Abstract
Microbialites are organosedimentary structures that are formed through the interaction of benthic microbial communities and sediments and include mineral precipitation. These lithifying microbial mat structures include stromatolites and thrombolites. Exuma Sound in the Bahamas, and Hamelin Pool in Shark Bay, Western Australia, are two locations where significant stands of modern microbialites exist. Although prokaryotic diversity in these structures is reasonably well documented, little is known about the eukaryotic component of these communities and their potential to influence sedimentary fabrics through grazing, binding and burrowing activities. Accordingly, comparisons of eukaryotic communities in modern stromatolitic and thrombolitic mats can potentially provide insight into the coexistence of both laminated and clotted mat structures in close proximity to one another. Here we examine this possibility by comparing eukaryotic diversity based on Sanger and high-throughput pyrosequencing of small subunit ribosomal RNA (18S rRNA) genes. Analyses were based on total RNA extracts as template to minimize input from inactive or deceased organisms. Results identified diverse eukaryotic communities particularly stramenopiles, Alveolata, Metazoa, Amoebozoa and Rhizaria within different mat types at both locations, as well as abundant and diverse signatures of eukaryotes with <80% sequence similarity to sequences in GenBank. This suggests the presence of significant novel eukaryotic diversity, particularly in hypersaline Hamelin Pool. There was evidence of vertical structuring of protist populations and foraminiferal diversity was highest in bioturbated/clotted thrombolite mats of Highborne Cay.
Keywords: protist, stromatolite, thrombolite, 18S rRNA, diversity, foraminifera
Introduction
Fossilized stromatolites, dating back to 3.45 billion years ago, comprise the earliest visible record of life on Earth (for example, Grotzinger and Knoll, 1999; Allwood et al., 2006; Walter, 1983). There are only a few locations in the world where modern analogues of marine stromatolites are actively forming, and these include the margin of Exuma Sound, with regular seawater salinity, Bahamas (Reid et al., 2000), and hypersaline Hamelin Pool, Australia (Jahnert and Collins, 2011). Stromatolites are laminated microbialites, that can be further classified according to degree of lithification and cell-taxonomic richness of different microbial groups (discussed in Baumgartner et al. (2009) and Foster et al. (2009)). Hamelin Pool and Exuma Sound are both sites of formation of oolitic sands and lithifying microbial mats. Additional carbonate occurs in these environments, including in the form of metazoan skeletal remains and micrite. These carbonates are trapped and bound by extracellular polymeric substances, produced by filamentous cyanobacteria and other bacteria (Reid et al., 2000; Stolz et al., 2001; Dupraz and Visscher, 2005). Thrombolites are microbialites that lack lamination, having instead a clotted fabric. Some attribute this to a specific microbial flora and contend thrombolites are essentially a lower Paleozoic phenomenon (Kennard and James, 1986). Others propose thrombolites result from ‘remodeling of a precursor fabric' by a combination of processes that include physical and metazoan disruption (Walter and Heys, 1985; Planavsky and Ginsburg, 2009). Regardless, both types of microbialites reflect the activities of complex microbial communities and their interactions with the environment. A better understanding of factors influencing microbialite structure has implications for interpretation of the paleorecord.
Although stromatolites are characterized by steep gradients of oxygen, sulfide and light (Visscher et al., 1998; Visscher and Stolz, 2005; summarized in Dupraz et al., 2009), thrombolites often lack a single oxygen maximum and the oxic zone typically extends to greater depth (Myshrall et al., 2010). Independent of the debate on the origins of thrombolites, it seems likely grazing, burrowing and sediment-binding activities of specific eukaryotes such as the foraminifera may significantly contribute to the more bioturbated mat structures (Bernhard et al., 2013). In alternative proposals, thrombolites might result from interactions between eukaryotic green algae and various coralline algae (Feldmann and McKenzie, 1998), the patchy calcification of the filamentous cyanobacterium Dichotrix (Planavsky et al., 2009), or by a combination of coccoid and filamentous cyanobacterial photosynthesis, as was proposed for Green Lake Fayetteville, NY, USA; (Thompson et al., 1990).
Flagellate protists have been identified in some modern stromatolites (Al-Qassab et al., 2002), as well as ciliates in stromatolite microbial nodules (Westphalen, 1993), and foraminiferal tests (shells) in some thrombolites (for example, Papineau et al., 2005; Mastandrea et al., 2006). These three groups are known to be successful in sulfide-enriched, oxygen-depleted environments (for example, Fenchel and Finlay, 1995; Bernhard, 2003; Bernhard et al., 2006). Myshrall et al. (2010) propose a relatively minor role for eukaryotes in thrombolites, and suggest based on the lower diversity and higher productivity they detected in thrombolites compared with stromatolites, that thrombolite communities are distinct from those in adjacent stromatolites, and not simply ‘bioturbated stromatolites'.
Owing to the limited depth of sequencing and methodological biases (for example, primer choice), numerous other potentially bioturbating eukaryotes may have been undetected by Myshrall et al. (2010). Here, we analyzed eukaryotic communities in different microbialite structures along the margin of Exuma Sound, Bahamas, and in Hamelin Pool, Australia, by applying high-throughput sequencing of eukaryotic small subunit ribosomal RNA (SSU rRNA) combined with multivariate statistical analyses. By using RNA, but not DNA, as template, we focus on the active fraction of the community in order to determine: (a) if the eukaryotic composition in different microbialite types is distinct, (b) if there are common communities in microbialites at both locations, and (c) if a greater diversity of SSU rRNA signatures of potential eukaryotic bioturbators other than metazoa can be found in more clotted microbialites that might help explain their structure.
Materials and methods
Field site and sample collection
Samples of microbialite mat types were collected from just below the water line at low tide on the windward side of Highborne Cay (24°43.5′ N, 76°49′ W), Bahamas, in March 2010 and from Hamelin Pool near Carbla Station on Hamelin Pool, Australia (26°15.4′ S, 114°13.5′ E), in June 2011. Highborne microbialite mat types were designated according to Reid et al. (2000) for stromatolite (that is, laminated) mats and according to Myshrall et al. (2010) for thrombolite (that is, clotted) mats. Samples collected at Highborne included stromatolite surface Type 1 mats, which result from binding and trapping of ooids by the filamentous cyanobacterium Schizothrix sp., surface Type 2 mats, which are biofilm structures comprised of a complex community that includes aerobic and anaerobic heterotrophic prokaryotes that precipitate a microcrystalline (ca 30 μm in diameter) CaCO3 crust, Incipient Type 2 mats (transitioning from Type 1 to Type 2, with crust firming, but not complete) and thrombolitic mats, referred to as button types by Myshrall et al. (2010), which are irregular, clotted structures.
Hamelin Pool microbialites, which were classified using nomenclature of Jahnert and Collins (2011, 2012) and Logan (1961), included pustular mats, which are irregular, clotted mats, colloform mats, which are coarse, laminoid wavy mats and smooth mats, which are fine, laminoid structures. In Hamelin Pool, we also sampled a smooth mat that was recently scoured (‘smooth scoured'). Only the 1–2 cm fraction of this was sampled because of disturbance to the top 1 cm. We also sampled the water column (500 ml) in the immediate vicinity of these microbialite types, as well as sulfidic waters (∼2 m below pool surface) of a nearby ‘blue hole' (a stratified shallow hypersaline (78 Practical Salinity Units (PSU)) pool on a platform on the western edge of Hamelin Pool ∼20 km from the microbialite sampling site) for comparison. Samples from both locations are described in Table 1. The salinity at the time of collection was 33–35.5 PSU at Highborne and 66–72 PSU at Hamelin Pool. Water temperature at Highborne during sampling was 25.4–25.9 °C and in Hamelin Pool was 14.1–15.0 °C. Maximum light intensity during midday at Highborne and Hamelin Pool during the time of sampling was between 2000 and 2100 μ Em−2 s−1 and 1100 and 1300 μ Em−2 s−1, respectively. Microbialite samples were collected using 50cc syringe cores (2.6 cm inner diameter), which were immediately sectioned into 1 cm intervals to 2 cm and preserved in RNALater (Ambion, Life Technologies, Austin, TX, USA).
Table 1. Temperature, salinity, oxygen, sulfide, temperature and salinity data for Hamelin Pool, Australia, and Highborne Cay, Bahamas, microbialite samples.
| Temperature (°C) | Salinity (PSU) | Max. % O2 saturation (depth in mm) | Sulfide (μM) (depth first observed in mm) | |
|---|---|---|---|---|
| Highborne Cay | ||||
| Type 1 mat 0–1 cm | 25.4–25.9 | 33–35.5 | 225–264 (2.2–2.4) | NA |
| Type 1 mat 1–2 cm | 25.4–25.9 | 33–35.5 | 0 (0) | 24 (12.25) |
| Type 2 mat 0–1 cm | 25.4–25.9 | 33–35.5 | 394–456 (5.8–6.4) | 76 (7.25) |
| Type 2 mat 1–2 cm | 25.4–25.9 | 33–35.5 | 0 (0) | 282 (throughout) |
| Incipient Type II mat 0–1 cm | 25.4–25.9 | 33–35.5 | 285–346 (7.4–8.2) | 17 (9.25) |
| Incipient Type II mat 1–2 cm | 25.4–25.9 | 33–35.5 | 0 (0) | 192 (throughout) |
| Thrombolite 0–1 cm | 25.4–25.9 | 33–35.5 | 536–622 (8.2–9.6) | 10 (9.75) |
| Thrombolite 1–2 cm | 25.4–25.9 | 33–35.5 | 0 (0) | 168 (throughout) |
| Highborne water | 25.4–25.9 | 33–35.5 | 100 | NA |
| Hamelin Pool | ||||
| Pustular mat 0–1 cm | 14–15 | 66–72 | 174–218 (5.8–6.4) | 38 (6.25) |
| Pustular mat 1–2 cm | 14–15 | 66–72 | 0 (0) | 81 (throughout) |
| Colloform mat 0–1 cm | 14–15 | 66–72 | 188–230 (5.8–6.2) | 19 (7.75) |
| Colloform mat 1–2 cm | 14–15 | 66–72 | 0 (0) | 189 (throughout) |
| Smooth mat 0–1 cm | 14–15 | 66–72 | 229–312 (7.4–8.2) | 26 (7.5) |
| Smooth mat 1–2 cm | 14–15 | 66–72 | 0 (0) | 170 (throughout) |
| Smooth Scoured mat 0–1 cm | 14–15 | 66–72 | 100–108 (2.6–3.4) | 32 (4.0) |
| Smooth Scoured mat 1–2 cm | 14–15 | 66–72 | 0 (0) | 76 (throughout) |
| Hamelin Pool water | 14–15 | 66–72 | 100 | NA |
| Blue Hole sulfidic water | 18 | 78 | 0 | NA |
Abbreviation: NA, not available.
For sulfide data, depth noted is where free sulfide (H2S/HS−/S2−) is first observed, and the maximum concentration during the day during sampling is based on single profiles (note: replicate profiles are very similar). Oxygen data are presented as the ranges of maximum depth and maximum % oxygen saturation in each sample.
Microelectrode measurements
Depth profiles of oxygen, sulfide and pH were determined using needle microelectrodes (Visscher et al., 1998, 2002; Myshrall et al., 2010) ex situ under ambient temperature and light intensity. Small samples (∼5 × 5 cm) were collected and submerged in 3 cm water collected from the site and pre-incubated for 12–24 h before the first measurement. Daytime electrode readings were carried out during the peak of photosynthesis between noon and 1400 hours, and nighttime measurements were made at the end of the dark period between 0300 and 0530 hours. Light measurements were done using a LiCor LI 250 meter (LiCor Biosciences, Lincoln, NE, USA) equipped with a SA190A quantum sensor, and salinity and temperature measurements were obtained with an Accumet AP75 temperature/conductivity meter (Fisher Scientific, Waltham, MA, USA).
RNA extraction and SSU rRNA gene sequencing
Samples stored in RNAlater (0.5 g of preserved material) were rinsed three times with RNAse-free and sterile 1 × phosphate-buffered saline before RNA extraction. RNA was extracted using the FastRNA Pro Soil-Direct Kit (MP Biomedical, LLC, Solon, OH, USA). The manufacturer's extraction protocol was modified to include the addition of 2 M sodium acetate following cell lysis and a Turbo DNase (Ambion) treatment before the RNA Matrix cleanup included in the extraction kit. DNA removal was confirmed by 45 cycles of PCR (see below) using extracted RNA as template. RNA was reverse transcribed and PCR amplified in one step using the Superscript One-Step RT-PCR kit (Invitrogen, Life Technologies, Austin, TX, USA) and either general eukaryotic SSU rRNA gene V4 hypervariable region primers and protocols (TAReuk454FWD1/ TAReukREV3, Stoeck et al., 2010) or general primers for foraminifera (S14F1/S17, Pawlowski, 2000). The foraminifera-specific amplifications were required because general V4 primers do not detect most foraminifera. Barcoded PCR products were purified from 1% agarose gels using the Zymoclean Gel DNA Recovery Kit (Zymo Research, Irvine, CA, USA). Foraminiferal PCR products were cloned into pCR4-TOPO using the TOPO TA Cloning Kit (Invitrogen) for Sanger sequencing (one 96-well plate per microbialite sample).
Pyrotags and clone sequences were processed for quality control and chimera removal using Bellerophon Chimera Check and the Check_Chimera utilities (Ribosomal Database Project; Cole et al., 2003). After denoising of the pyrosequencing data set using AmpliconNoise (Quince et al., 2009), sequences from clone libraries and pyrosequencing were clustered into operational taxonomic units (OTUs) at 97% similarity in QIIME (Caporaso et al., 2010), and taxonomy of OTU representatives was assigned using JAguc (Nebel et al., 2011). Taxonomy was linked to the QIIME OTU table via a PERL script available from the authors. As V4 primers did not encompass foraminifera, the V4 and clone library data sets were clustered separately and then 97% OTUs were combined from each library for each sample. We ran ordination and statistical analyses (described in detail below) on both foraminifera-only and combined data sets. For combined data sets, we ran analyses using non-normalized data sets as well as normalized data sets, where V4 libraries were resampled to generate data sets of similar size to our foraminiferal data sets. Normalization did not produce a different outcome, and so to retain the most detailed picture of eukaryotic diversity, we present the non-normalized analyses. An unweighted presentation of diversity data (stacked histograms) in each sample was used for foraminifera-only analyses, as clone libraries may not be saturated. Weighted analyses were used for total eukaryotes (combined foraminiferal clone and pyrosequencing data). We interpret fine-scale taxonomic assignments (species-level; sometimes genus-level assignments) with caution because BLAST-based assignments are complicated by the hypervariable nature of the V4 region of eukaryotic rRNA. Variable taxon representation in public databases means JAguc may make taxonomic assignments down to genus or species, but in others, only down to higher levels. Accordingly, our stacked histograms presenting snapshots of diversity typically do not present a single level of taxonomic resolution.
The BLASTn output and the OTU table were combined to calculate the number of sequences across each sample belonging to a specific taxonomic group. The relative abundance of each protistan group in a given sample was calculated as a percentage value by dividing the raw number of sequences associated with the specific taxon by the total number of sequences in the sample. This was used to generate heat maps in QIIME.
Cannonical correspondence analysis (CCA) was used to visualize relationships between community structure and concentrations of dissolved oxygen, sediment depth and sites. Multi-Response Permutation Procedure (MRPP) was used as a statistical test of significance for each of these factors on OTU distribution. A Monte Carlo test was also used to assess a null hypothesis of no relationship between OTU distribution and these variables. Ordination and multivariate statistics were performed on our data set clustered at 97% sequence identity threshold. MRPP, Monte Carlo tests and CCA analyses used the PC-ORD software package (MjM Software Design, Gleneden Beach, OR, USA). Non-metric multidimensional scaling (goodness of fit measured by stress values, with values <15 indicating a low probability of drawing the wrong inferences from the results) and Principal Component Analysis, as implemented within PC-ORD, were used to confirm CCA results (data not shown). An unweighted Unifrac analysis comparing the eukaryotic communities (beta diversity) in all Hamelin Pool and Highborne Cay samples was performed within QIIME.
Nucleotide sequences for foraminifera have been deposited in GenBank (accession numbers: JX872558–JX873273 (Hamelin Pool) and JX873274–JX873955 (Highborne Cay)). V4 pyrosequencing tag sequences for Highborne Cay and Hamelin Pool have been deposited in the GenBank (SRA061992 and SRA061825, respectively).
Results
After removal of low-quality and potentially chimeric sequences, pyrosequencing yielded a total of ∼150 000 eukaryotic tags and Sanger sequencing produced 733 and 820 foraminifera clone sequences for Hamelin Pool and Highborne Cay, respectively. The Hamelin Pool and Highborne data sets were clustered into 2388 (including 129 foraminiferal) and 1571 (including 96 foraminiferal) OTUs, respectively. These data were interpreted together with physicochemical data (Table 1).
Eukaryotic SSU rRNA diversity in Highborne Cay, Bahamas, microbialites
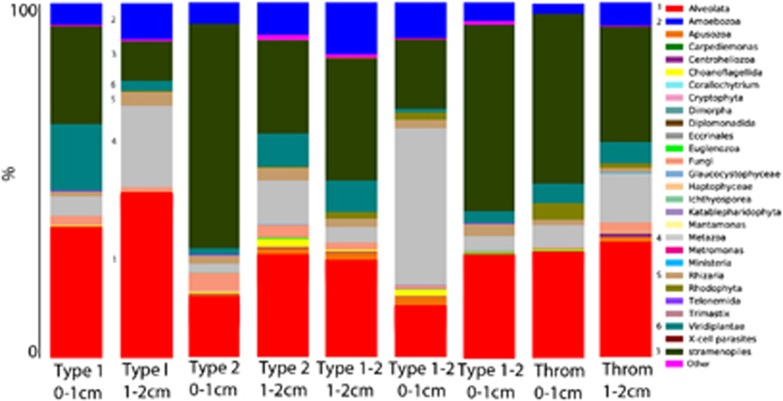
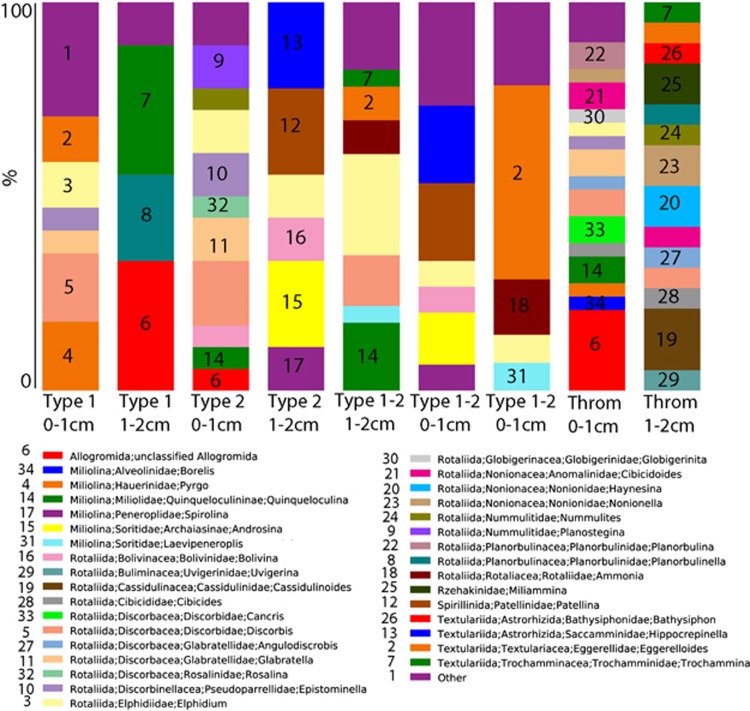
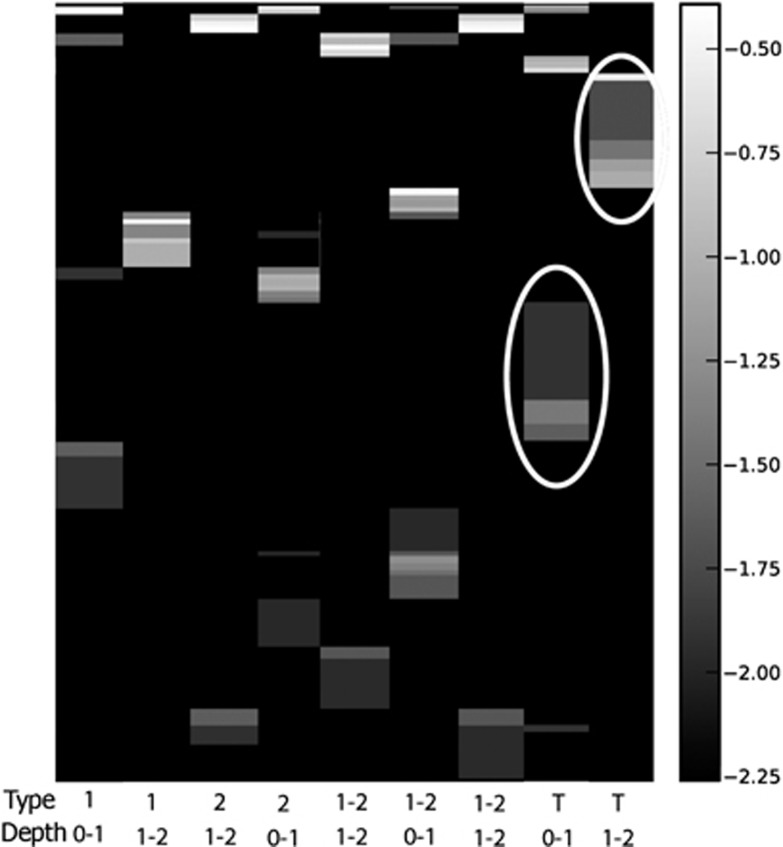
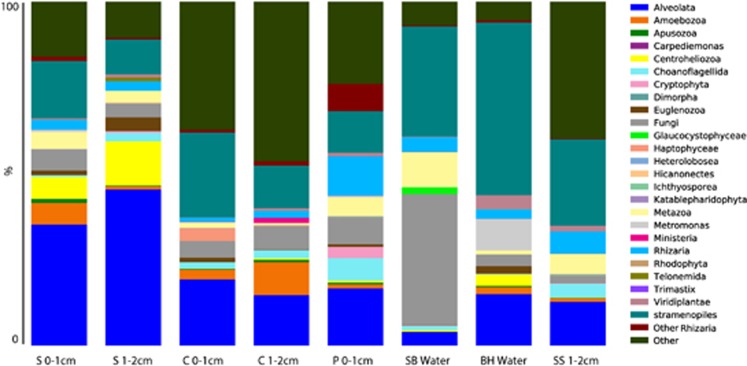
All mat types appear to support diverse eukaryotes, however, significant differences in SSU rRNA signatures are not generally observed between different mat types above order-level. Groups with the greatest representation in combined Sanger and pyrosequencing libraries were (in decreasing order): stramenopiles (for example, diatoms), Alveolata (ciliates and dinoflagellates), Metazoa (Annelids, Cnidaria, Gastrotricha, Nematoda, Platyhelminthes and Echinodermata), Amoebozoa and Rhizaria (Figure 1). However, within those dominant groups, there were differences in composition between mat types and between depths. For example, within Rhizaria (which includes foraminifera), Type 1 mat signatures in the 0–1 cm fraction affiliated with the foraminiferal taxa Texulariida, Miliolina, three types of Rotaliida and undescribed foraminifers with no close described relatives in GenBank (80% sequence similarity cutoff; Figure 2). In the 1–2 cm fractions, signatures of Allogromida were detected. Replicate 0–1 cm samples from two Incipient Type 2 mats were different in composition and relative proportions of foraminiferal OTUs, and different in composition from Type 1 mats, with more than half of OTUs coming from Textulariida (Figure 2). Differences may represent variations in the transitional state of one mat type to another. Thrombolite samples had the highest foraminiferal diversity; 16 OTUs in the 0–1 cm fraction and 14 OTUs in the 1–2 cm fraction (Figure 2), and differences between the 0–1 and 1–2 cm fractions. A heat map of foraminiferal OTUs indicates unique foraminiferal populations inhabiting thrombolites compared with stromatolites (Figure 3). Ciliate OTUs represented >50% of alveolate OTUs (Figure 1), with no clear difference in community composition between mat types and representatives from almost every ciliate class (10/12; Supplementary Figure 1).
Figure 1.
Stacked histogram of eukaryotic OTU composition of (97% sequence similarity, weighted data presentation) Highborne Cay, Bahamas, microbialite samples based on SSU rRNA signatures (cDNA template). Y axis corresponds to fraction of OTUs affiliating with each taxonomic grouping out of 100%. Throm, thrombolite.
Figure 2.
Stacked histogram of foraminiferal OTU composition of (97% sequence similarity, unweighted data presentation) Highborne Cay, Bahamas, microbialite samples based on SSU rRNA signatures (cDNA template). Y axis corresponds to fraction of OTUs affiliating with each taxonomic grouping out of 100%. Mat type noted for each sample. Type 1-2, Incipient Type 2 mat; Throm, thrombolite.
Figure 3.
Heat map of foraminiferal OTUs (97% sequence similarity) from Highborne Cay, Bahamas. Y axis represents log transformed abundance. Mat type noted for each sample. 1-2, Incipient Type 2 mat; T, thrombolite. Depths of fractions for each sample given in cm. White circles highlight examples of OTUs unique to thrombolite samples.
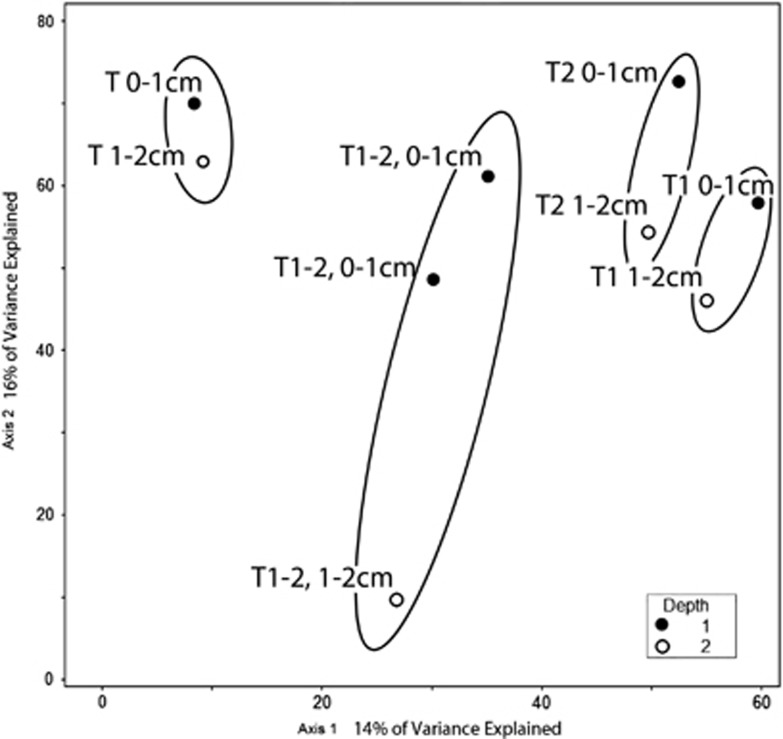
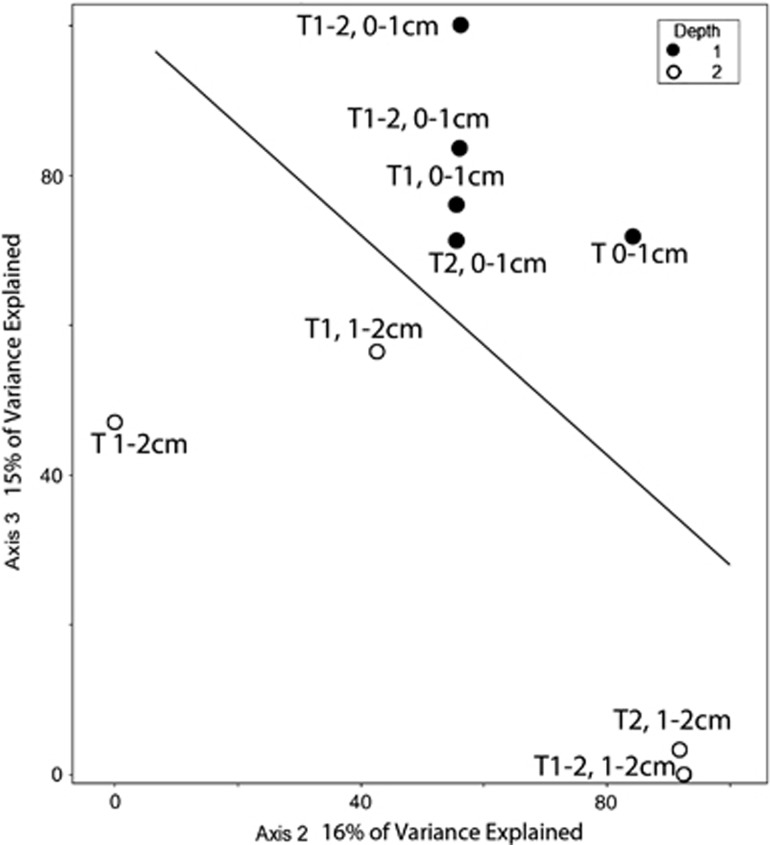
CCA of the Highborne data indicates that mat type and depth below the sediment–water interface explain much of the observed community structure (30% of the variation explained by axes 1 and 2 together). Values for MRPP tests of significance were combined for sediment depth and oxygen for all data sets because all 0–1 cm samples were at least partly oxic and all 1–2 cm samples were anoxic. The analysis shows a separation of protist communities within samples from different mat types and between the 0–1 and 1–2 cm fractions (Figure 4). CCA analysis considering depth and mat type also explains 31% of the foraminifera sequence variation, and shows a clear separation between the 0–1 cm fraction and the 1–2 cm fractions (Figure 5). Depth/oxygen was found to have a significant effect (P⩽0.05) on foraminifera OTU distributions in Highborne samples, and mat type was found to have a significant effect on whole eukaryotic community OTU distribution (Table 2).
Figure 4.
Biplot generated from Cannonical Correspondence Analysis of the 18S rRNA data set from Highborne Cay, Bahamas, for all eukaryotes clustered at the 97% sequence identity level. Sampled microbialite types are circled. Depths of each sample are noted. Filled circles represent 0–1 cm fractions and hollow circles represent 1–2 cm fractions. Mat type noted for each sample. (note: T1=Type 1, T2=Type 2, T 1-2=Incipient Type 2 mat, T=Thrombolite).
Figure 5.
Biplot generated from Cannonical Correspondence Analysis of our 18S rRNA data set from Highborne Cay, Bahamas, for foraminiferal sequences clustered at the 97% sequence identity level. Line separates samples from 0–1 cm depths for different microbialite types. Depths of each sample are noted. Filled circles represent 0–1 cm fractions and hollow circles represent 1–2 cm fractions. Mat type noted for each sample (note: T1=Type 1, T2=Type 2, T 1-2=Incipient Type 2 mat, T=Thrombolite).
Table 2. Multi-Response Permutation Procedure P-values for sample parameters for HC, HP and combined data sets.
| Depth/oxygen | Site | Mat type | Salinity | |
|---|---|---|---|---|
| HC-all eukaryotes | 0.455 | NA | 0.028 | NA |
| HC-foraminfera | 0.027 | NA | 0.776 | NA |
| HP-all eukaryotes | 0.817 | NA | 0.000 | NA |
| HP-foraminifera | 0.001 | NA | 0.000 | NA |
| Combined-all eukaryotes | 0.061 | 0.001 | 0.001 | <0.001 |
| Combined-foraminfera | 0.051 | 0.007 | 0.055 | NA |
Abbreviations: HC, Highborne Cay; HP, Hamelin Pool; NA, not applicable.
MRPP values are combined for depth and oxygen for all data sets because all 0-1 cm samples were oxic and all 1–2 cm samples were anoxic.
Eukaryotic SSU rRNA gene diversity in Hamelin Pool, Australia, microbialites
Eukaryotic rRNA sequences were dominated by Alveolata (10–50%), stramenopiles (10–30%) and unclassified eukaryotes (5–45% Figure 6). Alveolates were dominated by Heterocapsaceae and Protodinium (Dinophyceae) in colloform mats (with more diversity of alveolates in the 1–2 cm fraction), and Litostomatea (Ciliophora) in smooth mats. Pustular mats had the greatest variety of ciliate and dinoflagellate OTUs, most of which were also represented in the overlying water-column sample (Supplementary Figure 2).
Figure 6.
Stacked histogram of eukaryotic OTU composition of (97% sequence similarity, weighted data presentation) Hamelin Pool, Australia, microbialite and water samples based on SSU rRNA signatures (cDNA template). Y axis corresponds to fraction of OTUs affiliating with each grouping out of 100%. BH water, coastal enclosed sulfidic water body behind beach on Hamelin Pool, ∼20 km from microbialite sampling site; C, colloform; P, Pustular; S, smooth mat; SB water, Shark Bay (Hamelin Pool) waters at microbialite sampling site; SS, smooth scoured.
The Hamelin Pool (Carbla Beach) water sample collected from above the microbialites had almost 40% of total OTUs from fungi, 35% from stramenopiles and signatures of metazoa, choanoflagellates, rhizarians, glaucocystophytes (primarily a freshwater algae group) and low (<5%) contribution from unclassified eukaryotes. OTUs present in the sulfidic water sample from the shallow hypersaline ‘blue hole' at a distant location on the western shore of Hamelin Pool were distinct from those in Hamelin Pool, likely due to the presence/absence of sulfide.
Stramenopile OTUs were distinct between the local water samples and the microbialite samples (Supplementary Figure 3). The water samples were dominated by diatom stramenopiles, and microbialite stramenopiles were dominated by labyrinthulids. Between the two water samples, stramenopile OTUs in the sulfidic ‘blue hole' (90% Bacillariophyceae) were distinct from those in the sulfide-free seawater at Carbla Station (50% Coscinodiscophyceae and ∼40% silicoflagellates affiliating with Dictyochophyceae). Labyrinthulids affiliating to Thraustrochytridae represented between 60% and 85% of microbialite stramenopile sequences. Ten to fifty percent of OTUs were unassigned (defined as having <80% similarity to GenBank sequences), suggesting the presence of a novel eukaryotic community in Hamelin Pool, which will be the subject of a future study.
Smooth mats were dominated by Alveolata (50–90% ciliates), stramenopiles (∼20%) and unclassified eukaryotes (10–20% Figure 6). Dinoflagellate representatives in the 0–1 cm fraction included 40% Gymnodiniales, and several other taxa. In the 1–2 cm fraction, 90% of alveolate OTUs affiliated with Litostomatea (Ciliophora). The smooth mat that had been recently scoured (sample ‘SS') exhibited a shift in eukaryotic composition.
The colloform mats differed from the smooth mats by having a slightly lower contribution from alveolate signatures, and shifts within the alveolate signatures (Supplementary Figure 2), with OTUs affiliating with Dinophyceae dominating at both depth intervals. The pustular mat sample (only the 0–1 cm fraction was analyzed) differed from the others by having a greater representation of rhizarian signatures, new contributions from Cryptophyta and no signatures from Centroheliozoa.
When examining foraminiferal sequences from Hamelin Pool microbialites, differences in community composition are observed between microbialite types and between depths within a single mat core (Figure 7). Smooth mat samples were dominated by one OTU within Rotaliida. In the 0–1 cm fraction, there were additional OTU contributions from Textulariida, Rotaliida, Milliolina and Allogromida (that is, thecate or non-mineralized forms). Relative to the unscoured smooth mat, the scoured smooth mat had 8 vs 2 OTUs at the 1–2 cm depth, and mostly distinct OTUs, overall.
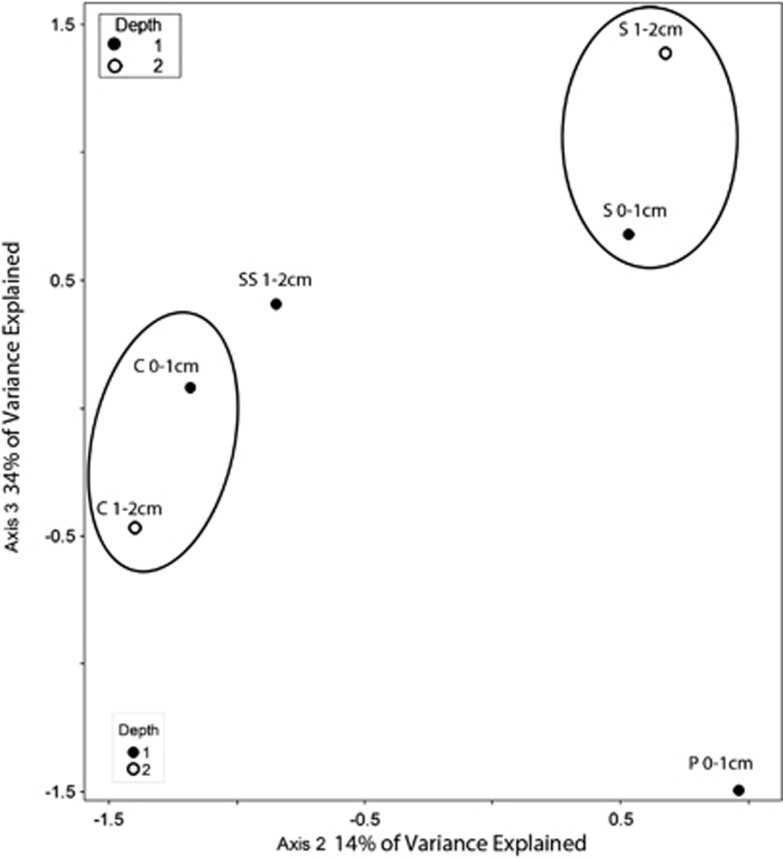
Figure 7.
Biplot generated from Cannonical Correspondence Analysis of our 18S rRNA data set from Hamelin Pool, Australia, for all eukaryotes clustered at the 97% sequence identity level. Sampled microbialite types are circled. Depths of each sample are noted. Filled circles represent 0–1 cm fractions and hollow circles represent 1–2 cm fractions. C, colloform mat; P, pustular mat; S, smooth mat; SS, smooth scoured mat.
Foraminiferal OTU compositions in the two depth fractions of the colloform and smooth mats were very different from one another (3 out of 8, and 3 out of 13 overlapped, respectively) (Supplementary Figure 4). The pustular mat type exhibited a distinct foraminiferal community composition in the 0–1 cm fraction, with 85% of the OTUs coming from Miliolina, and the rest from Allogromida (Supplementary Figure 4). A heat map of foraminiferal OTUs supports the unique nature of foraminiferal communities in the different mat types (minimal overlap) and depth intervals within individual microbialite types (Supplementary Figure 5).
CCA of our Hamelin Pool microbialite data identified mat type and depth as parameters that impact much of the observed protistan community distribution. This analysis supports the differentiation of eukaryote communities within different microbialite types and between different depths within individual microbialite types (Figure 7). As observed in our Highborne data, depth/oxygen was found to have a significant effect (P⩽0.05) on the observed distribution of OTUs for the foraminiferal data set only. Significantly different eukaryotic communities inhabit different mat types.
Combined CCA analysis of eukaryotic SSU rRNA diversity
Highborne and Hamelin Pool each contain distinct eukaryote communities (including foraminifera). Microbialite type and depth explained only 22.8% of the variation in a combined CCA analysis, implying other environmental factors such as salinity are driving most of the community differences between these two sites (Supplementary Figure 6). This notion is supported by MRPP analysis of this combined data set, where salinity is shown to be a significant influence (P⩽0.05) on eukaryotic OTU composition (Table 2). Site had a significant impact on distribution of OTUs for foraminifera separately. For the inclusive eukaryotic data set, mat type and site had a significant effect on distribution of OTUs.
Discussion
A new view of eukaryotic microbial diversity in microbialites
Microbialites in Hamelin Pool and Highborne Cay have been the subject of several recent investigations into their microbial communities and biogeochemistry (for example, Visscher et al., 1998; Reid et al., 2000; Burns et al., 2004; Allen et al., 2009; Baumgartner et al., 2009; Foster et al., 2009; Myshrall et al., 2010; Foster and Green, 2011). To our knowledge, only three studies have gathered information on microbial eukaryotes, one for Hamelin Pool (Allen et al., 2009) and two for Highborne (Baumgartner et al., 2009; Myshrall et al., 2010); and in comparison to this study, were more limited (and DNA-based) rRNA gene sequencing efforts. In the study of Hamelin Pool microbialites, only a limited number of signatures (11 unique clones from a pustular mat sample and 10 from a smooth mat) were detected, and most were from Nematodes (Allen et al., 2009). Fungal, tardigrade and microalgal OTUs were also detected. When using small sediment volumes and clone libraries, libraries can often be saturated by metazoan sequences. This was observed previously in microbialite samples, where nematode sequences dominated eukaryotic sequences in marine (Feazel et al., 2008; Baumgartner et al., 2007, 2009) and in freshwater microbialites (Couradeau et al., 2011).
In one of the studies of Highborne thrombolites, Myshrall et al. (2010) recovered low eukaryote diversity (only 26 unique eukaryotic ecotypes) relative to bacteria, with nematode sequences again figuring prominently in their clone libraries. Myshrall et al. (2010) also detected members of Alveolata and Chlorophyta, including sequences affiliating with those from hypersaline mats of Guerrero Negro (Feazel et al., 2008) and sequences in public databases from Hawaii Volcanoes National Park cave mats. They attributed their low recovered eukaryotic diversity in Highborne thrombolites at least in part, to the dynamic nature of microbialite environments, where they are periodically buried in oolitic sands, potentially for months (for example, Reid et al., 2000). PCR primers can also influence recovered diversity. Although no PCR primer set can be assumed to be inclusive for its target group, in the original description of the primers used by Myshrall et al. (Moon-van der Staay et al., 2001), it is acknowledged this set has biases against certain groups of eukaryotes including Chrysophyceae, some Chlorarachniophyceae, some Apicomplexa, some Ciliophora and deep-branching eukaryotes. In addition, we have found this set to be biased against many members of foraminifera, precluding its use in this study. The inclusion of foraminifera and other groups such as Ciliophora is particularly important for making valid comparisons of different microbialite communities.
Our analysis reveals diverse communities of eukaryotes in all microbialite types at both sites. The major difference in results (high vs low diversity) can be attributed to different nucleic acid extraction procedures, templates (RNA vs DNA), PCR primers and sequencing efforts/technologies. We attempted to minimize methodological biases by combining pyrosequencing of the V4 region of the SSU rRNA with Sanger clone libraries specifically targeting the foraminifera (missed by the employed V4 primers). In spite of the greater depth of sequencing in this study, for reasons explained above and because of minimal replication for specific microbialite types, we focus our observations on broad differences in taxonomic composition of higher groups (family and higher) between microbialite types that are likely representative of shifts in in-situ populations.
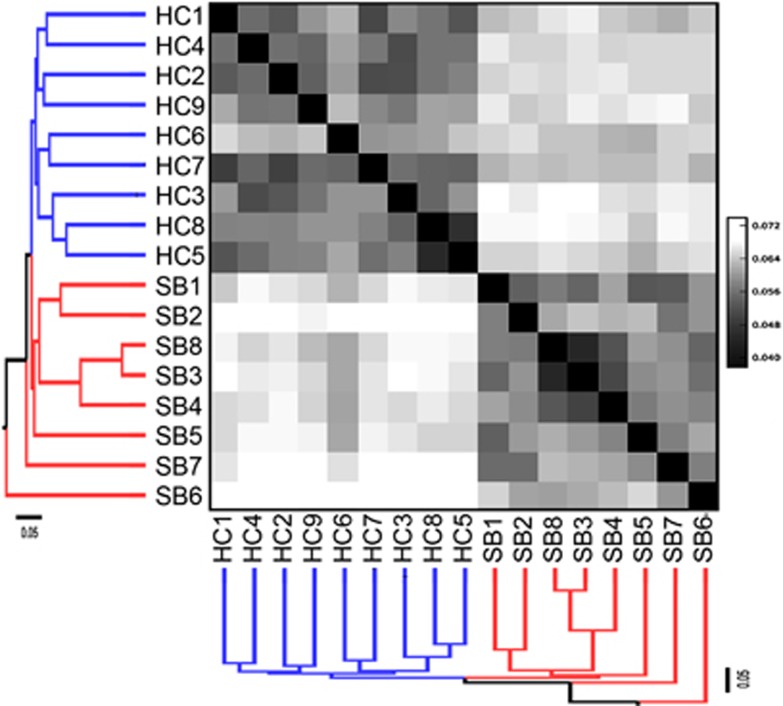
CCA indicates distinct eukaryotic communities in the different microbialite types and in different depth horizons within individual microbialite types in Highborne Cay and Hamelin Pool (Figures 4 and 8). It is notable that the water samples analyzed from Hamelin Pool (at the microbialite sampling site) and from the sulfidic ‘blue hole' (away from the sampling site) had eukaryotic communities largely distinct from the microbialite samples (Figure 6 and Supplementary Figure 2). The sulfidic ‘blue hole' water sample also contained stramenopile species that were distinct from those in the Hamelin Pool water sample, and dominated almost exclusively by diatoms, which are known to tolerate sulfide (for example, Admiral and Pelletier, 1979; Nagai and Imai, 1999). In contrast, stramenopile signatures from Hamelin Pool microbialites came primarily from Labyrinthulomycetes, which are known to degrade complex polysaccharides (Raghukumar, 2002). These organisms may be attracted to, and involved in, the degradation of the abundant polysaccharides in microbialite exopolymer matrices at this site.
Figure 8.
Unifrac analysis of Hamelin Pool and Highborne Cay samples. Hamelin Pool mat identifiers: 1 and 2=S, smooth mat, 0–1 and 1–2 cm depths; 3 and 4=C, colloform mat, 0–1 and 1–2 cm depths; 5=pustular mat, 0–1 cm depth; 6=Shark Bay (Hamelin Pool) water; 7=Blue Hole water; 8=smooth Scoured mat 0–1 cm depth. Highborne mat identifiers: 1 and 2=Type 2 mat, 0–1 and 1–2 cm depths; 3 and 4=Incipient Type 2 mat 0–1 cm and 1–2 cm depths; 5=thrombolite 0–1 cm depth; 6=Incipient Type 2 mat 0–1 cm depth; 7=Type 1 mat 0–1 cm depth; 8=thrombolite 1–2 cm depth; 9=Type 1 mat 1–2 cm depth.
The eukaryotic taxa that we report in microbialites at these two locations reflect modern microbialite ecoystems in our present oxic biosphere. The extent that taxonomic groups overlap with those in ancient microbialite ecoystems is unknown, primarily because of their poor fossilization potential. Exploration of preserved eukaryotic biosignatures in fossilized microbialites is an area of future investigation.
Potential metazoan bioturbators
Stable isotope data indicate that filaments of the cyanobacterium Dichotrix have a role in the formation of microbialite clots (Planavsky et al., 2009). Bioturbation by eukaryotes may also impact mat structures (Farmer, 1992), and includes churning of sediments caused by movements of fauna. Many eukaryotes, including metazoa (for example, Reichardt, 1988; Pike et al., 2001), foraminifera (Gross, 2002) and ciliates (for example, Glud and Fenchel, 1999) are known to bioturbate sediments. Such activities stimulate bacterial community activity (Reichardt, 1988) and have been proposed as an explanation for the formation and coexistence of clotted mat structures such as thrombolites in the same vicinity as laminated mat structures (for example, Type 1, 2, smooth and colloform mats; Walter and Heys, 1985).
On Highborne Cay, the dominant cyanobacteria in stromatolites (Type 1 and 2 mats) is Schizothrix (Reid et al., 2000) and in thrombolites is Dichothrix sp. (Planavsky et al., 2009; Myshrall et al., 2010). Previous studies have shown that nematodes may be attracted to volatile compounds produced by cyanobacteria via chemotaxis (Höckelmann et al., 2004). At Hamelin Pool, we detected signatures of diverse metazoa (Annelida, Cnidaria, Gastrotricha, Nematoda, Platyhelminthes and Echinodermata), including specific nematode signatures detected previously (Allen et al., 2009). Nematode signatures detected previously by Myshrall et al. (2011) and affiliating with Syringolaimus were found in Highborne microbialites.
Potential protist bioturbators
Bioturbation may occur as a result of feeding activities of ciliates and foraminifera (for example, Pusch et al., 1998; Gross, 2002) and biofilm degradation (for example, by Thraustochytrids (Labyrinthulomycetes); Raghukumar, 2002). Ciliate signatures figured prominently among alveolate OTUs in microbialites at both locations (Supplementary Figure 1). Alveolates in Hamelin Pool microbialites were dominated by dinoflagellate subgroups Heterocapsaceae and Protodinium in colloform mats, and by the ciliate class Litostomatea (includes taxa described from anoxic environments; Vd'ačny et al., 2011) in smooth mats. The 0–1 cm and 1–2 cm fractions of microbialite samples contained very different alveolate communities that correlated with oxygen concentration (Table 1 and Supplementary Figure 3).
As noted above, foraminifera are also capable of significantly altering sediment fabric (for example, Gross, 2002). The subgroup Allogromida feed on bacteria and smaller eukaryotes are known to rend bacterial biofilms (Bernhard and Bowser, 1992), and through their reticulopodial activities can disrupt the fine laminations of microbialites (Bernhard et al., 2013). Aside from some overlapping taxonomic groups, foraminiferal communities in different microbialite structures were distinct, and thrombolites harbored a greater family-level diversity than stromatolite types examined from Highborne (14–16 vs 4–11; Figure 2). Signatures affiliating with Allogromida were detected in the 1–2 cm fraction of Type 1 stromatolite mats, the 0–1 cm fraction of Type 2 stromatolite as well as thrombolite mats from Highborne Cay and the 0–1 cm fraction of smooth, colloform and pustular mats, and the 1–2 cm fraction of colloform mats from Hamelin Pool (Supplementary Figure 4). No signatures of Allogromida were recovered from the 1–2 cm fraction of smooth mat samples, consistent with their greater lamination. Heat maps of foraminifera OTU distribution show very little taxonomic overlap between foraminiferal communities from either site (Figure 3 and Supplementary Figure 5). The distinction of foraminiferal communities between different depth fractions within the same mat type was supported by CCA and MRPP analyses for both Highborne and Hamelin Pool samples (Figures 5 and 8 and Table 2). As foraminiferal populations can be patchy, the 0.5 g samples processed for molecular analyses may under-sample foraminifera (and possibly other eukaryotic groups). Our study also did not examine whether or not there was a seasonal component, so it is possible that there are seasonal differences in protist communities at both of these sites, particularly Hamelin Pool, where relatively greater seasonal fluctuations in temperature, salinity and light occur.
Comparison of Hamelin Pool and Highborne Cay eukaryotic communities
Unifrac analysis shows a clear distinction between communities at each site (Figure 8). Differences are likely impacted by salinity and temperature. Hamelin Pool is a hypersaline system within the wider Shark Bay environment, with salinities between 66 and 72 PSU compared with 33–33.5 PSU at Highborne Cay, and had lower temperatures at the time of sampling (14–15 °C vs 25.4–25.9 °C; Table 1). MRPP analyses of depth/oxygen, mat type and salinity indicate that salinity drives, in part, the structure of the eukaryotic community (P<0.001). It was impossible to differentiate between influences of depth and oxygen at the scale of our sectioning as all 1–2 cm fractions were anoxic and all 0–1 cm fractions were at least partly oxic. Eukaryotic communities are distinct between microbialite types and foraminiferal communities are distinct at different depths (Table 2), most likely the result of different types of habitat/substrate, and varying prokaryotic and algal populations. The observation that sediment depth is a more significant driver of foraminifera OTU composition than mat type in the combined analysis likely reflects sensitivity of different foraminiferal taxa to oxygen concentration and differences in ability to migrate in response to fluctuations in oxygen and sulfide occurring within microbialites during diurnal cycles. A similar pattern probably exists for other individual protistan taxonomic groups.
Conclusions
The sites sampled in this study are dynamic environments impacted by tides, storms and currents, all of which subject microbialites to a continual cycle of mat construction and deconstruction. Eukaryotes in the different microbialite types at Highborne and Hamelin Pool were more diverse than previously reported and distinct, suggesting they may shape or be shaped by different microbialite fabrics. Metazoa and protists are potential bioturbators of microbialite mat structure. Our analyses resulted in several hypotheses regarding the impact of eukaryotes on microbialite structures, including (a) eukaryotic bioturbation may contribute to the more clotted structures of several microbialite types, (b) eukaryotic communities transition in composition during mat rebuilding after scouring events, and as microbialites transition from one type to another and (c) protists such as Thraustochytrids may actively degrade/consume the mat extracellular matrix. These hypotheses can be tested using more refined sampling, time-course studies, laboratory-based experiments and comparisons with non-lithifying, soft, organic-rich mats.
Acknowledgments
We thank the captain and crew of the R/V Walton Smith, as well as the owners/operators of Carbla Station; World Heritage Site Ranger Ross Mack; Sabine Mehay, Sara Dilegge, Richard Sperduto, Anna McIntyre-Wressnig, Marti Jeglinski and Ricardo Jahnert for field assistance; Philip Forte for assistance transporting field samples, and Kliti Grice and Anais Pages for laboratory support in Australia. We thank Jan Pawlowski and Andrea Habura for helpful discussions about PCR primers for foraminifera. We also thank two anonymous reviewers for comments on an earlier draft. This work was funded by grant OCE-0926421 to JMB and VPE and OCE-0926372 to RES.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Admiraal W, Pelletier H. Sulphide tolerance of benthic ditoms in relation to their distribution in an estuary. Brit Phycol J. 1979;14:185–196. [Google Scholar]
- Al-Qassab S, Lee WJ, Murray S, Simpson AGB, Patterson DJ. Flagellates from stromatolites and surrounding sediments in Shark Bay, Western Australia. Acta Protozoologica. 2002;41:91–144. [Google Scholar]
- Allen M, Goh F, Burns BP, Neilan BA. Bacterial, archaeal and eukaryotic diversity of smooth and pustular microbial mat communities in the hypersaline lagoon of Shark Bay. Geobiology. 2009;7:82–96. doi: 10.1111/j.1472-4669.2008.00187.x. [DOI] [PubMed] [Google Scholar]
- Allwood AC, Walter MR, Kamber BS, Marshall CP, Burch IW. Stromatolite reef from the Early Archaean era of Australia. Nature. 2006;441:714–718. doi: 10.1038/nature04764. [DOI] [PubMed] [Google Scholar]
- Baumgartner LK.2007Diversity and Lithification in Microbial Mats and StromatolitesPhD dissertationUniversity of Connecticut; 55–74. [Google Scholar]
- Baumgartner LK, Spear JR, Buckley DH, Pace NR, Reid RP, Visscher PT. Microbial diversity in modern marine stromatolites, Highborne Cay, Bahamas. Environ Microbiol. 2009;11:2710–2719. doi: 10.1111/j.1462-2920.2009.01998.x. [DOI] [PubMed] [Google Scholar]
- Bernhard JM. Potential symbionts in bathyal foraminifera. Science. 2003;299:861–861. doi: 10.1126/science.1077314. [DOI] [PubMed] [Google Scholar]
- Bernhard JM, Bowser SS. Bacterial biofilms as a trophic resource for certain benthic foraminifera. Mar Ecol Prog Ser. 1992;83:263–272. [Google Scholar]
- Bernhard JM, Edgcomb VP, Visscher PT, McIntyre-Wressnig A, Summons RE, Bouxsein M, et al. Insights into foraminiferal influences on microfabrics of microbialites at Highborne Cay, Bahamas. Proc Natl Acad Sci USA. 2013;110:9830–9834. doi: 10.1073/pnas.1221721110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard JM, Habura A, Bowser SS.2006An endobiont-bearing allogromiid from the Santa Barbara Basin: Implications for the early diversification of foraminifera J Geophys Res Biogeosci 111doi: 10.1029/2005JG000158 [DOI] [Google Scholar]
- Burns BP, Goh F, Allen M, Neilan BA. Microbial diversity of extant stromatolites in the hypersaline marine environment of Shark Bay, Australia. Env Microbiol. 2004;6:1096–1101. doi: 10.1111/j.1462-2920.2004.00651.x. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Strombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, et al. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 2003;31:442–443. doi: 10.1093/nar/gkg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couradeau E, Benzerara K, Moreira D, Gerard E, Kazmierczak J, Tavara R, et al. Prokaryotic and eukaryotic community structure in field and cultured microbialites from the alkaline lake Alchichica (Mexico) PLoS One. 2011;6:e28767. doi: 10.1371/journal.pone.0028767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupraz C, Reid RP, Braissant O, Decho AW, Norman RS, Visscher PT. Processes of Carbonate Precipitation in Modern Microbial Mats. Earth Sci Rev. 2009;96:141–162. [Google Scholar]
- Dupraz C, Visscher PT. Microbial lithification in marine stromatolites and hypersaline mats. Trends Microbiol. 2005;13:429–438. doi: 10.1016/j.tim.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Farmer JD.1992Grazing and bioturbation in modern microbial matsIn: Schopf JW, Klein C, The Proterozoic Biosphere – A multidisciplinary Study. Cambridge University Press; New York, NY, pp295–297. [Google Scholar]
- Feazel LM, Spear JM, Berger AB, Harris JK, Frank DN, Ley RE, et al. Eucaryotic diversity in a hypersaline microbial mat. Appl Environ Microbiol. 2008;74:329–332. doi: 10.1128/AEM.01448-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann M, McKenzie JA. Stromatolite-thrombolite associations in a modern environment, Lee Stocking Island, Bahamas. Palaios. 1998;13:201–212. [Google Scholar]
- Fenchel T, Finlay B. Oxford University Press: Oxford; 1995. Ecology and Evolution in Anoxic Worlds. [Google Scholar]
- Foster JS, Green SJ.2011Microbial diversity in modern stromatolitesIn: Tewari VC, Seckbach J, (eds).Stromatolites: Interaction of Microbes with Sediments, Cellular Origin, Life in Extreme Habitats and Astrobiology,Vol 18. Springer Science and Business Media: Heidelberg, Germany, pp383–405.
- Foster JS, Green SJ, Ahrendt SR, Golubic S, Reid RP, Hetherington KL, et al. Molecular and morphological characterization of cyanobacterial diversity in the stromatolites of Highborne Cay, Bahamas. ISME J. 2009;3:573–587. doi: 10.1038/ismej.2008.129. [DOI] [PubMed] [Google Scholar]
- Glud RN, Fenchel T. The importance of ciliates for interstitial solute transport in marine sediments. Mar Ecol Prog Ser. 1999;186:87–93. [Google Scholar]
- Gross O. Sediment interactions of foraminifera: implications for food degradation and bioturbation processes. J Foraminiferal Res. 2002;32:414–424. [Google Scholar]
- Grotzinger JP, Knoll AH. Stromatolites in Precambrian carbonates: Evolutionary mileposts or environmental dipsticks. Annu Rev Earth Planetary Sci. 1999;27:313–358. doi: 10.1146/annurev.earth.27.1.313. [DOI] [PubMed] [Google Scholar]
- Höckelmann C, Moens T, Juttner F. Odor compounds from cyanobacterial biofilms acting as attractants and repellents for free-living nematodes. Limnol Oceanogr. 2004;49:1809–1819. [Google Scholar]
- Jahnert RJ, Collins LB. Characteristics, distribution and morphogenesis of subtidal microbial systems in Shark Bay, Australia. Mar Geol. 2012;303-306:115–136. [Google Scholar]
- Jahnert RL, Collins LB. Significance of subtidal microbial deposits in Shark Bay, Australia. Mar Geol. 2011;286:106–111. [Google Scholar]
- Kennard JM, James NP. Thrombolites and stromatolites: two distinct types of microbial structures. Palaios. 1986;1:492–503. [Google Scholar]
- Logan BW. Cryptozoon and associate stromatolites from the Recent, Shark Bay, Western Australia. J Geol. 1961;69:517–533. [Google Scholar]
- Mastandrea A, Perri E, Russo F, Spadafora A, Tucker M. Microbial primary dolomite from a Norian carbonate platform: northern Calabria, southern Italy. Sedimentology. 2006;53:465–480. [Google Scholar]
- Moon-van der Staay SY, De Wachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–610. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- Myshrall KL, Mobberley JM, Green SJ, Visscher PT, Havemann SA, Reid RP, et al. Biogeochemical cycling and microbial diversity in the thrombolitic microbialites of Highborne Cay, Bahamas. Geobiology. 2010;8:337–354. doi: 10.1111/j.1472-4669.2010.00245.x. [DOI] [PubMed] [Google Scholar]
- Nagai S, Imai I. Factors inducing resting-cell formation of Coscinodiscus wailesii Gran (Bacillariophyceae) in culture. Plankton Biol. 1999;46:94–103. [Google Scholar]
- Nebel ME, Wild S, Holzhauser M, Hüttenberger L, Reitzig R, Sperber M, et al. Jaguc—a software package for environmental diversity analyses. J Bioinformatics Comput Biol. 2011;9:749–773. doi: 10.1142/s0219720011005781. [DOI] [PubMed] [Google Scholar]
- Papineau D, Walker JJ, Mojzsis SJ, Pace NR. Composition and structure of microbial communities from stromatolites of Hamelin Pool in Shark Bay, Western Australia. Appl Environ Microbiol. 2005;71:4822–4832. doi: 10.1128/AEM.71.8.4822-4832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski J. Introduction to the molecular systematics of foraminifera. Micropaleontology. 2000;46 (Suppl 1:1–1. [Google Scholar]
- Pike J, Bernhard JM, Moreton SG, Butler IB. Microbioirrigation of marine sediments in dysoxic environments: implications for early sediment fabric formation and diagenetic processes. Geology. 2001;29:923–926. [Google Scholar]
- Planavsky N, Ginsburg RN. Taphonomy of modern marine Bahamian microbialites. Palaios. 2009;24:5–17. [Google Scholar]
- Planavsky N, Reid RP, Lyons TW, Myshrall PT, Visscher PT. Formation and diagenesis of modern marine calcified cyanobacteria. Geobiology. 2009;7:566–576. doi: 10.1111/j.1472-4669.2009.00216.x. [DOI] [PubMed] [Google Scholar]
- Pusch M, Fiebig D, Brettar I, Eisenmann H, Ellis BK, Kaplan LA, et al. The role of micro-organisms in the ecological connectivity of running waters. Freshw Biol. 1998;40:453–495. [Google Scholar]
- Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- Raghukumar S. Ecology of the marine protists, the Labyrinthulomycetes (Thraustochytrids and Labyrinthulids) Eur J Protistol. 2002;38:127–145. [Google Scholar]
- Reichardt W. Effect of bioturbation by Arenicola marina on microbiological parameters in intertidal sediments. Mar Ecol Prog Ser. 1988;44:149–158. [Google Scholar]
- Reid RP, Visscher PT, Decho AW, Stolz JF, Bebout BM, Dupraz C, et al. The role of microbes in accretion, lamination, and early lithification of modern marine stromatolites. Nature. 2000;406:989–992. doi: 10.1038/35023158. [DOI] [PubMed] [Google Scholar]
- Stoeck T, Bass D, Nebel M, Christen R, Jones MD, Breiner HW, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19 (Suppl 1:21–31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- Stolz JF, Feinstein TN, Salsi J, Visscher PT, Reid RP. TEM analysis of microbial mediated sedimentation and lithification in modern marine stromatolites. Am Minerol. 2001;86:826–833. [Google Scholar]
- Thompson JB, Ferris FG, Smith DA. Geomicrobiology and sedimentology of the mixolimnion and chemocline in Fayetteville Green Lake, New York. Palaios. 1990;5:52–75. [Google Scholar]
- Vd'ačny P, Bourland WA, Orsi W, Epstein SS, Foissner W. Phylogeny and classification of the Litostomatea (Protista, Ciliophora), with emphasis on free-living taxa and the 18S rRNA gene. Mol Phylogen Evol. 2011;59:510–522. doi: 10.1016/j.ympev.2011.02.016. [DOI] [PubMed] [Google Scholar]
- Visscher PT, Reid RP, Bebout BM, Hoeft SE, Macintyre IG, Thompson JA. Formation of lithified micritic laminae in modern marine stromatolites (Bahamas): the role of sulfur cycling. Am Mineral. 1998;83:1482–1493. [Google Scholar]
- Visscher PT, Stolz JF. Microbial mats as bioreactors: populations, processes and products. Palaeogeogr Palaeoclimatol Palaeoecol. 2005;219:87–100. [Google Scholar]
- Visscher PT, Surgeon TM, Hoeft SE, Bebout BM, Thompson J, Jr, Reid RP.2002Microelectrode studies in modern marine stromatolites: unraveling the Earth's pastIn: Taillefert M, Rozan T, (eds) Environmental Electrochemistry: Analyses of Trace Element BiogeochemistryAm Chem Soc Symp Ser 811 Oxford Univ Press: New York, NY; 265–282. [Google Scholar]
- Walter MR.1983Archean stromatolites: Evidence of Earth's earliest benthosIn: Schopf JW, (eds),Earth's Earliest Biosphere: It's Origin and Evolution Princeton University Press: Priceton, USA; 187–213. [Google Scholar]
- Walter MR, Heys GR. Links between the rise of the metazoa and the decline of stromatolites. Precambrian Res. 1985;29:149–174. [Google Scholar]
- Westphalen D. Stromatolitoid microbial nodules from Bermuda–A microhabitat for meiofauna. Mar Biol. 1993;117:145–157. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.