Abstract
Turnip crinkle virus (TCV) has been shown to interact with a NAC transcription factor, TIP, of Arabidopsis thaliana, via its coat protein (CP). This interaction correlates with the resistance response manifested in TCV-resistant Arabidopsis ecotype Di-17. We report that failure of a mutated CP to interact with TIP triggered the corresponding TCV mutant (R6A) to cause more severe symptoms in the TCV-susceptible ecotype Col-0. We hypothesized that TCV regulates antiviral basal immunity through TIP-CP interaction. Consistent with this hypothesis, we found that the rate of accumulation of R6A was measurably slower than wild-type TCV over the course of an infection. Notably, R6A was able to accumulate at similar rates as wild-type TCV in mutant plants with defects in salicylic acid (SA) signaling. Finally, plants with altered TIP expression provided evidence R6A's inability to evade the basal resistance response was likely associated with loss of ability for CP to bind TIP.
Keywords: Turnip crinkle virus, Arabidopsis, TIP, Coat protein, Resistance, Salicylic Acid, Basal Immunity, Defense signaling
Introduction
Plants employ multiple mechanisms to defend themselves against pathogens. A key element of plant defense is the recognition of the pathogen-encoded microbe-associated molecular patterns (PAMPs/MAMPs) and subsequent triggering of a mitogen-activated protein kinase (MAPK) signaling cascade that will induce the appropriate defense response (Antolín-Llovera et al., 2012). Multiple reviews have described the plant innate immune system as consisting of two layers (Bolton, 2009; Jones and Dangl, 2006; Schwessinger and Ronald, 2012). Layer one, PAMP-triggered immunity (PTI), uses transmembrane pattern recognition receptors (PRRs) to recognize PAMPs, such as flagellin (Felix et al., 1999). The other component referred to as effector triggered immunity (ETI) uses the polymorphic nucleotide binding-leucine rich repeat (NB-LRR) proteins encoded by most Resistance (R) genes to induce a more intense defense response (reviewed in Boller and Felix, 2009).
Salicylic acid (SA) is a small phenolic plant compound that plays a vital role in the defense responses against many pathogens in both branches of plant innate immunity (Vlot et al., 2009). Infections by biotrophic pathogens induce increased levels of SA, which in turn up-regulate the expression of many defense-related genes (Malamy et al., 1990; Sticher et al., 1997). Plants with defects in SA synthesis, signaling or accumulation exhibit enhanced susceptibility to pathogen infection (Glazebrook, 2001). In Arabidopsis and tobacco, SA is also crucial for the establishment of systemic acquired resistance (SAR; Durrant and Dong, 2004). SAR is accompanied by the induction of a set of SA dependent pathogenesis-related (PR) genes in inoculated and systemic tissue (Ryals et al., 1996) and senescence-associated genes (Morris et al., 2001). SA is also linked to parts of the senescence pathway like SEN1 (Schenk et al., 2005) which is one of the factors needed for regulating senescence (Morris et al., 2001).
NAC transcription factors are a plant specific group of proteins, which contain a highly conserved N-terminal DNA-binding domain and a variable C-terminal domain (Olsen et al., 2005). Previous analyses have identified over 100 NAC encoding genes in the genomes of Arabidopsis thaliana and Oryza sativa that have tissue and stress response specific expression (Fang et al., 2008; Ooka et al., 2003). Along with being involved with abiotic defense responses, NAC genes have been shown to be involved in defense signaling against viral pathogens (Selth et al., 2005; Xia et al., 2010; Yoshii et al., 2009). The Arabidopsis NAC transcription factor, ATAF2, is induced in response to a Tobacco mosaic virus (TMV) infection and TMV subsequently targets ATAF2 for degradation through an interaction with the viral 126 kDa replication protein (Wang et al., 2009). The NAC transcription factor, GRAB (Geminivirus RepA binding), alters Geminivirus DNA replication in connection to plant growth, development and senescence pathways. Another member of the NAC family, TIP (TCV-interacting protein), was shown to play a key role in the host defense response by interacting with TCV coat protein (CP; Ren et al., 2000). This study analyzed a series of single amino acid (aa) substitution mutants of TCV CP to assess the role of TIP binding in the R gene-mediated defense conditioned by an NB-LRR protein designated HRT in the TCV-resistant Arabidopsis ecotype Dijon 17 (Di-17). The ability of the R domain of TCV CP to bind to TIP, a NAC transcription factor, was shown to correlate with the observed variability in disease symptom severity in the susceptible Col-0 ecotype and the ability to confer resistance in the resistant Di-17 ecotype (Ren et al., 2000, 2005). It was suggested that the TIP-CP interaction was playing a functional role in the defense response of Arabidopsis to TCV and that its normal role was compromised by interaction with the invading viral CP.
A subsequent study by Jeong et al. (2008) demonstrated that the HRT response proceeded normally in plants with a T-DNA insertion in the promoter region of the TIP gene. They concluded from these results that TIP was not involved in this defense response (Jeong et al., 2008). They also revealed that the TIP-CP interaction was likely important in the basal defense response to both TCV and CMV (Jeong et al., 2008). However, this study left open the question as to why all of the non-TIP binding mutants reported by Ren et al (2000) failed to elicit a resistance response. To address this, we conducted a detailed study of one of the mutants, R6A, to further assess the role of TIP in basal resistance. Here we found that the ability of wild-type (wt) TCV CP to bind TIP correlated with a down-regulation of the SA pathway, which allowed TCV a clear replication advantage over R6A, leading to higher accumulation of wt TCV early in infection. We further showed a correlation between TCV accumulation levels and TIP availability in the susceptible Col-0 ecotype. This work confirms that TIP-CP binding does indeed play a role in the basal defense response to TCV infection in susceptible Col-0. We also show that a lack of TIP-CP interaction was correlated with an elevation of TCV symptom severity in the susceptible ecotype of Col-0, and this appeared to be a consequence of a specific alteration in SA pathway defense signaling.
Materials and Methods
Plant growth conditions
Plants lines of wt A. thaliana ecotype Col-0 and Di-17, and knockout (ko) lines in a Col-0 background of npr1, pad4, jar1 and ein2 were grown in growth chambers at 22°C with 12hr day cycles. Transgenic lines of antisense TIP (asTIP) and a constitutively up-regulated TIP (UpTIP) line that had an additional TIP gene under the control of a 35S promoter were initially grown on selective media to verify the presence of inserts prior to transplanting.
Plant inoculations, tissue collection, and RNA isolation
A total of 10 ng of virus transcript in a buffer solution containing 50 mM Na2HPO4 [pH 7.0] + 1% Celite 545 was inoculated to three leaves of 22 to 24 day old plants. Five to six leaves (apx 0.3g) from different plants treated with the same inoculum buffer were collected at each time point and flash frozen in liquid nitrogen. RNA was extracted as previously described (Chomczynski and Sacchi, 1987) and RNA samples were subsequently purified using RNeasy columns (Qiagen).
Virus detection and gene analysis
Detection of viral RNAs was conducted as previously described (Qu and Morris, 2000). CP was detected with the addition of 32P-γ-ATP end-labeled probes (Table 2). Virus titers were evaluated using ELISA as previously described (Lommel et al., 1982). The primers (Invitrogen) of the genes used for analysis for semi-quantitative PCR are listed in Table 1. Real-time PCR was also used to evaluate gene expression according to manufacturer's protocol (Applied Biosystems). Assay ID's are defined in Table 3.
Table 2. Probes for Northern Analysis.
| Gene | Sequence |
|---|---|
| TCV-CP | Rev: 5′-CAGGACCGAGAAGTCAGAGG-3′ |
| Rev: 5′-GGCCCACCCGACACCACTGG-3′ | |
| Rev: 5′-CTTGTCTTGACCGAGTTGGT-3′ |
Table 1. Sequences of probes used for semi-quantitative PCR.
| Gene | Sequence |
|---|---|
| ACT2 | Fwd: 5′-GTCTGAGATTTCTCCTGCCG-3′ |
| Rev: 5′-CACGGTTAGCCTTTGGGTTA-3′ | |
| EIN2 | Fwd: 5′-CTTGGCTTCATCGTGCTACA |
| Rev: 5′-CTTAAGCTGCGGAATGAAGG | |
| JAR1 | Fwd: 5′-ACTAGCGCAGGATGTTGGAG-3′ |
| Rev: 5′-AGCGTTTCCATTGAGACCAC-3′ | |
| NPR1 | Fwd: TGCATCAGAAGCAACTTTGG |
| Rev: GAGGCAAGAGTCTCACCGAC | |
| PAD4 | Fwd: 5′-TTGTCGATTCGAGACGAGTG-3′ |
| Rev: 5′-TTTTTAAATCACTTGGGCGG-3′ | |
| TIP | Fwd: 5′-CCGGCTCAAGATCAACGGTCACG-3′ |
| Rev: 5′- CTGCTCAGCACAACCCGGGG -3′ | |
| PDF1.2 | Fwd: 5′- TGCTTTCGACGCACCGGCAA |
| Rev: 5′- CCGCAAACCCCTGACCATGTCC | |
| WRKY70 | Fwd: 5′-TGAACCAACTCGTTGAAGGCCATGA |
| Rev: 5′-CAACGGCGGCGAGGGATGAG |
Table 3. Genes used for Time Course qRT-PCR.
| Gene Name/Description | Transcript ID | Assay ID | |
|---|---|---|---|
| 1 | TIP | AT5G24590 | At02185798_s1 |
| 2 | SEN1 | AT4G35770 | At02255940_g1 |
| 3 | WRKY6 | AT1G62300 | At02216109_gH |
| 4 | ACT2 | AT3G18780 | At02335270_gH |
Construction of Transgenic lines
Arabidopsis plants designed to up-regulated TIP (p35-UpTIP) were made by cloning the full length TIP cDNA into a plasmid pRTL2. This plasmid flanked the TIP cDNA insert with the CaMV 35S promoter and a polyA signal. The TIP cassette, including the 35S promoter, TIP cDNA, the 35S polyA signal (Fig. 4A) was cut out of pRTL2 and subcloned into the binary vector, pPZP212. The resulting construct was used to transform Agrobacterium tumefaciens train C58C1 as previously described (Bechtold et al., 1993). To down-regulate TIP expression, an antisenseTIP (asTIP) construct was made by cloning a partial fragment of the TIP cDNA (nt 1106 – 1637), in reverse orientation, into pER8 (between XhoI and SpeI sites), as previously described (Zuo et al., 2001). The resulting construct was then transformed into Agrobacterium and transformed as previously described. To induce the asTIP promoter, plants were sprayed with a 10μM estradiol solution once a day to induce the asTIP transcript for three days prior to virus inoculation and once a day throughout the time course.
Figure 4. Comparison of TCV and R6A accumulation in Arabidopsis lines with altered levels of TIP gene expression.
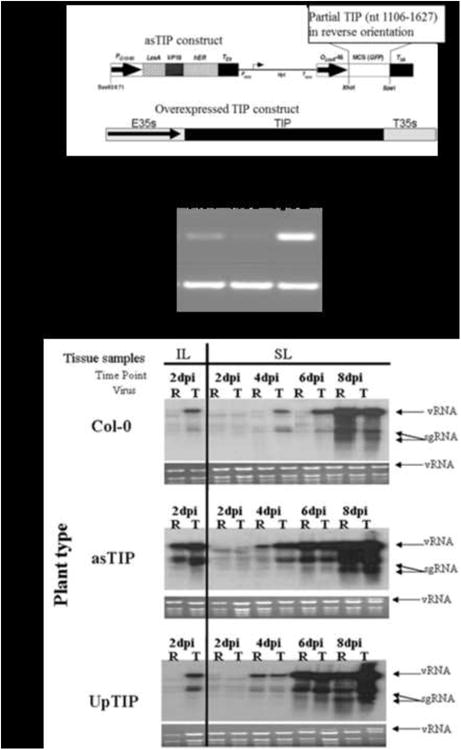
(A) Diagram showing the construction of two transgenic lines of A. thaliana ecotype Col-0. asTIP was designed to reduce TIP expression in mature plants by RNA silencing from an estradiole inducible promoter. TIPup was designed to overexpress TIP constitutively under the control of the CaMV 35S promoter (E35S). (B) Semi-quantitative PCR evaluation of TIP transcript expression in wild type and TIP altered transgenic lines of Col-0. ACT2 was used as the endogenous control. PCR cycle number used is shown on the right side of the figure. (C) Northern blots showing accumulation of viral RNAs of wt TCV (T) or mutant R6A (R) in inoculated leaves (IL) at 2 dpi and in systemic leaves (SL) at 2, 4, 6, and 8 dpi developed as described in Fig. 1. The panels show accumulation in the asTIP line after estradiole treatment and in the constitutively up-regulated TIP line. These are representative panels of three experiment which yielded consistent results.
Results
The mutant R6A accumulates more slowly than wt TCV in Arabidopsis
R6A, a TCV mutant containing an arginine-to-alanine (R-to-A) substitution at aa position 6 of CP, was found previously to break the HRT-mediated resistance in TCV-resistant Di-17 plants (Ren et al., 2000). We further observed that this mutant also caused more severe symptoms than wt TCV in susceptible Col-0 (Fig.1A). This observation suggested that the single aa change altered the ability of TCV to induce both the basal defense response in the susceptible host and the R gene-mediated defense in the resistant host. To provide an assessment of the basal defense response, virus accumulation levels were measured by collecting leaf tissues from both R6A and wt TCV-infected plants at multiple time points after inoculation.
Figure 1. Symptoms and virus accumulation of TCV and the R6A mutant in Arabidopsis thaliana.
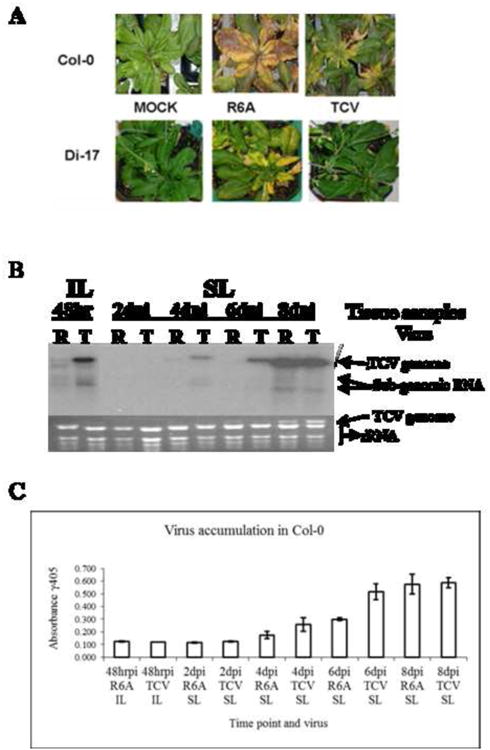
(A) Symptoms in susceptible Col-0 (top) and resistant Di-17 (bottom) at 14 days after inoculation (dpi). (B) Northern blot showing accumulation of viral RNAs of wt TCV (T) or mutant R6A (R) in inoculated leaves (IL) at 48 hr post inoculation (hrpi) and systemic leaves (SL) at 2, 4, 6, and 8 day post inoculation (dpi). The probe was specific for CP sequence as described in Materials and Methods. Arrows identify location of viral genome (vRNA) and subgenomic RNAs (sgRNA). The ethidium bromide (EtBr) stained gel is shown below as a loading control. Representative of three repeated experiments. (C) Virus particle accumulation measured by ELISA in inoculated (IL) and systemic leaves (SL) of A. thaliana ecotype Col-0 over the time course. Error bars represent standard deviation from the absorbance levels of ELISA readings for three independent experiments. Background reading of 0.200 OD units was subtracted as background.
In order to capture subtle changes in the levels of viral genomes and virions, we measured the levels of viral RNAs, as well as the titers of virus particles. We found that even though R6A caused more severe symptoms in Col-0 at late stages of infection, its viral RNA and virion titers were visibly lower than wt TCV at the early stage of infections, in both inoculated and systemic tissues (Fig. 1B, compare R and T lanes). This is most evident at four and six days post inoculation (4 and 6 dpi), when both genomic (g) and subgenomic (sg) RNAs were easily detected in wt TCV-infected samples, but not in those infected with R6A. These results were also confirmed by assessing virion accumulation with ELISA (Fig. 1C). Although these differences diminished by 8 dpi, their consistent detection at the early stage of infection suggests that R6A is more readily targeted than wt TCV by a basal defense response that is activated immediately after encountering the viruses. Furthermore, since the R6A CP lost the ability to interact with TIP, it can be speculated that this basal defense response could be compromised by TIP-CP interaction, thus suggesting a positive regulator role for TIP in the activation or execution of the basal defense.
R6A invasiveness was restored to wt TCV levels in mutant plants with defects in the SA signaling pathway
We speculated that the delayed accumulation of R6A could be caused by a more robust basal defense. To further elucidate the nature of this basal defense, we chose to first evaluate whether the SA signaling pathway plays any role in containing R6A. We thus acquired several Arabidopsis lines containing knockout mutations within key components of the SA and jasmonic acid (JA) signaling pathways (Huang et al., 2005). These included the SA defective npr1 and pad4 mutants, and the JA defective jar1mutant. The npr1 mutant contained a mutation within the NPR1 gene required for the induction of SAR, as well as pathogen-induced expression of PR genes (Cao et al., 1994; Dong et al., 1991). The pad4 mutant contained a defective PAD4 gene, making it unable to synthesize SA (Zhou et al., 1998). The jar1 mutant lacked a functional JAR1 gene responsible for JA biosynthesis and resistance to necrotrophic pathogens (Staswick et al., 1998).
All three mutant lines were first genotyped using RT-PCR to confirm the absence of transcripts for NPR1, PAD4 and JAR1 (Fig. 2A). We then subjected the mutant plants to R6A and wt TCV infections as described in the last section. In contrast to the wild type Col-0 plants, the npr1 and pad4 mutants permitted similar levels of R6A and wt TCV RNA accumulations (Fig. 2C, D and E). This result was also confirmed by measuring virion amounts using ELISA (Fig. 2B). These data suggest that a fully functional SA pathway likely played an important role in reducing the multiplication rate of R6A at the early stage of infections in Col-0. The observation that both R6A and wt TCV reached similar levels in npr1 and pad4 mutants further suggests that R6A was as replicatively competent as TCV, and that its primary defect was likely the inability to repress the SA signaling pathway. Interestingly, we noted that the wt TCV RNA levels were also elevated in npr1 and pad4 mutants, as evidenced by the more intense gRNA band in the ethidium bromide stained gel at 6 dpi (Fig. 2D and 2E. note the relative intensities of TCV gRNA and the rRNA bands in respective lanes). We also observed that npr1 and pad4 lines infected with TCV or R6A did not show any visual differences at 12 dpi (Figure 2G-J) though by 21 dpi the plants were 100% dead compared to the Col-0 control that still showed signs of life (data not shown). This suggests that, in wt TCV infections, the TIP-CP interaction only partially repressed the SA-mediated basal defense. We reason that this partial repression could be evolutionally selected to ensure the survival of both the virus and the host, as a fully activated basal defense as encountered by R6A could paradoxically cause more severe disease and possibly the death of the host plant, diminishing the chance for virus spread.
Figure 2. Comparison of TCV and R6A accumulation in Arabidopsis lines with compromised defense pathway signaling responses.
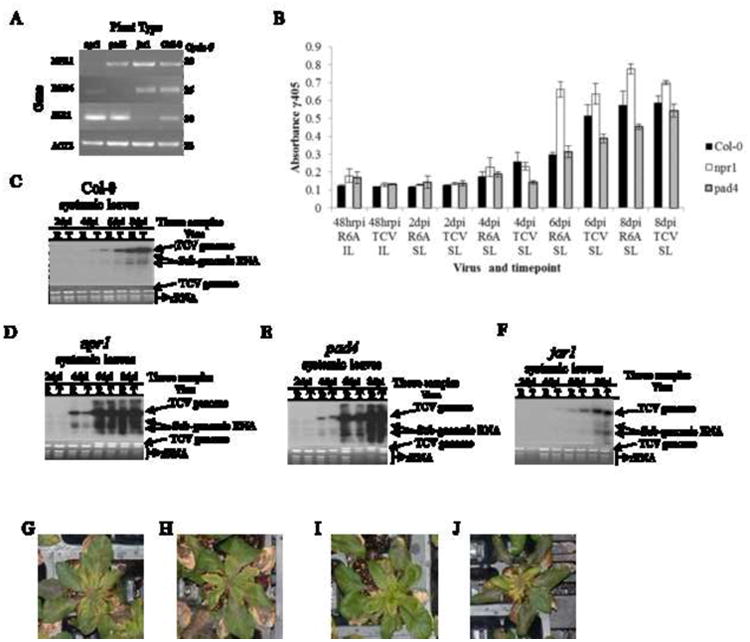
(A) PCR analysis confirming the absence of NPR1, PAD4, JAR1 and EIN2 in various knockout lines of Arabidopsis thaliana. Actin 2 was used as a control. The number of PCR cycles for each gene is shown on the right. Primer sequences are shown in Table 1. (B) Northern blots showing accumulation of viral RNAs of wt TCV (T) or mutant R6A (R) in systemic leaves (SL) at 2, 4, 6, and 8 day post inoculation (dpi) as described in Fig. 1. The panels show accumulation in each of the specific knockout lines identified in panel A. This experiment was repeated 3 times with similar results. (C-E) Virus particle accumulation measured by ELISA in inoculated (IL) and systemic leaves (SL) of the NPR1, PAD4 and JAR1 KO lines. Error bars indicate standard deviation for 3 independent experiments. (G-J) NPR KO lines at 12 dpi, infected with TCV (G) or R6A (H). PAD4 KO lines at 12 dpi infected with TCV (I) or R6A (J).
By contrast, the jar1 mutant was similar to wild-type Col-0 in that it facilitated slower accumulation of R6A than wt TCV (Fig. 2F). However, both viruses accumulated more slowly in jar1 than in Col-0, as demonstrated by both viral RNA levels and virion amounts (Fig. 2F; data not shown). Taken together, these observations reinforce our contention that a functional SA pathway is important for defense against viral pathogens like TCV, and the role of the JA and ET signaling pathway is not essential for defense against these viruses.
Differential effect of TCV and R6A infections in Col-0 on TIP expression
In order to assess the effect that virus infections might have on the levels of TIP, we used the real-time PCR assays to examine TIP expression during the time course. After 48 hpi, we were unable to detect any significant gene expression differences in IL in the viral infections relative to the mock inoculated controls (Fig. 3A). However, we were consistently able to observe a 5-6 fold increase at 2 dpi and 2-3 fold increase at 4 dpi in levels of TIP in SL in both wt and mutant virus infections. These results, in comparison to the mock infection, suggest that TIP levels, although induced somewhat by a wound response, show enhanced induction in response to systemic virus infection by both viruses (Fig. 3A). R6A infections induced TIP expression significantly more than wt TCV infections at 2 dpi and 4 dpi time points in SL and then declined at 6 dpi. The temporal pattern of TIP induction during virus infection was consistent in 3 separate experiments. Most interesting was the 8 dpi time point where it is evident that TIP levels declined in TCV infections but appeared to increase a second time in R6A infections at time of onset of increasing symptom severity. We hypothesize that the ability of TCV CP to bind TIP may allow TCV to more effectively control TIP expression and, as a consequence, more effectively regulate the PTI response.
Figure 3. Comparison of expression of defense related genes in TCV and R6A infections over the time course of infection using Real-time PCR.
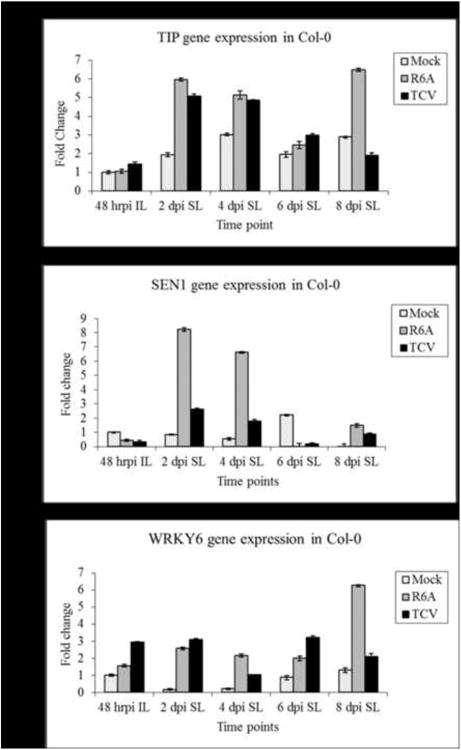
A. thaliana ecotype Col-0 was inoculated with either TCV or R6A transcripts and total RNA was extracted for each time point and analyzed using One-step Real-time PCR as described in Materials and Methods. The relative fold change of (A) TIP, (B) SEN1 and (C) WRKY6 are plotted. Fold change is calculated relative to the mock infection for each gene at 48hrpi. ACT2 was used as the endogenous control. This graph is the average of results obtained for 2 independent experiments.
R6A infections caused enhanced expression of senescence pathway genes
One of the most characteristic features of R6A infections was the enhanced symptom severity compared to TCV (Fig. 1A). The systemic symptoms induced by R6A infections also resembled that of a prematurely senescent plant. To further assess this possible link between senescence and increased symptom severity, we evaluated SEN1 gene expression to see if the senescence pathway was differentially affected in R6A versus TCV infections. SEN1 regulates signals that link plant defense and senescence responses, and hence is a useful marker to study in crosstalk between the two responses (Schenk et al., 2005). Our results showed that SEN1 gene expression was elevated 8 fold at 2 dpi and 6 fold at 4 dpi in systemic tissue compared to 2-2.5 fold for TCV infections at the same time points (Fig. 3B). This temporal pattern of expression, which was consistent in three separate experiments, mirrored the pattern of TIP induction. Although our data does not directly link TIP and SEN gene expression differences to the differences observed in virus accumulation and symptom severity, it does support the idea that defense induction and senescence responses are connected. Therefore we propose that TCV's ability to bind TIP not only allows it to suppress the SA response and hence evade basal immunity, but it also prevents the senescence pathway from being induced, resulting in milder symptoms.
Differential induction of WRKY family transcription factors
We also evaluated WRKY6 gene expression in response the different virus infections (Fig. 3C). WRKY6 is a transcription factor that belongs to a plant specific transcription factor family with members that have roles as both activators and repressors of defense responses. WRKY6 negatively regulates itself and WRKY42, however it is also a positive regulator of PR1, NPR1 and SIRK, which are also involved in senescence pathway signaling (Robatzek and Somssich, 2002). We did observe a slight induction of WRKY6 in IL of R6A infections (1.5 fold compared to mock) compared to a 3 fold induction in TCV IL. In TCV SL the levels varied between 1 and 3 fold over mock for the duration of the time course. Interestingly, WRKY6 levels were induced 6 fold in R6A over mock at 8 dpi (Fig. 3C), a time point coincident with elevated TIP expression and the timing of increased symptom severity.
Making a connection between viral accumulation, TIP-CP interaction, and basal resistance
To determine if TIP was playing a direct role in basal defense, we attempted to manipulate the TIP expression levels by making two types of transgenic plants. In the first, we tried to down-regulate the expression of TIP mRNA by producing transgenic plants that had inducible expression an antisense transcript of TIP, referred to as asTIP plants. In the other, we produced transgenic plants that constitutively overexpress TIP mRNA, referred to as UpTIP plants (Fig. 4A). The level of TIP mRNA in these lines was verified by semi-quantitative PCR (Fig. 4B). Over expression of TIP did not cause any dramatic alteration in plant phenotype. However, we did notice that over-production of TIP caused plants to flower earlier while antisense TIP plants showed a delay in flower development (results not shown).
A time course experiment was implemented to compare rates of virus accumulation of TCV and R6A in these transgenic plants. We observed consistent changes in the accumulation of the R6A mutant in both the TIP up-regulated and down-regulated transgenic plants. The data presented in Fig. 5c are representative panels of three independent experiments. In the asTIP lines, in which TIP levels were reduced transiently, R6A accumulated to equivalent levels as TCV in inoculated leaves and to almost the same levels as TCV at the 4 dpi time point. This is in contrast to the results in Col-0 where R6A is barely detectable in IL and 4 and 6 dpi SL (compare panel 1 and 2 in Fig. 4c). This result demonstrated that down-regulation of TIP appeared to eliminate the replicative advantage displayed by TCV over R6A in Col-0. This observation provided additional evidence that the binding of TCV CP to TIP was indeed responsible for the down-regulation of basal defense that permitted the more rapid accumulation of TCV compared to the R6A mutant in Col-0 susceptible plants.
Infections in the TIP overexpression plants (UpTIP line) showed similar results for the systemic leaf infections where we clearly observed that both R6A and TCV accumulate to similar levels through the course of the infection (Fig. 4C, panel 3). It is evident in the blot that in SL, R6A accumulated to equivalent levels as TCV at the 4 and 6 dpi time points. These results demonstrate that elevated TIP levels also eliminated the advantage that TCV displayed over R6A in Col-0 infections. A likely explanation is that under conditions of excess TIP, the ability of TCV CP to sequester a sufficient quantity of TIP to compromise the resistance response is negated. Curiously, this effect was only observed in the systemic, but not the inoculated leaves in the infections in TIP over-expressing plants. These results provide additional confirmation for a direct role of TIP in modulating basal defense.
Discussion
In this study, we analyzed the potential mechanisms associated with the differential ability of TCV and the mutant R6A to accumulate in susceptible and resistant ecotypes of Arabidopsis thaliana. We were able to connect the observed phenotype of slower accumulation of R6A directly with the innate defense response of the host by identifying two knock out lines that had dysfunctional SA pathways. In pad4 and npr1 mutants, R6A accumulation recovered and/or exceeded wt TCV virus accumulation levels. This provided corroboration that not only was R6A as robust as wild type TCV, but also that the R6A deficiency was due to its inability to turn off or elude the SA defense pathway and the subsequent SAR defense response that is characteristic of wt TCV infections. The recovery of the virus accumulation by the mutant was attributed to both wt TCV and R6A eliciting a similar level of silencing suppressor activity encoded in their CP (Choi et al., 2004). Therefore, we concluded that TIP-CP interaction was associated with an earlier basal resistance such as the induction of SAR or the activation of the senescence pathway.
These experiments support our contention that TCV has evolved a mechanism to specifically down-regulate the SA defense pathway thus giving wild type virus a distinct advantage over the non-TIP binding mutants. This also raises the unexpected possibility that the increased symptom severity associated with R6A infection may be linked to over-activation of the SA defense response. This phenomenon would be analogous to an inflammation response in animal systems due to the over-stimulation of the innate immune response (Ausubel, 2005; Nürnberger et al., 2004). Together these data also suggest that the SA signaling in association with the PTI response is important in partially repressing viral invasion.
We also assessed accumulation of TCV and R6A transcript levels in plants where levels of TIP were reduced (asTIP) and elevated (UpTIP) provided additional evidence that linked TIP-CP interaction more directly with the differential activation of the basal resistance response. These experiments uncovered that the down-regulation of TIP may not play as much of a role in the defense against TCV infection as was previously hypothesized (Ren et al., 2000, 2005). This could be a consequence of genetic redundancy within the NAC transcription factor family which has over 100 members. Therefore, eliminating one NAC gene might not necessarily abolish the resistance modulating function due to compensation by another related gene (Briggs et al., 2006; Pickett and Meeks-Wagner, 1995). However, constitutive overexpression of TIP did affect the virus accumulation (Fig. 4C). When we infected the overexpressing TIP plants with TCV or R6A, we observed both viruses were able to accumulate to equivalent levels similar to what we observed in the knockout lines with dysfunctional SA pathways (compare Fig. 2D-E and 4C). This suggested that the increased fitness of TCV over the mutant R6A was less dependent on the complete absence of TIP in the cell, but more affected by the total amount of TIP present during an infection. In addition, because TIP has a membrane localization signal along with a nuclear localization signal (Kang, unpublished data), we suggest that it localizes outside the nucleus in a cellular membrane from which it undergoes controlled released by cleavage when the plant is not infected to maintain negative regulation of the innate defense responses (Ren et al., 2005). We propose that TCV CP interaction with TIP could either positively or negatively alter the rate of its nuclear localization which would then subsequently alter defense signaling to favor enhanced TCV invasion. In contrast, because the mutant R6A CP has lost the ability to interact with TIP, the consequence is a more robust PTI response.
Jeong et al. (2008) described a tip knockout line in Col-0 background that also expressed the R gene, HRT. This transgenic line was found to be resistant to TCV infection. Therefore, they concluded that TIP did not have a direct role in the R-gene resistance to TCV infection (Jeong et al., 2008). However, our results support the conclusion that TIP binding is a key factor in the basal defense response. It is possible that our studies suggest the possibility that basal level defense and HR induction are distinct and separately controlled defense responses (Jones and Dangl, 2006). However, more studies will need to be conducted to more convincingly show the separation of these defense mechanisms in Arabidopsis infected with TCV.
It is well known that plants have the capacity to recognize pathogens and in many cases there is functional redundancy within multigene families that often complicates genetic attempts to define the role of individual genes (Bouché and Bouchez, 2001). One example of this was found with a wrky6 knockout mutation, which resulted in no obvious mutant phenotype yet overexpression of WRKY6 caused a stunted phenotype and a significant increase in SA pathway associated genes like PR1 and NPR1 (Robatzek and Somssich, 2002). It is possible that we observed a similar effect because adjusting TIP levels appeared to impact the plant's defense system to a similar extent as seen in the SA pathway defective mutant knockout lines of pad4 and npr1 (compare Fig. 2 and 4). In this case, we observed that defective defense signaling permitted virus to accumulate at a faster rate than what was observed in wt Col-0. Interestingly, reduced levels of TIP in the asTIP plants permitted both mutant and wild type TCV to accumulate equally rapidly in both inoculated and systemic leaves. This result supports the conclusion that TIP may be a positive regulator of the anti-viral basal defense response. However, in the case where there was excess TIP, there were no evident differences in the levels of wild type and mutant virus in systemic leaves. Interestingly, under conditions of excess TIP, wild type virus accumulated more rapidly in inoculated leaves suggesting that the TIP-CP interaction resulting in reduced basal resistance was more transient in inoculated leaves. The data presented here demonstrates the importance of the TCV CP binding TIP to suppress the PTI response and how parts of the SA signaling pathway are key components in TCV defense.
Other major factors affecting the ability for virulent viruses to successfully infect their host are suppression of the RNA silencing pathway and manipulating the ability to control the expression of endogenous genes which regulate pathogen invasion. Recently, it was discovered that the TCV CP mimics host-encoded glycine/tryptophan (GW)-containing proteins normally required for RNA induced silencing complex (RISC) assembly and function (Azevedo et al., 2010). TCV coat protein GW residues bind directly and specifically to Arabidopsis AGO1, which has been identified as a major effector of TCV-derived siRNAs (Azevedo et al., 2010). The observation that one of the two functional GW motifs in the CP is in close physical proximity (N terminal aa 25-26) to the TIP binding domain (N-terminal aa 1-25) raised the possibility of a connection between TIP-CP interaction and silencing suppressor function. It has, however, been demonstrated that the silencing suppressor activity of TCV CP is not altered in the non-TIP binding mutants R6A and R8A (Choi et al., 2004). However, it remains an open question if the basal defense response modulated by TIP-CP binding might not be a consequence of altering the expression of one to several components of the RNA silencing pathway.
In summary, our data demonstrates that TIP expression is important in regulating the basal resistance response which then impacts the rate of TCV accumulation in A. thaliana. We have shown that the level of TIP expression affects proper signaling of the SA pathway and other defense responses. Therefore we conclude that TIP is a key player in the basal defense response against TCV. Further research is needed to examine what other proteins may have similar or redundant functions to TIP that could possibly mask the effects of its absence in the knockout lines as Jeong (2008) previously described. Recent work from our lab provides some evidence that TIP, although not required for HR induction, could be playing a role in fine tuning the HR defense response to TCV infection (Kang, 2012).
Highlights.
TCV CP ability to bind TIP correlates with a down-regulation of the SA pathway.
Correlation with TCV accumulation and the ability to bind TIP in suspectible Col-0.
Lack of TIP-CP interaction was linked with an increase disease symptoms.
Mutatnt TCV unable to evade SA pathway signaling.
Acknowledgments
This publication was made possible by grants from the National Center for Research Resources (5P20RR016469) and the National Institute for General Medical Science (NIGMS; 5P20GM103427), a component of the National Institutes of Health (NIH) and its contents are the sole responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antolín-Llovera M, Ried MK, Binder A, Parniske M. Receptor Kinase Signaling Pathways in Plant-Microbe Interactions. Annual Review of Phytopathology. 2012;50:451–473. doi: 10.1146/annurev-phyto-081211-173002. [DOI] [PubMed] [Google Scholar]
- Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nature immunology. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- Azevedo J, Garcia D, Pontier D, Ohnesorge S, Yu A, Garcia S, Braun L, Bergdoll M, Hakimi MA, Lagrange T. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes & development. 2010;24:904–915. doi: 10.1101/gad.1908710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. Comptes rendus de l'Académie des sciences. Série 3, Sciences de la vie. 1993;316:1194–1199. [Google Scholar]
- Boller T, Felix G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annual Review of Plant Biology. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- Bolton MD. Primary metabolism and plant defense-Fuel for the fire. Molecular Plant-Microbe Interactions. 2009;22:487–497. doi: 10.1094/MPMI-22-5-0487. [DOI] [PubMed] [Google Scholar]
- Bouché N, Bouchez D. Arabidopsis gene knockout: phenotypes wanted. Current opinion in plant biology. 2001;4:111–117. doi: 10.1016/s1369-5266(00)00145-x. [DOI] [PubMed] [Google Scholar]
- Briggs GC, Osmont KS, Shindo C, Sibout R, Hardtke CS. Unequal genetic redundancies in Arabidopsis–a neglected phenomenon? Trends in plant science. 2006;11:492–498. doi: 10.1016/j.tplants.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. The Plant Cell Online. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CW, Qu F, Ren T, Ye X, Morris TJ. RNA silencing-suppressor function of Turnip crinkle virus coat protein cannot be attributed to its interaction with the Arabidopsis protein TIP. Journal of general virology. 2004;85:3415–3420. doi: 10.1099/vir.0.80326-0. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step RNA isolation from cultured cells or tissues. Anal Biochem. 1987;162:156–259. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM. Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. The Plant Cell Online. 1991;3:61–72. doi: 10.1105/tpc.3.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant W, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- Fang Y, You J, Xie K, Xie W, Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Molecular Genetics and Genomics. 2008;280:547–563. doi: 10.1007/s00438-008-0386-6. [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. The Plant Journal. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Genes controlling expression of defense responses in Arabidopsis—2001 status. Current opinion in plant biology. 2001;4:301–308. doi: 10.1016/s1369-5266(00)00177-1. [DOI] [PubMed] [Google Scholar]
- Huang Z, Yeakley JM, Garcia EW, Holdridge JD, Fan JB, Whitham SA. Salicylic acid-dependent expression of host genes in compatible Arabidopsis-virus interactions. Plant physiology. 2005;137:1147–1159. doi: 10.1104/pp.104.056028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong RD, Chandra-Shekara A, Kachroo A, Klessig DF, Kachroo P. HRT- mediated hypersensitive response and resistance to Turnip crinkle virus in Arabidopsis does not require the function of TIP, the presumed guardee protein. Molecular Plant-Microbe Interactions. 2008;21:1316–1324. doi: 10.1094/MPMI-21-10-1316. [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Lommel S, McCain A, Morris T. Evaluation of indirect enzyme-linked immunosorbent assay for the detection of plant viruses. Phytopathology. 1982;72:1018–1022. [Google Scholar]
- Malamy J, Carr JP, Klessig DF, Raskin I. Salicylic Acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science (New York, N.Y. ) 1990;250:1002–1004. doi: 10.1126/science.250.4983.1002. [DOI] [PubMed] [Google Scholar]
- Morris K, Mackerness SAH, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V. Salicylic acid has a role in regulating gene expression during leaf senescence. The Plant Journal. 2001;23:677–685. doi: 10.1046/j.1365-313x.2000.00836.x. [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Brunner F, Kemmerling B, Piater L. Innate immunity in plants and animals: striking similarities and obvious differences. Immunological reviews. 2004;198:249–266. doi: 10.1111/j.0105-2896.2004.0119.x. [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends in plant science. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA research. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- Pickett FB, Meeks-Wagner DR. Seeing double: appreciating genetic redundancy. The Plant Cell. 1995;7:1347. doi: 10.1105/tpc.7.9.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Morris TJ. Cap-independent translational enhancement of turnip crinkle virus genomic and subgenomic RNAs. Journal of virology. 2000;74:1085–1093. doi: 10.1128/jvi.74.3.1085-1093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T, Qu F, Morris TJ. HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. The Plant Cell Online. 2000;12:1917–1925. doi: 10.1105/tpc.12.10.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T, Qu F, Morris TJ. The nuclear localization of the Arabidopsis transcription factor TIP is blocked by its interaction with the coat protein of Turnip crinkle virus. Virology. 2005;331:316–324. doi: 10.1016/j.virol.2004.10.039. [DOI] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes & development. 2002;16:1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD. Systemic acquired resistance. The plant cell. 1996;8:1809. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Rusu AG, Manners JM, Maclean DJ. The SEN1 gene of Arabidopsis is regulated by signals that link plant defence responses and senescence. Plant Physiology and Biochemistry. 2005;43:997–1005. doi: 10.1016/j.plaphy.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Schwessinger B, Ronald PC. Plant innate immunity: perception of conserved microbial signatures. Annual review of plant biology. 2012;63:451–482. doi: 10.1146/annurev-arplant-042811-105518. [DOI] [PubMed] [Google Scholar]
- Selth LA, Dogra SC, Rasheed MS, Healy H, Randles JW, Rezaian MA. A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication. The Plant Cell Online. 2005;17:311–325. doi: 10.1105/tpc.104.027235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P, Yuen G, Lehman C. Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant Journal. 1998;15:747–754. doi: 10.1046/j.1365-313x.1998.00265.x. [DOI] [PubMed] [Google Scholar]
- Sticher L, Mauch-Mani B, Métraux JP. Systemic acquired resistance. Annual review of phytopathology. 1997;35:235–270. doi: 10.1146/annurev.phyto.35.1.235. [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DMA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annual review of phytopathology. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- Wang X, Goregaoker SP, Culver JN. Interaction of the Tobacco mosaic virus replicase protein with a NAC domain transcription factor is associated with the suppression of systemic host defenses. Journal of virology. 2009;83:9720–9730. doi: 10.1128/JVI.00941-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia N, Zhang G, Liu XY, Deng L, Cai GL, Zhang Y, Wang XJ, Zhao J, Huang LL, Kang ZS. Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Molecular biology reports. 2010;37:3703–3712. doi: 10.1007/s11033-010-0023-4. [DOI] [PubMed] [Google Scholar]
- Yoshii M, Shimizu T, Yamazaki M, Higashi T, Miyao A, Hirochika H, Omura T. Disruption of a novel gene for a NAC-domain protein in rice confers resistance to Rice dwarf virus. The Plant Journal. 2009;57:615–625. doi: 10.1111/j.1365-313X.2008.03712.x. [DOI] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. The Plant Cell Online. 1998;10:1021–1030. doi: 10.1105/tpc.10.6.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. The Plant Journal. 2001;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]


