Abstract
UVR8 is a recently discovered UV-B photoreceptor with a homodimer as the active state. UV-B perception of an interfacial tryptophan (W285) causes dissociation of the dimer into two functional monomers. Here, we investigate the molecular mechanism behind UV perception by W285 in UVR8. We observed a significant quenching dynamics in about 150 ps within the interfacial four-tryptophan cluster and an unusual resonance energy transfer from the other ten tryptophans to the tryptophan cluster in 1–2 nanoseconds to enhance functional efficiency. With mutation of W285 to F, the quenching dynamics is highly suppressed in this intact mutant dimer and the overall fluorescence intensity dramatically increases by a factor of 6, indicating W285 as a dominant quencher. These results reveal a unique energy transfer mechanism for efficient UV perception and the critical functional role of W285 for primary quenching dynamics for initiating dimer dissociation to trigger the function.
Keywords: UV-B Photoreceptor, Intrinsic Chromophore Tryptophan, Active-State Homodimer, Primary Photochemistry, Resonance Energy Transfer, Ultrafast Dynamics
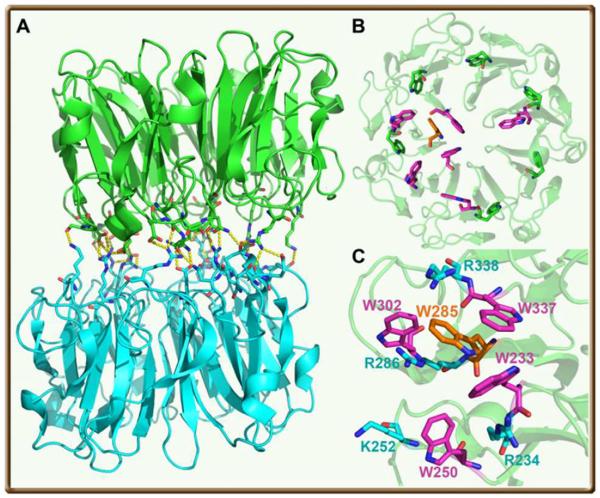
Sunlight is essential to sessile plants, both as an energy source to fuel photosynthesis for chemical energy and as a biological signal to control growth and development in their life cycle. Several families of plant photoreceptors have been discovered,1 including the blue-light sensing proteins of phototropins (a light-oxygen-voltage LOV domain),2 xanthopsins,3 cryptochromes,4 rhodopsins5 and blue-light utilizing flavin adenine dinucleotide sensors (BLUF)6 as well as phytochromes for detecting red and far-red light.7 These six photoreceptors have evolved to mainly sense visible light (400–700 nm), the major portion of sunlight, through absorption of their chromophores to trigger various biological signaling for regulating all major developmental and physiological processes.8 However, the ultraviolet (UV) radiation in sunlight, especially after recent ozone depletion, is detrimental to biosphere by causing DNA damage and cell dysfunction. Humans do not possess a substantial defense mechanism against UV radiation but within plants certain levels of UV-B radiation (280–315 nm) induce gene regulation that provides protective measures against such radiation.9–11 A UV-B photoreceptor in Arabidopsis,10 UV RESISTANCE LOCUS 8 (UVR8), was recently discovered and its high-resolution X-ray structure has even been solved.12,13 UV-B perception is by intrinsic amino-acid tryptophan (W) of UVR8, not any external chromophores like other photoreceptors. The active form is shown to be a homodimer consisting of two UVR8 molecules (Fig. 1A).
Figure 1.
(A) Overall architecture of UVR8 homodimer with numerous salt bridges and hydrogen bonds across the dimer interface. (B) A view of one monomer from the dimer interface with 13 tryptophans highlighted in different colors (orange-W285 and 6 magenta-tryptophan residues at the interface; 6 green-tryptophan residues buried in the middle of the blades). The tryptophan residue (W400) at the C-terminus is not shown. (C) The critical tryptophan cluster consists of W285, W233 and W337 from one monomer and W94 from the other monomer (not shown). The surrounding residues of the tryptophans and important positive amino acids are also shown.
Each core UVR8 domain forms a 7-bladed β-propeller and each blade contains 3 β-strands (Fig. 1B). The two monomers are linked together by non-covalent electrostatic interactions and 32 intermolecular hydrogen bonds. The two molecules of UVR8 consist of two acidic and basic patches of complementary charges of amino acids at interface. Significantly, there are 14 tryptophan residues in UVR8; 7 tryptophan molecules from each monomer lie at the interface of UVR8 dimer and 6 with one tyrosine are aligned around the middle of the 7-bladed β-propeller, one for each blade, along with one tryptophan at the C-terminal of the protein (Fig. 1B). At the interface of the dimer, two symmetric clusters of tryptophan residues, each containing 4 tryptophans, with 3 (W233, W285 and W337) from one monomer and one (W94, not shown) from the other monomer, are condensed all within van der Waals contacts. Around these tryptophans, several arginine residues interact with tryptophans to form strong cation-pi interactions (Figure 1C). With UV-B radiation, the dimer dissociates into two monomers and the resulting monomer interacts with downstream protein(s) to initiate transcriptional responses.10 Surprisingly, a mutation of W285, the centered tryptophan in the cluster (Fig. 1C), totally abolishes the function,10i.e., the dimer would not dissociate into two monomers to interact with signaling partner protein(s). Thus, the hypothesis is that UV-B is sensed by the interfacial tryptophan(s), leading to the breakdown of interfacial electrostatic interactions to form two monomers. W285 was proposed to be the key chromophore of UVR8 to sense UV-B radiation. However, how does UV-B perception by W285 trigger dimer dissociation and what is the functional role of other tryptophan residues?
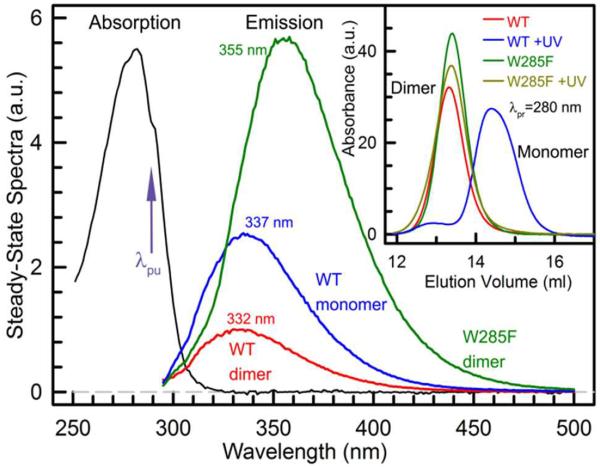
Here, we present our initial attempt to characterize the dynamics of tryptophan(s) after UV-B perception and with site-directed mutagenesis to differentiate the contributions of the interfacial and middle tryptophans. All samples were prepared by following the standard protocols.12,13 Figure 2 shows the steady-state absorption (~5 μM) and fluorescence spectra (~1 μM) of the UVR8 wild type (WT) and W285F mutant. In inset of Fig. 2, the mutant shows no effect on UV radiation and stays the dimer configuration.12,13 Both dimers absorb light at λmax=282 nm with a profile that is similar to the tryptophan absorption, further confirming that tryptophan is the chromophore for UV photoreception, not other external chromophore molecules. Surprisingly, the fluorescence emissions of the two dimers show dramatic difference in intensities and positions. The WT dimer has a fluorescence peak at 332 nm while the W285F dimer is at 355 nm, even longer than the emission peak of tryptophan in water at 350 nm.14,15 However, the 6 tryptophans in the middle of the β-propeller are buried and in the more hydrophobic environment. The emission peak should be more blue-shifted around 330 nm. Thus, the excitation of 6 tryptophans are either locally quenched or transferred into the interfacial tryptophans through resonance energy transfer (RET) and the 7 interfacial tryptophans should be in a highly hydrophilic environment with many salt bridges and 32 hydrogen bonds and/or a lot of interfacial water molecules.
Figure 2.
Steady-state absorption and relative emission spectra of WT dimer, WT monomer (WT+UV) and W285F. These three protein samples have a similar absorption spectrum, but exhibit dramatically different fluorescence emissions (λpu=290 nm) in their intensities and peaks. Size exclusion chromatography in the inset confirms that W285F mutant is a constitutive dimer and does not dissociate into two monomers under UV light.
More significantly, the intensity of the WT dimer is more than 6 times weaker than the mutant dimer, i.e., only 16.7% of the mutant intensity. This observation is striking and shows the dramatic effect of W285 during UV-B perception. There are 14 tryptophans in UVR8 and if one tryptophan (W285) is mutated, the total intensity would slightly decrease but we observed a huge increase in intensity by a factor of more than 6. We have measured the averaged fluorescence quantum yield of 0.42 for the W285F mutant. If the 6 tryptophans in the middle of the β-propeller have no RET with the interfacial tryptophans and their excitations are locally quenched, the observed intensity from the 7 interfacial tryptophans in the mutant would be too strong with an unusual emission quantum yield (>0.7). Thus, the middle 6 tryptophans transfer excitation energy to the interfacial tryptophans for enhancement of UV perception and they have a function role. Furthermore, if only the W285 excitation is quenched, the total intensity would not change considerably. We observed here that the fluorescence was greatly quenched by W285, resulting in the emission being blue-shifted before the dimer is fully relaxed to give an emission peak around 355 nm.16–18 Thus, W285 must interact with other tryptophans at the interface (also see below), consistent with the tryptophan cluster at the interface and the related exciton model proposed recently by Christie et al.,13i.e., the tryptophan cluster (Fig. 1C) may form delocalized exciton excitation. With W285 in the center of the cluster, the huge quenching is not only from W285 but instead comes from the entire exciton formed with at least the three neighboring tryptophans. By mutation of W285, other tryptophans can emit fluorescence and thus the total fluorescence intensity increases significantly.
After UV-B radiation, the WT dimer dissociates into two monomers (inset in Fig. 2). The fluorescence intensity only increases by a factor of ~2.5 with the emission peak red-shifted by 5 nm, indicating that the quenching in the dimer is related to its interfacial chemical and structural properties constituted from both two monomers. However, the monomer intensity is still much less than that of the mutant dimer, reflecting that in the WT monomer the excited tryptophan is also quenched. These results signify that the tryptophan quenching at the dimer interface must be related to the neighboring residues in both monomers.
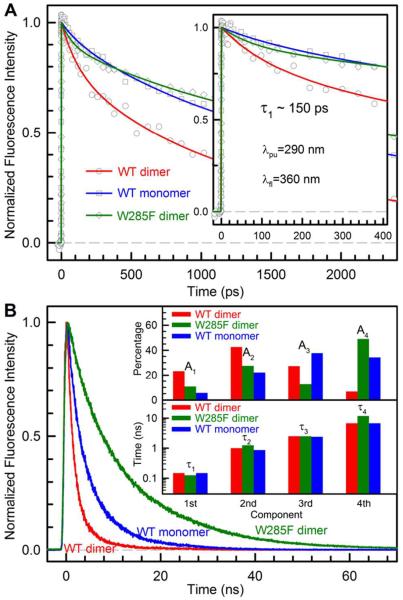
To examine the quenching dynamics and determine the quenching time scales, we characterized the time-resolved fluorescence dynamics of both WT and W285F mutant proteins from femtoseconds to nanoseconds. Figure 3A shows the femtosecond-resolved fluorescence transients of the WT dimer, WT monomer, and W285F mutant dimer gated at 360 nm with a dimer concentration of ~20 μM (monomer is ~40 μM) and Figure 3B gives the complete longtime behaviors obtained by using the time-resolved single-photon counting method at the dimer concentration of ~5 μM. The results are striking and the three transients show dramatically different decay behaviors, especially in Fig. 3B. The dynamics of the WT dimer and monomer become much shorter than that of the W285F mutant dimer, clearly showing the significant quenching in the WT proteins. Without the functional quenching in the mutant dimer, the fluorescence transient at the red side of peak (360 nm) has the negligible solvation contributions16–18 and mostly reflects the dynamics of effective lifetime emissions. Among thirteen tryptophans, any quenching through RET between tryptophans will be observed in the transient at 360 nm.
Figure 3.
Time-resolved fluorescence signals of WT dimer, WT monomer and W285F at 360 nm. (A) Femtosecond-resolved fluorescence transients show that WT dimer has significantly faster quenching than WT monomer and W285F (inset). For WT dimer, the transient was averaged only by about 10 scans due to the dimer dissociation. For each scan, each data point has a 30 laser-shot average. (B) Longtime fluorescence decays on nanosecond time scales. The percentages and the corresponding time scales of four exponential components are shown in inset. Note the logarithmic scale in inset for 4 decay times.
We did not observe any ultrafast decay in less than 100 ps (inset of Fig. 3A). Within our temporal resolution (~350 fs), we can resolve any ultrafast dynamics longer than 150 fs. The quenching dynamics with excited tryptophan should not be super ultrafast in less than 150 fs and thus must be longer than 100 ps. As we reported before,19 excited tryptophan is easily quenched through electron transfer with carbonyl- and sulfur-containing residues and possible peptide bonds. Typically, these quenchers are at van der Waals contacts with tryptophan and the quenching dynamics are on picosecond time scales.19 Thus, no quenchers within 3–4 Å of W285 were found, otherwise there would be evidence of ultrafast quenching in a few to tens of picoseconds in the initial transients. It was proposed that arginine residues may quench excited tryptophan,20 but no such quenching dynamics has been reported. The analyses of both femtosecond-resolved fluorescence transients (Fig. 3A) and longtime fluorescence signals (Fig. 3B) at 360 nm found that four decay components are needed to completely describe the dynamic behaviors. The resulting amplitude percentages and corresponding time scales are displayed in inset of Fig. 3B.
Overall, for the WT, the decay signal is dominated by the first three components: 150 ps (23%), 1 ns (43%), 2.5 ns (27%) and 6.8 ns (7%). For the mutant W285F, the dominant components are the second and fourth ones, 125 ps (11%), 1.25 ns (28%), 2.5 ns (12%) and 11.8 ns (49%). Since the fluorescence intensity profiles clearly show the significant quenching from WT to the mutant (Fig. 2) but we did not observe the ultrafast decay dynamics, the quenching in WT must occur on a longer timescale among the dominant three components. Thus, we propose that the initial ~150 ps component represents the fast quenching reactions among the interfacial cluster of 4 tryptophans and the percentage of 23% is roughly equivalent to 3–4 tryptophans out of the total 14 tryptophans in the protein. The components in 1–2 ns are the RET dynamics from the middle excited tryptophans (green in Fig. 1B) or the 4 distant interfacial tryptophans (3 purple ones at the bottom left in Fig. 1B and one not shown at the C-terminus) to the interfacial tryptophan cluster. These RET dynamics depend on the orientation factors and the center distances and due to the small overlap of absorption and emission spectra,14,21 the Trp-Trp RET is expected to be slow. After energy transfer to the cluster, the quenching reaction can occur immediately in ~150 ps and such reverse kinetics cannot accumulate significant immediate signals. With the mutation of W285, the key quenching reaction is nearly eliminated and all transferred excitations give rise to fluorescence emissions, leading to a huge increase of fluorescence intensity (Fig. 2). It is surprising that some interfacial tryptophans have a lifetime of ~12 ns, an unusual case never reported in proteins. The tryptophans in WT monomer after dissociation seem to have a usual time behavior with some longtime quenching.
We reported here the systematic study of UV perception of the UVR8 monomer, dimer, and key mutant (W285F) dimer by examining their emission properties and temporal behaviors. We observed that the fluorescence emission of the WT dimer is only ~17% of the W285F mutant (with 1 less tryptophan). This observation not only showed that W285 plays a crucial role in the initial quenching dynamics but also revealed the unique resonance energy transfer from all other tryptophans to the interfacial critical tryptophan cluster for enhancing UV-B perception in the homodimer. The primary quenching dynamics for initiating dimer dissociation occurs in about 150 ps and the resonance energy transfer for efficient UV harvesting takes about 1–2 ns. The primary quenching observed here in the interfacial tryptophan cluster could be the exciton evolution to a charge-separated state to induce the disruption of the salt bridges at the interface and lead to monomer formation for binding downstream proteins to form signal transduction for biological function.13 Various mutation experiments are under way to further elucidate the complete molecular mechanism of UV-B perception in UVR8.
ACKNOWLEDGMENT
We thank Dr. Di Wu (Tsinghua University) for his help during our sample preparation. The work was supported in part by the National Science Foundation (Grant CHE0748358) and the National Institute of Health (Grant GM095997).
Footnotes
The authors declare no competing financial interests.
REFERENCES
- (1).Moglich A, Yang XJ, Ayers RA, Moffat K. Structure and Function of Plant Photoreceptors. Annu. Rev. Plant Biol. 2010;61:21–47. doi: 10.1146/annurev-arplant-042809-112259. [DOI] [PubMed] [Google Scholar]
- (2).Christie JM. Phototropin Blue-light Receptors. Annu. Rev. Plant Biol. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- (3).Genick UK, Borgstahl GEO, Ng K, Ren Z, Pradervand C, Burke PM, Srajer V, Teng TY, Schildkamp W, McRee DE, Moffat K, Getzoff ED. Structure of a Protein Photocycle Intermediate by Millisecond Time-resolved Crystallography. Science. 1997;275:1471–1475. doi: 10.1126/science.275.5305.1471. [DOI] [PubMed] [Google Scholar]
- (4).Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GTJ, Batschauer A, Ahmad M. The Cryptochromes: Blue Light Photoreceptors in Plants and Animals. Annu. Rev. Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- (5).Okada T, Ernst OP, Palczewski K, Hofmann KP. Activation of Rhodopsin: New Insights from Structural and Biochemical Studies. Trends Biochem. Sci. 2001;26:318–324. doi: 10.1016/s0968-0004(01)01799-6. [DOI] [PubMed] [Google Scholar]
- (6).Barends TRM, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. Structure and Mechanism of a Bacterial Light-regulated Cyclic Nucleotide Phosphodiesterase. Nature. 2009;459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- (7).Ulijasz AT, Cornilescu G, Cornilescu CC, Zhang JR, Rivera M, Markley JL, Vierstra RD. Structural Basis for the Photoconversion of a Phytochrome to the Activated Pfr Form. Nature. 2010;463:250–254. doi: 10.1038/nature08671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Jiao YL, Lau OS, Deng XW. Light-regulated Transcriptional Networks in Higher Plants. Nat. Rev. Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- (9).Jenkins GI. Signal Transduction in Responses to UV-B Radiation. Annu. Rev. Plant Biol. 2009;60:407–431. doi: 10.1146/annurev.arplant.59.032607.092953. [DOI] [PubMed] [Google Scholar]
- (10).Rizzini L, Favory JJ, Cloix C, Faggionato D, O'Hara A, Kaiserli E, Baumeister R, Schafer E, Nagy F, Jenkins GI, Ulm R. Perception of UV-B by the Arabidopsis UVR8 Protein. Science. 2011;332:103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- (11).Heijde M, Ulm R. UV-B Photoreceptor-mediated Signalling in Plants. Trends Plant Sci. 2012;17:230–237. doi: 10.1016/j.tplants.2012.01.007. [DOI] [PubMed] [Google Scholar]
- (12).Wu D, Hu Q, Yan Z, Chen W, Yan CY, Huang X, Zhang J, Yang PY, Deng HT, Wang JW, Deng XW, Shi YG. Structural Basis of Ultraviolet-B Perception by UVR8. Nature. 2012;484:214–219. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- (13).Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O'Ha-ra A, Kelly SM, Hothorn M, Smith BO, Hitomi K, Jenkins GI, Getzoff ED. Plant UVR8 Photoreceptor Senses UV-B by Tryptophan-Mediated Disruption of Cross-Dimer Salt Bridges. Science. 2012;335:1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Lakowicz J. Principles of Fluorescence Spectroscopy. 3rd ed. Springer; New York: 2006. pp. 63–65. [Google Scholar]
- (15).Lu W, Qiu W, Kim J, Okobiah O, Hu J, Gokel GW, Zhong D. Femtosecond Studies of Crown Ethers: Supramolecular Solvation, Local Solvent Structure and Cation-π Interaction. Chem. Phys. Lett. 2004;394:415–422. [Google Scholar]
- (16).Zhong D. Advances in Chemical Physics. John Wiley & Sons, Inc.; 2009. Hydration Dynamics and Coupled Water-Protein Fluctuations Probed by Intrinsic Tryptophan; pp. 83–149. [Google Scholar]
- (17).Zhang L, Yang Y, Kao Y-T, Wang L, Zhong D. Protein Hydration Dynamics and Molecular Mechanism of Coupled Water-protein Fluctuations. J. Am. Chem. Soc. 2009;131:10677–10691. doi: 10.1021/ja902918p. [DOI] [PubMed] [Google Scholar]
- (18).Qin Y, Chang C-W, Wang L, Zhong D. Validation of Response Function Construction and Probing Heterogeneous Protein Hydration by Intrinsic Tryptophan. J. Phys. Chem. B. 2012;116:13320–13330. doi: 10.1021/jp305118n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Qiu W, Li T, Zhang L, Yang Y, Kao Y-T, Wang L, Zhong D. Ultrafast Quenching of Tryptophan Fluorescence in Proteins: Interresidue and Intrahelical Electron Transfer. Chem. Phys. 2008;350:154–164. [Google Scholar]
- (20).Siemiarczuk A, Petersen C, Ha C-E, Yang J, Bhagavan N. Analysis of Tryptophan Fluorescence Lifetimes in a Series of Human Serum Albumin Mutants with Substitutions in Subdomain 2A. Cell Bio-chem. Biophys. 2004;40:115–122. doi: 10.1385/CBB:40:2:115. [DOI] [PubMed] [Google Scholar]
- (21).Andrews DL, Demidov AA, editors. Resonance Energy Transfer. John Wiley and Sons; Chichester, England: 1999. [Google Scholar]