Abstract
The development of efficient and biocompatible non-viral vectors for gene therapy remains a great challenge, and exploiting the properties of both nanoparticle carriers and cationic polymers is an attractive approach. In this work, we have developed gold nanoparticle (AuNP) polyamidoamine (PAMAM) conjugates for use as non-viral transfection agents. AuPAMAM conjugates were prepared by crosslinking PAMAM dendrimers to carboxylic-terminated AuNPs via EDC and sulfo-NHS chemistry. EDC and sulfo-NHS have been utilized widely and in numerous applications such as amino acid coupling; however, their use in the coupling of PAMAM dendrimers to AuNPs presents new challenges to form effective and stable constructs for delivery that have not yet been examined. Enhanced colloidal stability and DNA condensation ability was established by probing two critical synthetic parameters: the reaction rate of the PAMAM crosslinking step, and the amine to carboxyl ratio. Based on this work, increasing the amine to carboxyl ratio during conjugation of PAMAM onto AuNPs yielded the optimal vector with respect to colloidal stability and transfection efficiency in vitro. AuPAMAM conjugates present attractive candidates for non-viral gene delivery due to their commercial availability, ease of fabrication and scale-up, high yield, high transfection efficiency and low cytotoxicity.
Keywords: Gold, Nanoparticle, Nanocomposite, Gene therapy, Dendrimer, DNA
1. Introduction
An improved understanding of the genes involved in the development of cancer and other diseases in recent years has led to the expansion of gene therapy approaches [1]. Although gene therapy represents an attractive alternative for cancer therapy, the development of effective and safe delivery platforms is the limiting step in the implementation of therapeutic plasmids in the clinic [2]. All vectors must overcome multiple cellular hurdles for effective transfection including cellular uptake, endosomal escape, cytoplasmic trafficking, DNA release, and nuclear uptake [3]. While viral vectors have traditionally been effective DNA delivery vehicles, they present issues of immunogenicity, oncogenicity, small DNA load, and expensive and complex production procedures [1,4]. As such, recent research has focused on the development of non-viral vectors for gene delivery [1,5].
Cationic polymers are one of the main classes of non-viral vectors, and they boast benefits such as low immunogenicity and ease of chemical modification [6]. Poly(ethyleneimine) (PEI) is among the most common and widely studied cationic polymers and has been used as a gold standard for in vitro gene delivery experiments. However, due to its highly cationic nature and non-biodegradable structure, PEI's toxicity has limited its in vivo applications [1,7]. Dendrimers are a commercially available class of cationic, branched polymers that are increasingly being investigated for delivery applications [8–11]. The degree of branching in a dendrimer is referred to as its ‘generation’, with size increasing linearly and terminal groups increasing exponentially as a function of the generation. Polyamidoamine (PAMAM) dendrimers are among the most commonly studied class of dendrimers due to their high transfection efficiency in vitro with generations 6 or 7 [4]. However, transfection at high generations (>6) results in cytotoxic effects due to their highly cationic nature [1].
Gold nanoparticles (AuNPs) provide particularly attractive scaffolds for the development of non-viral vectors due to their size tunability, ease of functionalization, and biocompatibility [12–14]. Several groups have used gold nanoparticles for not only in vitro oligonucleotide transfection but also in vivo oligonucleotide delivery for gene regulation [14–20]. Recently, groups have begun to exploit the properties of both nanoparticles and dendrimers for applications in gene delivery. For example, Chen et al. combined polypropylenimine (PPI) dendrimers with AuNPs using simple electrostatic attraction to form efficient transfection vectors [21]. An additional benefit of this approach is that combining nanoparticles with dendrimers has the potential to alleviate issues of dendrimer cytotoxicity [19,22–24]. However, electrostatic binding of dendrimers and AuNPs may not be strong enough to withstand changes in pH, temperature and ionic strength in the biological environment and may decompose prior to reaching the cells of interest and delivering the DNA cargo [25].
Therefore, we designed a facile bottom-up covalent conjugation method to generate PAMAM conjugated gold nanoparticles (AuPAMAM). In short, lower generation PAMAM dendrimers (<6) were conjugated to 5 nm AuNPs with a self-assembled monolayer (SAM) of 11-mercaptoundecanoic acid (MUA) using crosslinking agents 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and N-hydroxy sulfosuccinimide (sulfo-NHS) in 2-(N-morpholino)ethanesulfonic acid (MES) buffer. EDC, a carbodiimide, catalyzes the formation of amide bonds between the MUA carboxyl and PAMAM amine groups, while sulfo-NHS stabilizes the coupling reaction via formation of amine reactive esters on the carboxylate. EDC and sulfo-NHS coupling to bind amine-containing molecules onto carboxylated gold nanoparticles has been used in several occasions [26–29]. Although preparation of AuPAMAM particles is not chemically tedious and can be fabricated and washed in less than 6 h, the use of EDC and sulfo-NHS in the coupling of PAMAM to carboxylic-terminated AuNPs presents new challenges to form effective and stable vectors. In this study, the reaction rate of the EDC/sulfo-NHS conjugation and the PAMAM/MUA-AuNP amine to carboxyl ratio were altered to elucidate their effect on AuPAMAM colloidal stability and transfection efficacy. Finally, the AuPAMAM particles synthesized by two different schemes were evaluated in a human breast adenocarcinoma (SkBr3) cell line for their potential as gene therapy vectors.
2. Materials and methods
2.1. Materials
All chemicals were purchased from Sigma Aldrich (St. Lewis, MO) or Fisher Scientific (Waltham, MA) unless otherwise stated. 1-Ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) and N-hydroxy sulfosuccinimide (Sulfo-NHS) were purchased from Thermo Scientific (Waltham, MA). 5 nm citrate stabilized colloidal gold nanoparticles (AuNPs) were purchased from Ted Pella (Redding, CA). AlamarBlue was purchased from Invitrogen (Carlsbad, CA). Plasmid DNA with cytomegalovirus (CMV) promoter and enhanced green fluorescent protein (eGFP) as the reporter gene (pCMV-eGFP, 4.7 kb) were obtained from Clark Needham at Rice University [30]. SK-BR-3 cells, cell culture medium and phosphate buffered saline (PBS) were purchased from ATCC (Manassas, VA).
2.2. AuPAMAM synthesis
11-mercaptoundecanoic acid (MUA) were added to 5 nm gold nanoparticles (AuNPs) (5 × 1013 particles/ml) to a final concentration of 83.33μM in 12 ml. After 24 h, the solution was raised to 0.1 M NaCl, 100 mM sodium phosphate, and 0.1% v/v Tween 20 and incubated for another 24 h. Next, excessive MUA was removed by using a centrifuge filter (10,000 molecular weight cutoff) at 2500 g for 20 min and washed 3 × with PBS. Total organic carbon (TOC) analysis of the supernatant was conducted to verify that 3 centrifuge washes was sufficient to remove the unbound MUA from the MUA-AuNPs (Fig. S1). After the last centrifugation, the MUA-AuNPs were resuspended in pH 6 MES buffer. EDC and sulfo-NHS linkers were added to a final concentration of 0.44 mM and 0.59 mM for 15 min. Then, the particles were added to 4 ml of ethylene diamine core PAMAM dendrimers (generation 0–5) in PBS (Table S1). After 1 h, 2 ml of 50 mM hydroxylamine (pH 7) in PBS was added to the solution for another hour to backfill any unbound sulfo-NHS esters. Last, the solution was washed 3 × using a centrifuge filter (50,000 MWCO) with sterile DNase free deionized water (Fig. S1). The AuPAMAM was resuspended in DI water and stored at 4 °C until further use. To estimate the amount of dendrimer needed for the conjugation, a surface packing model was used. A 10× excess concentration of the maximal dendrimer binding number was used for the conjugation process. The following alterations were made for the pH 4.7 AuPAMAM synthesis protocol: the MUA-AuNPs were resuspended in pH 4.5–4.7 MES buffer, the PAMAM conjugation time was extended to 2 h, and the hydroxylamine backfilling duration was extended to 24 h. The following alterations were made for the sMUA AuPAMAM synthesis protocol: various ratios of MUA and MUOH were added during the MUA-AuNP self-assembling process, and the sMUA-AuNPs were resuspended in pH 6 MES buffer.
2.3. Transmission electron microscopy
For characterization of the AuPAMAM/DNA complexes, samples were dispersed in ultrapure water and deposited onto a 300-mesh carbon copper grid. After the excess water had evaporated at room temperature, 1 drop of 3% uranyl acetate was added and filter paper was used to wick away excess moisture. The copper grid was air dried overnight and observed by TEM (JEOL 2100 Field Emission Gun Transmission Electron Microscope) at 120 kV accelerating voltage.
2.4. Gel electropheresis
AuPAMAM/DNA complexes were freshly prepared in water by mixing equal volumes of GFP plasmid (4 nM) and AuPAMAM in DI water at molar ratios (MR) of 1:5, 1:15, 1:30, 1:50, 1:100, and 1:250. Following incubation for 20 min at room temperature, the complexes were loaded into wells in a 1% agarose gel. Gel electrophoresis was carried out in 1 × TBE buffer at 80 V. The gels were visualized with a Bio-Rad UV transilluminator.
2.5. Cell culture
SK-BR3 cells were cultured in a humidified incubator (5% CO2, 37 °C). The cells were suspended in McCoy's 5A medium and supplemented with 10% Fetal Bovine Serum (FBS) and 1% Penicillin-Streptomycin, with the exception of transfection experiments where cells were grown in complete media without antibiotics.
2.6. Cell transfection experiments
For transfection assays, 75,000 cells/well was added to 24 well plates and grown overnight. Complexes were first prepared at room temperature as follows: 50 μl solutions of AuPAMAM at various concentrations and GFP plasmid (0.8 ug) were prepared in DI water separately. Water was used as the solvent in order to prevent charge screening effects prior to complex formation. The vectors and plasmids were mixed and incubated for 20 min. The resulting complexes were added directly into wells. Six hours later, wells were rinsed with PBS and complete media was added and 48 h later the medium was changed. At 48 h, GFP expression of all conditions was visualized using a Zeiss Axio Observer inverted microscope. Transfection efficacy was measured using flow cytometry (BD FACSCanto II).
2.7. Reporter gene expression
GFP activity was observed visually using Zeiss Axio Observer inverted fluorescence microscope. Flow cytometry of fixed cells was used to quantify the amount of expressed protein on a BD Canto FACS machine at the end of the transfection experiment.
2.8. AlamarBlue assay
All cytotoxicity experiments were conducted in clear bottom black 96 well plates. For toxicity assays, 15,000 cells/well were added to 96 well plates and grown overnight. Complexes were first prepared at room temperature as follows: 10 μl solutions of AuPAMAM at various concentrations and plasmid (0.16 μg) were prepared in water separately. The vectors and plasmids were mixed and incubated for 20 min. The resulting complexes were added directly into wells. Six hours later, wells were rinsed with PBS and complete media was added, and 48 h later the medium was changed. At 48 h, cells were treated with 20 μl of alamarBlue and after 2 h, fluorescence was measured on a Fluorolog-3 micromax plate reader (emission at 585 nm, excitation at 570 nm).
2.9. Statistical analysis
All data are expressed as mean ± standard error of the mean. Statistical differences were evaluated using ANOVA and Tukey's HSD and considered significant at p < 0.05. All figures shown were obtained from three independent experiments. Any images shown are representative of the entire experiment.
3. Results & discussion
3.1. AuPAMAM synthesis and characterization
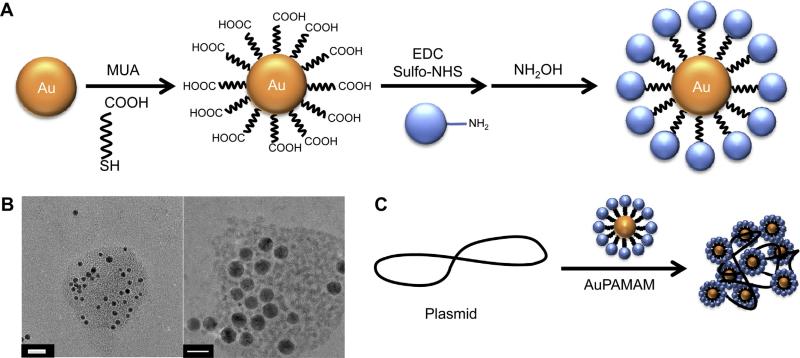
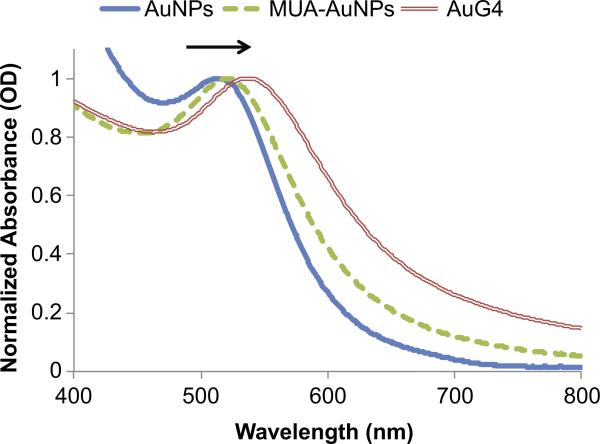
The AuPAMAM synthesis process (Fig. 1) was modified from the original EDC/sulfo-NHS coupling protocol described by Grabarek and Gergely (1990) [31]. Briefly, MUA was self-assembled onto 5 nm citrate stabilized AuNPs. EDC and sulfo-NHS were then added to the particles in MES buffer at pH 6 [32]. After 15 min, PAMAM dendrimers (G0-G5) were added to the solution and incubated for 1 h. Then, the reaction was quenched by the addition of hydroxylamine for 1 h. The particles were collected via several centrifuge steps and washed with sterile water. Successful conjugation of PAMAM onto MUA-AuNPs was confirmed by measuring the absorbance spectra and observing the plasmon peak position. As the refractive index of the immediate environment surrounding the AuNP changes, the peak absorbance of the complexes red shifts towards longer wavelengths (Fig. 2). When MUA was conjugated to the surface of the AuNP, a SAM was formed on the surface and a 3 nm peak shift from 517 to 520 nm was observed. Following conjugation of PAMAM to the surface of the MUA-AuNPs, another peak shift towards longer wavelengths was observed. AuPAMAM generation 0 (AuG0) to AuG5 shifted an average of 16.5 nm compared to MUA-AuNPs (Fig. S2). The observed peak shift and broadening can be attributed to the change in the local and chemical interface dampening [33], and verified that the pH 6 MES buffer based EDC/sulfo-NHS coupling method successfully formed AuPAMAM conjugates on the surface of the AuNP.
Fig. 1.
(A) Schematic illustration showing AuPAMAM NPs synthesized using the original pH 6 method. (B) TEM micrographs of generation 4 AuPAMAM (AuG4) complexed with GFP plasmid at a 1:30 MR. DNA was stained with Uranyl Acetate. Scale bar represents 20 nm. (C) Schematic illustration showing the complexation of AuPAMAM NPs with plasmid DNA.
Fig. 2.
UV–vis spectra obtained for AuG4 synthesized by the pH 6 method showing red shift (arrow) after formation of an 11-MUA SAM on the AuNP and after conjugation of PAMAM. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
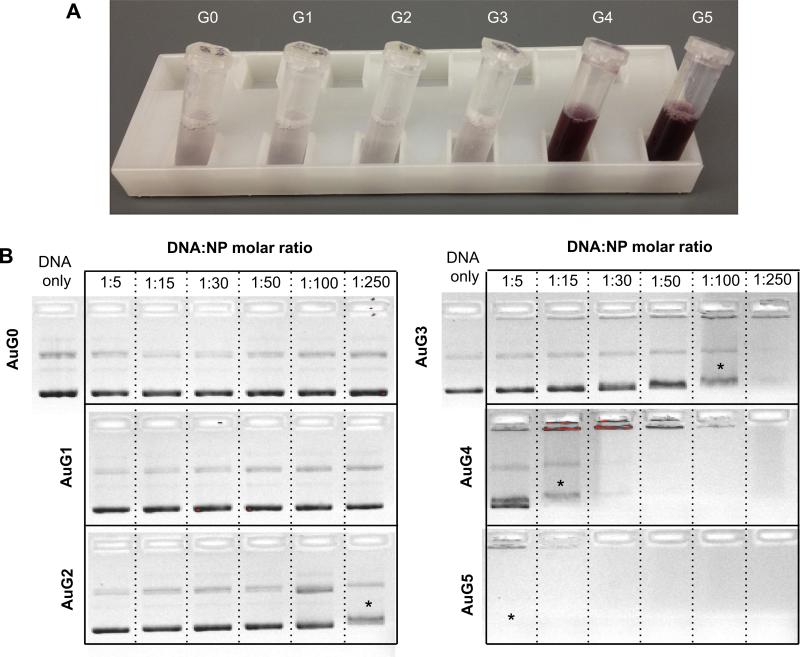
Although the conjugation process was the same for all PAMAM generations, agglomeration of AuPAMAM occurred with AuG0 to AuG3 but not for AuG4 and AuG5 (Fig. 3a). This phenomenon was likely influenced by the generation-dependent flexibility of the PAMAM dendrimer [10,34]. PAMAM dendrimers of generation 3 and below are flexible and possess an extended conformation in contrast to higher generations, which are slightly rigid and possess a spherical structure due to steric hindrance. However, the flexibility of low generation dendrimers alone is not sufficient to explain the dramatic differences in agglomeration seen in AuG3 and AuG4. Another factor is the EDC/sulfo-NHS conjugation process. When the terminal amine of a dendrimer couples with a MUA molecule on the AuNP, its neighboring amines can compete with unbound dendrimers for binding to adjacent carboxyl groups. If the dendrimer is flexible (G0–G3), these amines possess a proximity advantage to bind to the carboxyls, which not only lowers the number of dendrimers bound per AuNP, but can also decrease the total charge of the AuPAMAM complexes by reducing the number of primary amines. The decrease in primary amines and overall charge of the AuPAMAM thus causes the complexes to agglomerate. Conversely, since G4 and G5 PAMAM are stiffer, a bound PAMAM dendrimer is less likely to bind multiple carboxyls, and therefore the complexes maintain their positive charge and do not agglomerate.
Fig. 3.
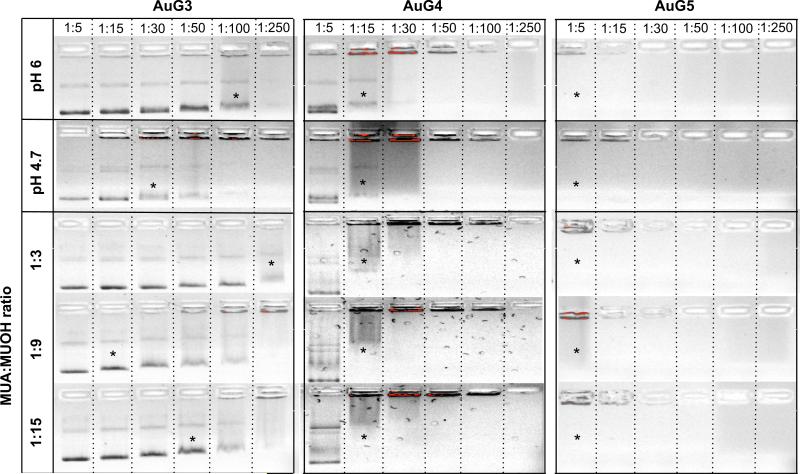
(A) Photograph of AuPAMAM synthesized by the pH 6 method at the end of the synthesis without sonication. (B) Agarose gel electrophoresis of pH 6 AuPAMAM complexed with GFP plasmid at various molar ratios (MR). Asterisks represent first indication of band smearing.
Changes in charge and stability can alter the ability of AuPAMAM constructs to condense DNA, a critical criteria for effective plasmid delivery. To evaluate the capability of the AuPAMAM particles to condense DNA, a green fluorescent protein (GFP) plasmid was used. Transmission electron microscopy (TEM) was used to visualize the AuPAMAM/DNA complexes and the stained plasmid formed a complex with numerous AuPAMAM particles (Fig. 1b). Dynamic light scattering (DLS) analysis was not successful due to the complicated nature of these complexes. The GFP plasmids were complexed with the AuPAMAM constructs at varying molar ratios (1:5–1:250 DNA:AuPAMAM) and the ability of the AuPAMAM particles to condense DNA was evaluated by gel electrophoresis (Fig. 3b). AuG0 and AuG1 particles were not able to successfully retain the plasmid in the well chamber even at high nanoparticle concentrations. However, they did bind to DNA loosely, evidenced by visible faint red particle smears as the AuPAMAM complexes were dragged through the gel (data not shown). AuG3 constructs exhibited smearing of the plasmid band at 1:100 M ratio (MR) while 1:250 resulted in complete retention of the complexes in the well. In comparison, AuG4 and AuG5 were able to condense DNA at much lower ratios (1:15 and 1:5, respectively). These results indicate that the AuPAMAM particle stability and DNA condensation ability improved with higher generations.
3.2. Optimization of colloidal stability: decreased reaction rate and increased amine to carboxyl ratio
To further evaluate the aforementioned agglomeration issues, two variables were examined: the reaction rate of the PAMAM conjugation and the carboxyl to amine ratio. As mentioned previously, the rapid conjugation of amines to carboxyls may favor the binding of proximal amines from already bound dendrimers. Therefore in the first approach, the conjugation rate was decreased so that free dendrimers would have more time to diffuse toward the activated carboxyls and compete with bound dendrimers. Activation of carboxyls occurs during a 15 min incubation of EDC and sulfo NHS with the MUA-AuNPs. During this incubation, carboxyl groups are converted to sulfo-NHS ester groups. This process is preferred at lower pH because the NHS groups are more stable at lower pH [31]. At higher pH, the NHS ester groups are more reactive but also more likely to be hydrolyzed by water. Thus, to slow the reaction rate, pH 4.5–4.7 MES buffer replaced pH 6 MES buffer in the conjugation process [31]. The conjugation time was increased to reflect the decreased reaction rate.
Another approach was pursued to investigate the agglomeration of lower generation AuPAMAM particles. In this second approach, the number of carboxyls on the AuNP was decreased, subsequently increasing the amine to carboxyl ratio and reducing the probability of the same dendrimer binding to multiple carboxyl groups. The foot space of a thiol molecule is approximately 0.35 nm2; therefore, a maximum of 224 carboxyls are estimated to be present on the AuNP surface without taking steric hindrance into account [35]. To approximate the amine to carboxyl ratio, we developed a model to estimate the maximum coverage of PAMAM on MUA-AuNPs (Fig. S3). In this model, the cross sectional area of a PAMAM dendrimer is projected onto the surface area of the MUA-AuNPs and the number of bound PAMAM dendrimers is determined by two-dimensional hexagonal packing of circles. The radius (R) of the binding area for PAMAM was dependent on the AuNP diameter (5 nm) and the height of MUA, which was estimated to be 1.5 nm [36]. Therefore, the total PAMAM binding surface area was calculated to be 201 nm2. Assuming the dendrimers are spheres, the effective area for maximal hexagonal packaging was approximately 91% of the overall binding area [37]. The ratio of the effective binding area and the cross-sectional area of the dendrimers results in a maximum number of dendrimers per MUA-AuNP, which ranged from 22 to 146 depending on the generation (Table 1). The amine to carboxyl ratio was greater than 1 for all generations, suggesting that all MUA can be occupied by amines from the PAMAM dendrimers, neglecting steric effects. The amine to carboxyl ratio increased with the PAMAM generation, as did the stability of the AuPAMAM constructs. Therefore, to increase amine to carboxyl ratios, 11-mercaptoundecanol (MUOH) molecules were used to space out the MUA on the AuNP surface. The spacing of the carboxyl groups created a more favorable environment for free dendrimers to bind and decreased the probability of multiple binding events by one dendrimer. Various MUA to MUOH molar ratios were used to test the effects of spacing. For the spaced MUA (sMUA) constructs, pH 6 MES buffer and the original conjugation scheme were used in order to isolate the effects of this method. It is worth noting that the limitation of using the MUOH spacer is that it decreases the amount of secondary amines from hydroxylamine, which are also involved in DNA binding and condensation [38].
Table 1.
Estimated density of AuNP surface modifications calculated by the maximum hexagonal packing model.
| Generation | PAMAM/MUA-AuNP | Amines/MUA-AuNPs | Amine to carboxyl ratio |
|---|---|---|---|
| 0 | 146 | 584 | 2.61 |
| 1 | 78 | 626 | 2.79 |
| 2 | 51 | 823 | 3.67 |
| 3 | 38 | 1209 | 5.40 |
| 4 | 28 | 1798 | 8.02 |
| 5 | 22 | 2869 | 12.81 |
3.3. Characterization of pH 4.7 based AuPAMAM constructs
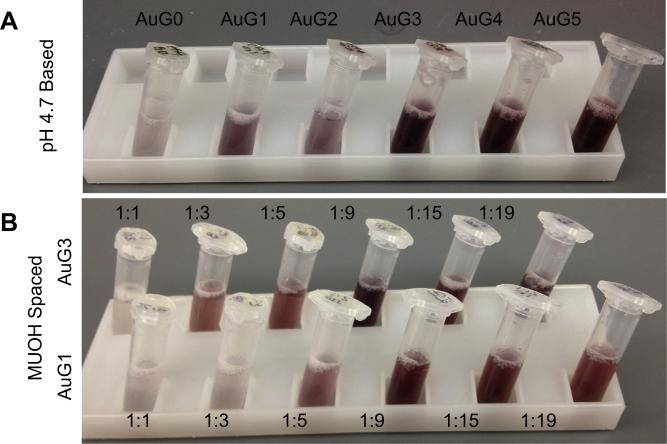
As with the pH 6 based AuPAMAM constructs, successful conjugation using the pH 4.7 and sMUA methods were evaluated for peak shifts in their absorbance spectra. The pH 4.7 AuG0–AuG5 particles had an average red shift of 10.5 nm, verifying the successful conjugation of PAMAM (Fig. S4a). Most importantly, the stability of the pH 4.7 particles improved for lower generations compared to the original pH 6 method (Fig. 4a). As shown in Fig. 4a, the pH 4.7 AuG3 particles were comparable to the pH 6 AuG4 and AuG5 particles (Fig. 3a) in terms of colloidal stability. The higher generation pH 4.7 AuPAMAM particles (AuG4 and AuG5) did not exhibit differences compared to the pH 6 method. These results support the theory that slowing the reaction rate of the conjugation helps improve particle stability.
Fig. 4.
Photographs of AuPAMAM aqueous solutions following synthesis and without sonication. (A) AuPAMAM synthesized by the pH 4.7 method. (B) AuG1 and AuG3 synthesized by the sMUA method. Ratios represent the molar ratio between 11-MUA and MUOH.
In addition, pH 4.7 based AuPAMAM constructs outperformed their pH 6 counterparts in condensing DNA. While AuG0 particles synthesized by both methods still did not condense DNA, pH 4.7 AuG1 showed a less intense band at a 1:250 MR, and pH 4.7 AuG2 exhibited band smearing at a 1:100 MR (Fig. S5). Furthermore, the pH 4.7 AuG3 constructs were able to condense DNA at a 1:30 ratio in comparison to 1:100 for the pH 6 method (Fig. 5). The pH 4.7 method did not visibly alter the condensability of the AuG4 and AuG5 constructs, which paralleled the colloidal stability results.
Fig. 5.
Agarose gel electrophoresis assay comparing DNA condensation by the pH 6, pH 4.7 and sMUA synthesis methods for AuG3 to AuG5. Asterisks represent first indication of band smearing. Top bar shows DNA:AuPAMAM MR.
3.4. Characterization of sMUA AuPAMAM constructs
sMUA AuPAMAM constructs also showed improved stability compared to the pH 6 synthesis method. For initial testing, AuG1 and AuG3 were synthesized with MUA:MUOH molar ratios varying from 1:1–1:19 (Fig. 4b). Increasing the amine to carboxyl ratio with MUOH spacers decreased the agglomeration of the AuPAMAM complexes. As expected, a higher MUOH ratio was needed for AuG1 (1:5) for improved stability in contrast to AuG3 (1:3), which has a higher amine to carboxyl ratio than AuG1 (Table 1). The conjugation yield of 1:19 sMUA AuPAMAM was significantly less compared to the yield of lower MUA:MUOH ratios. Thus, ratios of 1:3, 1:9 and 1:15 were chosen for further optimization experiments. The absorbance peak of 1:3, 1:9, and 1:15 sMUA AuPAMAM constructs shifted an average of 14.8, 5.2, and 5.7 nm respectively (Fig. S4b), suggesting successful conjugation of the PAMAM onto the sMUA AuNPs. The improved stability of the sMUA AuPAMAM supported the theory that the amine to carboxyl ratio influenced colloidal stability for the lower generation dendrimers.
Similar to the pH 4.7 AuPAMAM constructs, the sMUA AuPAMAM constructs condensed DNA more efficiently than the original pH 6 AuPAMAM constructs. Again, while there was not significant DNA band retention for the lower generations (AuG0 and AuG1), the DNA bands appeared more faint as the DNA:AuPAMAM MR increased (Fig. S5). However, the gel electrophoresis results for sMUA AuG3 showed visible improvement compared to the pH 6 AuG3, particularly at a 1:9 MUA:MUOH MR (Fig. 5). Analogous to the visual stability results, sMUA AuG4 and AuG5 performed similarly to pH 4.7 and pH 6 AuG4 and AuG5.
These results revealed a correlation between particle stability and DNA condensation. The decreased DNA condensation ability observed by the less stable constructs may be due to decreased electrostatic interaction between AuPAMAM particles and DNA as a result of AuPAMAM agglomeration. Furthermore, the less cationic AuPAMAM agglomerates may result in the formation of larger DNA/ AuPAMAM complexes that are not readily endocytosed. AuG3 constructs showed the largest variability in particle stability and DNA condensation, as expected based on the structural differences between G3 and G4 PAMAM. Therefore, the improvements from slowing the reaction rate and decreasing the amine to carboxyl ratio supported our hypothesis that the flexibility of PAMAM was a major factor in the stability and effectiveness of AuPAMAM constructs as gene delivery vectors.
3.5. Transfection efficacy of pH 4.7 and sMUA AuPAMAM constructs
Following characterization of DNA condensation by the pH 4.7 and sMUA AuPAMAM constructs, the transfection efficacy was examined by conducting in vitro transfection studies using a green fluorescent protein (GFP) plasmid and human breast adenocarcinoma cells (SkBr3). The vectors were tested at various DNA to AuPAMAM MR ranging from 1:5 to 1:50, and transfection efficiency was investigated by fluorescence microscopy and flow cytometry via GFP expression after 48 h. All experiments were performed in triplicate and in parallel with a negative control and a PEI control at N:P = 7.5, where the N:P ratio is moles of polymer amine groups to moles of DNA phosphate groups. It is worth noting that all cellular experiments were conducted in the presence of serum.
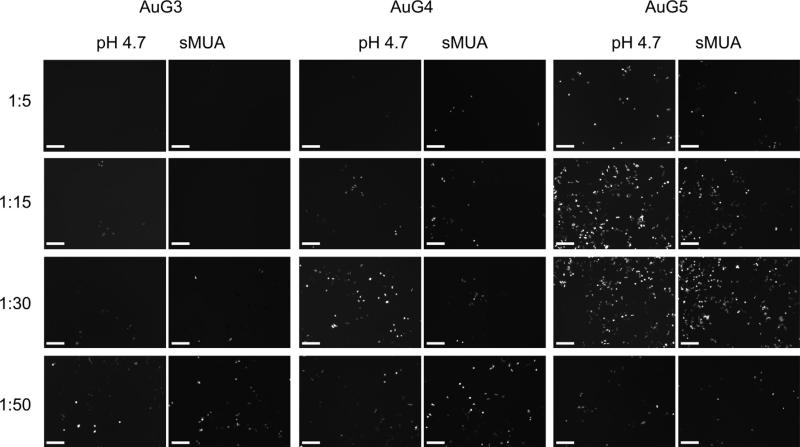
In Fig. 6, fluorescent microscope images are shown for AuG3 to AuG5 at the end of the 48-h transfection experiment. As anticipated from the agarose gel electrophoresis assay results, AuG0, AuG1 and AuG2 did not transfect cells appreciably (data not shown). However, at AuG3 and above, GFP expressing cells were visualized. Qualitatively, AuG4 and AuG5 constructs synthesized by both methods appear the most efficient. Next, we quantified transfection efficiency by flow cytometry analysis of GFP expressing cells.
Fig. 6.
Fluorescence microscopic images (10 ×) of GFP gene expression in SkBr3 cells using AuG3 to AuG5 synthesized by the pH 4.7 and sMUA methods. Left column represents DNA:AuPAMAM MR. Images were taken 48 h after transfection. Scale bars represent 200 microns.
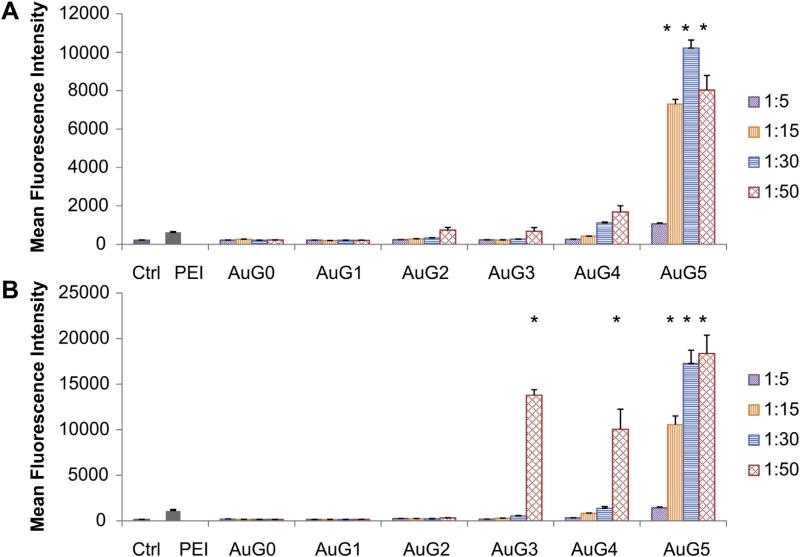
In Fig. 7, the average mean fluorescence intensity (MFI) of each tested condition is shown. The most efficient vector was AuG5 in both the pH 4.7 and sMUA constructs, with the sMUA vectors exhibiting larger MFI values than the pH 4.7 vectors. Of the pH 4.7 constructs, AuG5 1:15 to 1:50 were significantly better than PEI, and AuG4 1:50 had a significantly higher MFI than the untreated control but was comparable to PEI. Among the sMUA constructs, AuG3 1:50, AuG4 1:50, and AuG5 1:15 to 1:50 were all significantly better transfection agents than PEI.
Fig. 7.
Transfection efficiency of (A) pH 4.7 and (B) sMUA AuPAMAM at different DNA:AuPAMAM MRs. Asterisks represent significance from both the control and PEI (p < 0.05).
These results mirror those illustrated by fluorescence microscopy (Fig. 6).
Based on the imaging and flow transfection results, the gene transfection efficiency is dependent on both the generation of the PAMAM dendrimer and the selected DNA:AuPAMAM MR. Generation 3 to 5 AuPAMAM particles were the most efficient at transfecting the cells. Further studies are necessary to determine the mechanism for improved transfection by the sMUA AuPAMAM constructs. Next, in order to isolate the optimal AuPAMAM synthesis method biocompatibility was assessed.
3.6. Viability of pH 4.7 and sMUA AuPAMAM constructs
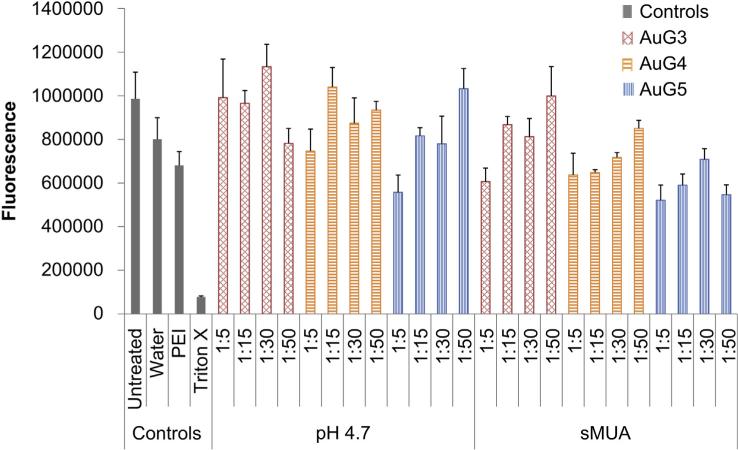
We assessed cellular metabolic activity as an indicator of cell health using the alamarBlue assay to evaluate toxicity that may arise from AuPAMAM nanoparticles during transfection. For these experiments, the pH 4.7 and sMUA AuPAMAM vectors complexed with DNA were evaluated in comparison with a control and PEI/ DNA complexes. AlamarBlue assays showed some decrease in viability by PEI which is in agreement with other published results at the chosen N:P of 7.5 [39], and minimal cell death by the AuPAMAM/DNA complexes in SkBr3 cells (Fig. 8). In general, increasing PAMAM generation was associated with more cell death. However, the majority of complexes were not significantly cytotoxic compared to the controls. Of the pH 4.7 constructs, only AuG5 at a 1:5 MR was significantly less viable from the controls. Similarly, the only statistically less viable sMUA construct was AuG5 at a 1:5, 1:15 and 1:50 MR. These results suggest that AuPAMAM nanoparticles are able to maintain satisfactory cell viabilities, and in the case of AuG5 further optimization is needed to yield improved biocompatibility (Fig. 9).
Fig. 8.
AlamarBlue metabolic activity assay of SkBr3 cells treated with pH 4.7 and sMUA AuPAMAM particles complexed with DNA at various MRs.
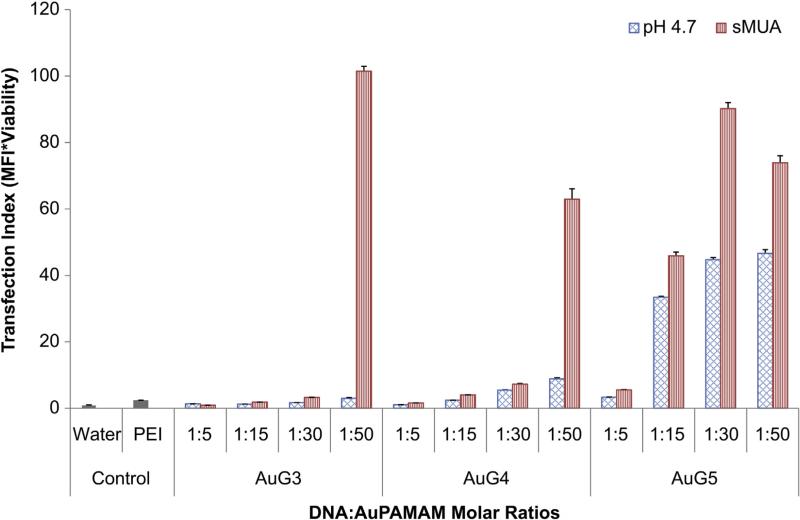
Fig. 9.
Transfection Index (TI) of pH 4.7 and sMUA AuPAMAM measures the product of gene expression and viability normalized to the control.
3.7. Transfection index
The ratio of transfection efficiency and cell viability was used as an indicator of overall vector performance by calculating the product of the normalized MFI and viability. This ratio is referred to as the Transfection Index (TI), and will be larger for more biocompatible and efficient vectors [40]. Thus, the TI was used to determine the optimal synthetic method that combines efficient transfection with low cytotoxicity. The control had a TI of 1 as all data was normalized to it. PEI had a TI of 2.4.
Of the pH 4.7 vectors, all ratios of AuG0 through AuG2 were not significantly better than PEI or the control (data not shown). However, at a 1:50 MR, AuG4 (TI: 8.9) was significantly more efficient than both control and PEI. For AuG5, a complex ratio as low as 1:5 (TI: 3.3) was able to surpass the control and match PEI efficacy while a MR of 1:15, 1:30 and 1:50 largely surpassed PEI (TI: 33.4, 44.7, 46.6, respectively).
Of the sMUA complexes, all ratios of AuG0 through AuG2 were no different than PEI or the control as was the case with the pH 4.7 AuPAMAM constructs (data not shown). However, unlike the pH 4.7 constructs, AuG3 at a 1:50 MR resulted in the highest transfection index of all the conditions, surpassing both the control and PEI (TI: 101.5). This difference was also seen in the DNA condensation results (Fig. 5). Similarly, sMUA AuG4 was statistically comparable to PEI at 1:15 and 1:30 MRs (TI: 4.0, 7.2, respectively) and superior to PEI at a 1:50 MR (TI: 62.9). Finally, sMUA AuG5 surpassed PEI at 1:15, 1:30 and 1:50 MRs (TI: 45.9, 90.1, 73.9).
For both the pH 4.7 and sMUA methods, the TI results are consistent with the electrophoresis data. AuG0 to AuG2 did not condense DNA and did not differ significantly from the control transfection index, while AuG3, which had better DNA condensation results, showed improved TI. AuG4 and AuG5 showed the best colloidal stability as well as DNA condensation capabilities, which translated into improved transfection index. Comparing the two synthesis routes, it is clear that the sMUA method yielded more optimal delivery vectors than the pH 4.7 method. Thus, future work in optimization of synthesis parameters will be carried out with the sMUA constructs for AuG3 to AuG5.
4. Conclusion
In the present study, a new class of non-viral AuNP-based gene delivery vectors was synthesized and characterized in vitro. Generation dependent AuPAMAM colloidal stability was investigated and improved by altering the dynamics of AuNP-PAMAM conjugation via the EDC/sulfo-NHS crosslinking reaction. We also show that the colloidal stability is an important factor for DNA condensation ability. Furthermore, the reported approach of functionalizing AuNPs using a mixed 11-MUA/MUOH SAM and EDC/sulfo-NHS dendrimer chemistry may be applicable for various NPs and ligands, thereby providing a general strategy to fabricating diverse NPs for a range of biological and therapeutic applications. Efficient transfection by non-viral vectors is achieved as a balance between low toxicity and sufficient DNA transfection. The sMUA AuPAMAM constructs showed the highest stability, gene transfection efficacy and a reasonable cytotoxicity profile. With further optimization of synthesis parameters to improve the overall transfection index, sMUA AuPAMAM particles may be used as a new family of non-viral gene delivery vectors.
Supplementary Material
Acknowledgments
This work was supported by a pre-doctoral fellowship from the Ford Foundation, a training fellowship from the Keck Center of the Gulf Coast Consortia, on the Nanobiology Interdisciplinary Graduate Training Program, National Institute of Biomedical Imaging and Bioengineering (NIBIB) (T32EB009379, contact PI Rebekah Drezek), the Medical Scientist Training Program at Baylor College of Medicine, the Edward and Josephine Hudson Scholarship, the Ruth L. Kirschstein National Research Service Awards for Individual Predoctoral MD/PhD Fellows (F30CA165686-01A1) by the National Institute of Health (NIH) and National Cancer Institute (NCI), the Welch Foundation (C-1598), and the NIH (R01 CA172836). We would like to thank Clark Needham and Ryan Schweller for donating plasmids and for their advice, Veronica Leautaud and the Richards-Kortum Lab for use of their UV transilluminator, and Denise Benoit and the Colvin Lab for training and for use of their TOC instrument. The authors confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found online at http://dx.doi.org/10.1016/j.biomaterials.2013.11.026.
References
- 1.Gascón AR, del Pozo-Rodríguez A, Solinís MÁ. Non-viral delivery systems in gene therapy. In: Martin F, editor. Gene ther.–tools potential appl. InTech; 2013. pp. 3–34. [Google Scholar]
- 2.Klinman DM, Takeno M, Ichino M, Gu M, Yamshchikov G, Mor G, et al. DNA vaccines: safety and efficacy issues. Springer Semin Immunopathol. 1997;19:245–56. doi: 10.1007/BF00870272. [DOI] [PubMed] [Google Scholar]
- 3.Van Gaal EVB, van Eijk R, Oosting RS, Kok RJ, Hennink WE, Crommelin DJA, et al. How to screen non-viral gene delivery systems in vitro? J Control Release. 2011;154:218–32. doi: 10.1016/j.jconrel.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Hahn P, Scanlan E. Gene delivery into mammalian cells: an overview on existing approaches employed in vitro and in vivo. Top Curr Chem. 2010;296:1–13. doi: 10.1007/128_2010_71. [DOI] [PubMed] [Google Scholar]
- 5.De Laporte L, Cruz Rea J, Shea LD. Design of modular non-viral gene therapy vectors. Biomaterials. 2006;27:947–54. doi: 10.1016/j.biomaterials.2005.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morille M, Passirani C, Vonarbourg A, Clavreul A, Benoit J-P. Progress in developing cationic vectors for non-viral systemic gene therapy against cancer. Biomaterials. 2008;29:3477–96. doi: 10.1016/j.biomaterials.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 7.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11:990–5. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Biricova V, Laznickova A. Dendrimers: analytical characterization and applications. Bioorg Chem. 2009;37:185–92. doi: 10.1016/j.bioorg.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Tomalia DA, Reyna LA, Svenson S. Dendrimers as multi-purpose nanodevices for oncology drug delivery and diagnostic imaging. Biochem Soc Trans. 2007;35:61–7. doi: 10.1042/BST0350061. [DOI] [PubMed] [Google Scholar]
- 10.Svenson S, Tomalia DA. Dendrimers in biomedical applications—reflections on the field. Adv Drug Deliv Rev. 2012;64:102–15. doi: 10.1016/j.addr.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Zhu J, Shi X. Dendrimer-based nanodevices for targeted drug delivery applications. J Mater Chem B. 2013;1:4199. doi: 10.1039/c3tb20724b. [DOI] [PubMed] [Google Scholar]
- 12.Han G, Ghosh P, De M, Rotello VM. Drug and gene delivery using gold nanoparticles. NanoBiotechnology. 2007;3:40–5. [Google Scholar]
- 13.Lee J-S, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Gold, poly(beta-amino ester) nanoparticles for small interfering RNA delivery. Nano Lett. 2009;9:2402–6. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh PS, Kim C-K, Han G, Forbes NS, Rotello VM. Efficient gene delivery vectors by tuning the surface charge density of amino acid-functionalized gold nanoparticles. ACS Nano. 2008;2:2213–8. doi: 10.1021/nn800507t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi C a, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–70. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas M, Klibanov AM. Conjugation to gold nanoparticles enhances polyethylenimine's transfer of plasmid DNA into mammalian cells. Proc Natl Acad Sci U S A. 2003;100:9138–43. doi: 10.1073/pnas.1233634100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim ST, Chompoosor A, Yeh Y-C, Agasti SS, Solfiell DJ, Rotello VM. Dendronized gold nanoparticles for siRNA delivery. Small. 2012;8:3253–6. doi: 10.1002/smll.201201141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan X, Blacklock J, Li J, Möhwald H. One-pot synthesis of polypeptide-gold nanoconjugates for in vitro gene transfection. ACS Nano. 2012;6:111–7. doi: 10.1021/nn202939s. [DOI] [PubMed] [Google Scholar]
- 19.Shan Y, Luo T, Peng C, Sheng R, Cao A, Cao X, et al. Gene delivery using dendrimer-entrapped gold nanoparticles as nonviral vectors. Biomaterials. 2012;33:3025–35. doi: 10.1016/j.biomaterials.2011.12.045. [DOI] [PubMed] [Google Scholar]
- 20.Li P, Li D, Zhang L, Li G, Wang E. Cationic lipid bilayer coated gold nanoparticles-mediated transfection of mammalian cells. Biomaterials. 2008;29:3617–24. doi: 10.1016/j.biomaterials.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 21.Chen AM, Taratula O, Wei D, Yen H-I, Thomas T, Thomas TJ, et al. Labile catalytic packaging of DNA/siRNA: control of gold nanoparticles “out” of DNA/siRNA complexes. ACS Nano. 2010;4:3679–88. doi: 10.1021/nn901796n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin W, Yang K, Tang H, Tan L, Xie Q, Ma M, et al. Improved GFP gene transfection mediated by polyamidoamine dendrimer-functionalized multi-walled carbon nanotubes with high biocompatibility. Colloids Surf B Biointerfaces. 2011;84:206–13. doi: 10.1016/j.colsurfb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Liu M, Chen B, Xue Y, Huang J, Zhang L, Huang S, et al. Polyamidoamine-grafted multiwalled carbon nanotubes for gene delivery: synthesis, transfection and intracellular trafficking. Bioconjug Chem. 2011;22:2237–43. doi: 10.1021/bc200189f. [DOI] [PubMed] [Google Scholar]
- 24.Kasturirangan V, Nair BM, Kariapper MTS, Lesniak WG, Tan W, Bizimungu R, et al. In vivo toxicity evaluation of gold-dendrimer composite nanodevices with different surface charges. Nanotoxicology. 2013;7:441–51. doi: 10.3109/17435390.2012.668570. [DOI] [PubMed] [Google Scholar]
- 25.Worden JG, Dai Q, Huo Q. A nanoparticle-dendrimer conjugate prepared from a one-step chemical coupling of monofunctional nanoparticles with a dendrimer. Chem Commun (Camb) 2006:1536–8. doi: 10.1039/b600641h. [DOI] [PubMed] [Google Scholar]
- 26.De la Fuente JM, Berry CC. Tat peptide as an efficient molecule to translocate gold nanoparticles into the cell nucleus. Bioconjug Chem. 2005;16:1176–80. doi: 10.1021/bc050033+. [DOI] [PubMed] [Google Scholar]
- 27.Bartczak D, Kanaras AG. Preparation of peptide-functionalized gold nano-particles using one pot EDC/sulfo-NHS coupling. Langmuir. 2011;27:10119–23. doi: 10.1021/la2022177. [DOI] [PubMed] [Google Scholar]
- 28.Lee S, Pérez-Luna VH. Dextran-gold nanoparticle hybrid material for biomolecule immobilization and detection. Anal Chem. 2005;77:7204–11. doi: 10.1021/ac050484n. [DOI] [PubMed] [Google Scholar]
- 29.Lin AY, Lunsford J, Bear AS, Young JK, Eckels P, Luo L, et al. High-density sub-100-nm peptide-gold nanoparticle complexes improve vaccine presentation by dendritic cells in vitro. Nanoscale Res Lett. 2013;8:72. doi: 10.1186/1556-276X-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Needham CJ, Williams AK, Chew SA, Kasper FK, Mikos AG. Engineering a polymeric gene delivery vector based on poly(ethylenimine) and hyaluronic acid. Biomacromolecules. 2012;13:1429–37. doi: 10.1021/bm300145q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grabarek Z, Gergely J. Zero-length crosslinking procedure with the use of active esters. Anal Biochem. 1990;185:131–5. doi: 10.1016/0003-2697(90)90267-d. [DOI] [PubMed] [Google Scholar]
- 32.Hermanson GT. Bioconjugate techniques. 2nd ed. Academic Press; 2008. [Google Scholar]
- 33.Jain PK, Qian W, El-Sayed MA. Ultrafast cooling of photoexcited electrons in gold nanoparticle-thiolated DNA conjugates involves the dissociation of the gold-thiol bond. J Am Chem Soc. 2006;128:2426–33. doi: 10.1021/ja056769z. [DOI] [PubMed] [Google Scholar]
- 34.Tomalia DA. The dendritic state. Mater Today. 2005;8:34–46. [Google Scholar]
- 35.Qian X, Peng X-H, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 36.Srinivasan C. Hybrid strategies for nanolithography and chemical patterning. BiblioBazaar. 2012 [Google Scholar]
- 37.Chang H-C, Wang L-C. A simple proof of Thue's theorem on circle packing. 2010.
- 38.Reinemann C, Stoltenburg R, Strehlitz B. Investigations on the specificity of DNA aptamers binding to ethanolamine. Anal Chem. 2009;81:3973–8. doi: 10.1021/ac900305y. [DOI] [PubMed] [Google Scholar]
- 39.Kwok A, Eggimann GA, Reymond J-L, Darbre T, Hollfelder F. Peptide dendrimer/lipid hybrid systems are efficient DNA transfection reagents: structure–activity relationships highlight the role of charge distribution across dendrimer generations. ACS Nano. 2013;7:4668–82. doi: 10.1021/nn400343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vozza-Brown L, Fan J, Vasu S, Yu X, Wang B, Lakshmipathy U, et al. Lipofectamine™ LTX: a new transfection reagent for effective transfection of primary cells, hard-to-transfect cells and sensitive established cell lines. Invit Corp. n.d. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.