560–571. doi: 10.1016/j.str.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lynch M, Bobay LM, Catania F, Gout JF, Rho M. The repatterning of eukaryotic genomes by random genetic drift. Annu Rev Genomics Hum Genet. 2011;12:347–366. doi: 10.1146/annurev-genom-082410-101412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lukeš J, Archibald JM, Keeling PJ, Doolittle WF, Gray MW. How a neutral evolutionary ratchet can build cellular complexity. IUBMB Life. 2011;63:528–537. doi: 10.1002/iub.489. [DOI] [PubMed] [Google Scholar]
- 80.Curtis BA, Tanifuji G, Burki F, Gruber A, Irimia M, Maruyama S, Arias MC, Ball SG, Gile GH, Hirakawa Y, Hopkins JF, Kuo A, Rensing SA, Schmutz J, Symeonidi A, et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature. 2012;492:59–65. doi: 10.1038/nature11681. [DOI] [PubMed] [Google Scholar]