Abstract
White plague (WP)-like diseases of tropical corals are implicated in reef decline worldwide, although their etiological cause is generally unknown. Studies thus far have focused on bacterial or eukaryotic pathogens as the source of these diseases; no studies have examined the role of viruses. Using a combination of transmission electron microscopy (TEM) and 454 pyrosequencing, we compared 24 viral metagenomes generated from Montastraea annularis corals showing signs of WP-like disease and/or bleaching, control conspecific corals, and adjacent seawater. TEM was used for visual inspection of diseased coral tissue. No bacteria were visually identified within diseased coral tissues, but viral particles and sequence similarities to eukaryotic circular Rep-encoding single-stranded DNA viruses and their associated satellites (SCSDVs) were abundant in WP diseased tissues. In contrast, sequence similarities to SCSDVs were not found in any healthy coral tissues, suggesting SCSDVs might have a role in WP disease. Furthermore, Herpesviridae gene signatures dominated healthy tissues, corroborating reports that herpes-like viruses infect all corals. Nucleocytoplasmic large DNA virus (NCLDV) sequences, similar to those recently identified in cultures of Symbiodinium (the algal symbionts of corals), were most common in bleached corals. This finding further implicates that these NCLDV viruses may have a role in bleaching, as suggested in previous studies. This study determined that a specific group of viruses is associated with diseased Caribbean corals and highlights the potential for viral disease in regional coral reef decline.
Keywords: viral metagenomics, eukaryotic ssDNA virus, satellite DNAs, nucleocytolasmic large DNA virus (NCLDV), coral bleaching
Introduction
Difficulties associated with isolating, culturing and manipulating potential pathogens as well as in quantitatively and temporally connecting the prevalence of a given microbe or virus to specific signs of infection have limited our understanding of the etiology of many wildlife diseases. Given these challenges, non-culture-based methods are increasingly used in research to characterize disease in novel environments or organisms. For example, surveys such as amplicon-based 16S ribosomal DNA profiling, can compare whole microbial assemblages from organisms of different health states, but to comprehensively survey viruses, shotgun-based metagenomic sequencing approaches are often required (Mokili et al., 2012). This is because viruses lack a common phylogenetic marker, and therefore marker-based amplicon sequencing (for example, 16S or 18S ribosomal RNA surveys) cannot necessarily be applied to diverse viral consortia. Some examples of the successful application of viral metagenomics to elucidate the pathogens associated with a disease include the identification of: an astrovirus as the probable cause for shaking mink syndrome (Blomström et al., 2010), an anellovirus associated with a California sea lion mortality event (Ng et al., 2009) and a coronavirus that causes human respiratory disease (Bermingham et al., 2012).
Marine wildlife diseases are increasing in prevalence and incidence (Ward and Lafferty, 2004; Harvell et al., 2004). Reports of non-bleaching coral diseases have increased over 50-fold from 1965 to 2005 (Sokolow, 2009). Such epizootics are of concern because they have contributed to declines in coral abundance and cover (Gardener et al., 2003; Rogers, 2009). However, of the >20 described global coral diseases, only 6–8 have pathogens ascribed to their etiology (Green and Bruckner, 2000; Sutherland et al., 2004; Rosenberg et al., 2007; Bourne et al., 2009; Pollack et al., 2011). One coral disease of particular ecological importance is white plague (WP), a rapid tissue loss disease that affects multiple species of Caribbean corals, including dominant reef-building Montastraea species. Caribbean WP is characterized by lesions that begin basally or peripherally on a colony, and then progress rapidly (mm to cm per day) across the colony surface resulting in partial to total colony mortality (Richardson et al., 2001; Miller et al., 2006; Weil et al., 2006). Three types of WP (I, II and III) are differentiated based on tissue loss progression rates, with type I progressing the slowest (a few mm per day), type II progressing at a maximum rate of 2 cm per day and type III progressing the fastest (>2 cm per day; Dustan, 1977; Richardson et al., 1998). Although an infectious agent, the bacterium Aurantimonas coralicida, was hypothesized to be the cause of WP type II in the coral Dichocoenia stokesii (Richardson et al., 1998; Denner et al., 2003), A. coralicida is not always associated with WP-infected coral colonies. For example, this bacterium was associated with healthy but not WP-infected Montastraea annularis colonies (Pantos et al., 2003) or WP-infected colonies from different geographic regions and/or host species (Siderastrea, Diploria; Sunagawa et al., 2009; Cárdenas et al., 2012).
Given the limited success in identifying the causative agents of WP diseases, controversy exists as to whether similar disease signs represent unique etiologies. It is possible that the different WP types are either the same disease variably manifested or alternatively, WP signs may represent a variety of different diseases, even in cases where progression rates are similar (Pollock et al., 2011). For example, a WP-like disease of corals in the Red Sea has been shown to be caused by the bacterial pathogen, Thalassomonas loyana, and not A. coralicida. Phage therapy reduces signs of this disease in situ and in aquaria, confirming the bacterial nature of the pathogen (Thompson et al., 2006; Efrony et al., 2009; Atad et al., 2012). Alternatively, an additional hypothesis is that a virus and not a bacterium might be the cause of some of these WP signs. To test this hypothesis, we determined if various viral types are differentially associated with a WP type I-like outbreak that occurred in the US Virgin Islands in 2010.
Currently little is known about viral disease in corals. Studies have characterized the viruses associated with bleached and healthy corals, corals exposed to different environmental stressors, and within the algal symbionts (Symbiodinium spp.) of corals (Wilson et al., 2004; Wegley et al., 2007; Marhaver et al., 2008; Vega Thurber et al., 2008; Correa et al., 2013). Based on these works, four major groups of viruses are predicted to infect corals and their associated microbes: bacteriophages, enveloped herpes-like viruses, nucleocytoplasmic large DNA viruses (NCLDVs, including members of the Phycodnaviridae, Mimiviridae and Iridoviridae), and small circular single-stranded DNA (ssDNA) viruses (for a review, see Vega Thurber and Correa, 2011). For example, herpes-like sequences were found in stressed Porites compressa corals as well as in other cnidarians such as the starlet anemone, Nematostella, and the medusozoan, Hydra (Vega Thurber et al., 2008). Genomic and microscopic evidence indicate that phycodnavirus-like particles can be present within bleached and/or heat-stressed corals, and these virus-like particles (VLPs) have been suggested to target Symbiodinium (Patten et al., 2008; Wilson et al., 2009; Correa et al., 2013). Active infection of Symbiodinium by members of the Phycodnaviridae was indirectly demonstrated based on the identification of phycodnavirus-like complementary DNAs in cultures of two different clades of Symbiodinium (Correa et al., 2013). Finally, sequence similarities to and viral-like particles reminiscent of nano-, circo- and geminiviruses, all of which are eukaryotic circular Rep-encoding ssDNA (CRESS-DNA) viruses and their associated satellites (collectively referred to ‘SCSDVs' in this paper) also have been found in corals (Davy and Patten, 2007; Wegley et al., 2007; Marhaver et al., 2008; Patten et al., 2008; Vega Thurber et al., 2008; Littman et al., 2011; Rosario et al., 2012).
Although these previous works laid the foundation for our understanding of viruses associated with diseased corals, the potential contribution of these viruses to colony WP disease signs is unknown, and their role in bleaching is poorly understood. Henceforth, we refer to coral colonies exhibiting signs of WP as ‘diseased' while colonies showing signs of symbiont loss, which may or may not be due to a pathogen, are described as ‘bleached.' To determine if viruses contribute to either bleaching and/or WP signs, we applied transmission electron microscopy (TEM) and replicated metagenomics to characterize viruses from Caribbean M. annularis corals that appeared healthy, bleached and/or affected by a WP-like disease. Here, we present evidence that (1) the viral consortia among Healthy (H), Bleached (B), WP Diseased (D) and WP Diseased-Bleached (BD) coral tissues are significantly different, and (2) SCSDVs dominate WP Diseased corals but are absent from healthy corals. These data suggest SCSDVs may have a role in some manifestations of WP-like disease in corals.
Materials and methods
Reef site and specimen collection
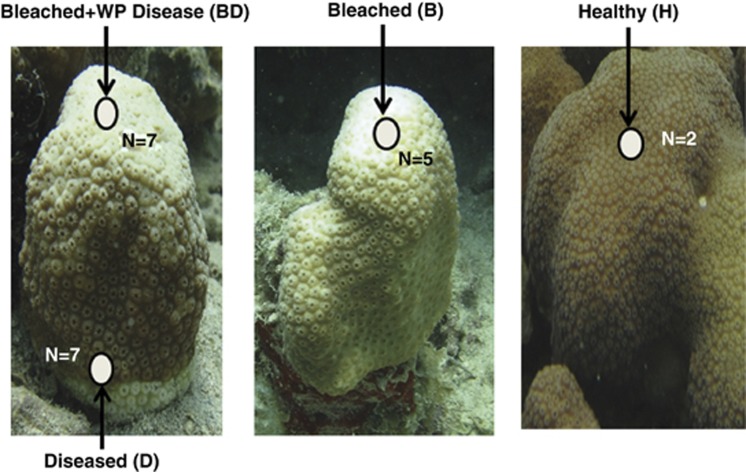
Sampling of the scleractinian coral, M. annularis, was conducted at Brewers Bay, in St Thomas, US Virgin Islands, during a concurrent WP outbreak and bleaching event in September 2010. During this event, up to 90% of corals on the examined reef were bleached, while a maximum of 7% showed signs of WP and bleaching (Brandt et al., 2013). Collections took place over 2 days at depths of 5.5–7.6 m. Temperature at depth was ∼29.0 °C. All M. annularis colonies used in this study were located within ∼75 m of each other. In addition to being affected with WP disease signs, all WP Diseased colonies showed signs of bleaching (Figure 1). Therefore, bleached coral colonies closest to WP Diseased colonies were selected as controls. All apparently healthy M. annularis colonies (N=2) that could be located during specimen collection dives were sampled.
Figure 1.
Locations of sampled tissue from M. annularis colonies of different health states. BD tissue was taken from ∼5 cm from the D tissue, where disease was progressing from the base of the coral colony. Seawater samples were taken ∼16 cm above BD and B coral colonies.
WP infections were defined using the following signs: (1) lesions consisted of an area of recent tissue loss where denuded skeleton with little to no algal colonization was delineated from living tissue by a smooth, undulating margin, (2) lesions were located peripherally or basally on the colony and (3) lesions expanded at an mean rate of 0.23±0.12 cm per day (Figure 1). These disease signs and epidemiological properties are consistent with those previously reported for WP (reviewed in Bythell et al., 2004; Sutherland et al., 2004; Brandt et al., 2013), and tissue loss rates were most similar to reports of WP type I disease (Dustan, 1977; Sutherland et al., 2004).
Coral tissue collections
Coral tissue samples were collected using SCUBA. Two to three plugs of tissue and skeleton were removed from each M. annularis colony (Figure 1) using a 2-cm diameter corer and hammer (USVI Department of Planning and Natural Resources permit #STT-050–10). From the seven WP Diseased colonies, tissues were cored from both the bleached top ‘Bleached+WP Diseased (BD)' and the margin of lesions ‘WP Diseased (D),' which did not show signs of bleaching. Tissue from an additional five ‘Bleached (B)' non-WP Diseased corals and two ‘Healthy (H)' corals (normal pigmentation, non-diseased) were taken as controls (Figure 1). Tissue samples were placed in individual sterile bags underwater, stored on ice and processed on shore within 4 h. All sampled colonies were examined for diseases signs and progression (if applicable) 2 days after sample collections. In all sampled WP Diseased colonies lesions had expanded, indicating that the disease was active when samples were taken. In all sampled non-WP Diseased colonies, no lesions had developed.
Cored specimens were rinsed with 0.02 μm filtered seawater. Tissue was removed by airbrushing with ∼40 ml of 0.02 μm filtered phosphate buffer saline solution (pH 7.3). Tissue homogenates were 0.22 μm filtered, preserved in molecular biology grade chloroform (2% final concentration) and stored at 4 °C until viral metagenome generation processing.
Seawater sampling, viral concentration and purification
Three liters of seawater were collected ∼16 cm above three diseased coral colonies using sterile containers. Replicate samples were pooled into one 9 l sample from which the ‘seawater Bleached+WP Diseased (SWBD)' virome was generated. Seawater also was collected and pooled from above three Bleached colonies for the ‘seawater Bleached (SWB)' virome.
VLPs were concentrated using a 100 kD tangential flow filter and purified via passage through a 0.22 μm sterivex (Vega Thurber et al., 2009). The resulting viral concentrate was preserved in molecular biology grade chloroform (2% final concentration), and stored at 4 °C until DNA extraction was performed as described below. The cesium chloride gradient ultracentrifugation steps were not conducted before extracting DNA from the seawater samples.
Viral metagenome generation
VLPs were isolated from coral tissues using cesium chloride density gradient ultracentrifugation, with buoyant densities ranging from 1.2 to 1.7 g ml−1 before addition of samples (Vega Thurber et al., 2009). After comparing all cesium chloride density layers from each tissue type for the presence of viral particles, viruses were isolated from the 1.2 g ml−1 density layer, where the majority of VLPs were present. To confirm the presence of VLPs and that bacterial/eukaryotic contamination was minimal, an aliquot of every viral fraction from all samples and density layers were stained with SYBR Gold (Invitrogen, Carlsband, CA, USA) and imaged using an epifluorescent microscope. Following this, all samples were again filtered through 0.22 μm Sterivex (Billerica, MA, USA) to remove residual bacteria.
DNA was isolated using an organic extraction protocol (Vega Thurber et al., 2009) and amplified using nonspecific multiple displacement amplification (MDA) according to the manufacturer's protocol (GenomPhi, GE Healthcare, Pittsburgh, PA, USA). PCR reactions using 16S and 18S primer sets were performed to determine whether bacterial or eukaryotic DNA contaminated the viromes. No contamination was detected.
The coral viromes (24 samples total, 1 plate) were barcoded and pyrosequenced on a Titanium 454 platform from Roche at EnGencore (San Francisco, CA, USA) (University of South Carolina). The final numbers of replicate libraries for each coral health state or seawater type were: H (n=2), B (n=5), BD (n=7), D (n=7), SWB (n=1) and SWBD (n=1).
Virome processing and bioinformatic analyses
Sequence reads underwent several preliminary bioinformatic steps. SFF files were converted to FASTA/FASTQ files and de-replicated using the program GALAXY (Goecks et al., 2010). Low quality reads (that is, those<100 bp in length and/or with quality scores<Q20) were removed. To eliminate any potential non-viral sequences from the data sets, the program DeconSeq was used to identify and remove reads with nucleic acid homology (based on⩾60% identity and 94% similarity) to eukaryotes (mouse, fish, human and mosquito), bacteria and/or archaea (Schmieder and Edwards, 2011). These conservative quality and similarity parameters may have underestimated viral sequence abundance because viral genomes commonly contain gene sequences homologous to bacteria, archaea and eukaryotes, but importantly, they reduced the likelihood that sequence similarities in this study were false positives. Sequences designated as contamination by Deconseq also were run through CAMERA (see below), in order to confirm that they were in fact not similarities to possible coral-infecting viruses (that is, were truly human or bacterial contamination).
Using the CAMERA (Community Cyberinfrastructure for Advanced Microbial Ecology Research and Analysis) platform, the tBLASTx algorithm (e values ⩽1 × 10−5) was used to find similarities to sequences in the National Center for Biotechnology Information (NCBI) non-redundant viral database (Altschul et al., 1990; Sun et al., 2011). Hierarchical taxonomic information (for example, viral order and family membership) was manually assigned for the strongest similarity identified to a known viral genome. Similarities to viruses that lacked any phylogenetic information (for example, ‘unclassified virus' or ‘fecal metagenome') or those describing paraphyletic groupings (for example, ‘chimp virus') were removed from the data sets. Sequence similarities at the viral family or satellite level were normalized to the total number of viral similarities identified in each library by dividing significant hits to known viral types by total known viral hits.
Viral family diversity analyses
Sequence similarity richness and evenness were determined using the Shannon Index diversity measure, H'=−∑i (pi)(logepi), where pi is the proportion of the total sample belonging to the ith viral family/satellites (Clarke and Warwick, 2001). The index was calculated for all 24 viromes and averaged for each virome type where applicable.
Statistical analyses
To compare differences among the ratios of phage and eukaryotic viral similarities as well as among Shannon diversity values, one-way analysis of variance followed by Tukey's HSD post hoc (95% confidence) tests (when statistical differences of P⩽0.05 were found) were performed using Analyse-it V2.2 Ltd (Leeds, UK). In addition, significant differences in viral taxa among the virome types were determined using a Kruskal–Wallis test followed by pairwise comparisons with Bonferroni corrections, when statistical differences (P⩽0.05) were found.
Viral consortia variability was determined using Bray–Curtis similarity matrices and non-metric multidimensional scaling plots of all normalized eukaryotic viral family similarities with 25 iterations. To test for statistical differences among the resulting multidimensional scaling clusters, analysis of similarity tests were performed while similarity of percentage analyses were executed to determine which viral types drove dissimilarity among samples (Clarke and Warwick, 2001). To further examine the difference in viral sequence similarities between the paired WP Diseased and WP Diseased-Bleached tissues isolated from individual colonies, a cluster analysis was performed only on the WP Diseased coral colonies (D vs DB). Shannon Diversity, multidimensional scaling, analysis of similarity, similarity of percentage and cluster analyses were all performed in PRIMER v.6 (Clarke and Gorley, 2006).
SCSDV replication gene phylogenetic analysis
Phylogenetic analysis of the SCSDVs was performed using the replication (Rep) initiator protein genes (rep) found in all WP Diseased and both Seawater viromes (SWB and SWBD) using the Metavir tool with the ‘compute phylogenic trees from multiple viromes' option (Roux et al., 2011). The Rep initiator protein was selected as a marker because of its evolutionary conservation in all SCSDVs and their associated satellite DNAs (Ilyina and Koonin, 1992; Gibbs and Weiller, 1999; Martin et al., 2011; Delwart and Li, 2012). METAVIR conducts the following steps to construct phylogenetic comparisons: (1) assembly of reads using CAP3, (2) sequence translation of un-assembled reads and (3) determination of reads with similarities to Rep proteins (BLASTx to the non-redundant database, e value<10−3) to SCSDV genomes in the PFAM database. Using the Metavir FASTA output, GENEIOUS VR6 (Biomatters, http://www.geneious.com/) was used to construct a neighbor-joining tree using the Jukes–Cantor model with 10 000 bootstraps. Only branches with>60% support were retained.
Viral metagenomic sequence alignments to SCSDV genomes and verification of SCSDV features
To confirm that our libraries contained sequences similar to all regions of SCSDV genomes, contigs from each individual library were constructed in Newbler using default settings (Margulies et al., 2005). Contigs from WP Diseased libraries were analyzed in Metavir's ‘Contig Map' feature. Representative contigs of 1000–3000 bp length that had similarities to SCSDVs were compared with annotated genomes from NCBI in GENEIOUS VR6 (Biomatters) by generating multiple alignments in MUSCLE (1000 iterations; Edgar, 2004). Average nucleotide identities (percentages) were derived from multiple nucleotide alignments of WP Diseased contigs and the genomes of known circoviruses, nanoviruses and satellite DNAs. These percentages were compared across each major category (for example, known circoviruses to known nanoviruses or putative circoviurses to known circoviruses) using univariate nonparametric statistics. Various SCSDV features also were annotated in GENEIOUS R6 (confirmed by tBLASTx to the non-redundant database), such as Rep and capsid genes, as well as stem loops, to further confirm that these sequences were similar to known SCSDVs (data not shown).
Transmission electron microscopy
Coral tissue and skeleton samples (∼2 mm) for TEM were collected in tandem with all samples obtained for viral metagenome generation. Samples were preserved in 0.02 μm filtered 2% gluteraldehyde and 0.05 M sodium cacodylate-buffered seawater and stored at 4 °C. Samples were post-fixed in osmium tetroxide, dehydrated in ethanol, embedded in Spurr resin, sectioned with a diamond knife in a Sorval MT-2 ultra-microtome and mounted onto copper grids. Sections were stained with lead citrate and uranyl acetate as needed and then imaged on a JOEL 1400 (Tokyo, Japan) transmission electron microscope at the University of Miami Center for Advanced Microscopy (Miller et al., 2011). Two individuals from each health state were examined for the presence of VLPs.
Results
To determine if specific viruses are associated with the documented St Thomas, USVI WP-like Disease outbreak, we generated 24 pyrosequenced shotgun viromes from H, B, BD and D coral tissues, as well as from seawater adjacent to Bleached (SWB) and Bleached+Diseased colonies (SWBD). These libraries totaled over 1 million reads, of which >360 000 passed quality control and in silico contaminant screening. Over 21% (∼78 000) of the curated sequences had similarity to known annotated viral genomes (Supplementary Table S1).
A majority of these sequences (averaged from all coral and seawater samples) were similar to phages (72.0%±16.8) in the Order Caudovirales (Myo-, Sipho- and Podoviridae). Within only the coral tissues libraries, similarities to large eukaryotic viruses in the families Herpesviridae (30.0%±18.2) and Phycodnaviridae (13.8%±11.8) were the most abundant eukaryotic viral similarities. These Phycodnaviridae and Herpesviridae similarities were found to have homology to genes (that is, capsid and topoisomerase genes) located across multiple regions of the known genomes (data not shown). Of eukaryotic viral comparisons, percentage of similarities to Circoviridae (15.2±17), Nanoviridae (7.9±13) and satellite DNAs (alpha satellites; 8.6±16) ranged from 0% (in Healthy tissue) to 64% (WP Diseased tissue).
Ratios of phage to eukaryotic viral similarities vary among coral tissue health states
Theoretically, eukaryotic viruses should increase in abundance relative to bacteriophages in coral tissues or seawater when an active eukaryotic viral infection is occurring. Although bacteriophage similarities dominated all libraries, the percentage of sequences similar to eukaryotic viruses was significantly higher (df=3) in the Bleached (34.47±17.0%) and Bleached+WP Diseased (32.61±18.8%) samples relative to the Healthy ones (11.11±8.3% Table 1). In addition, the ratio of phage to eukaryotic viral sequence similarities trended toward a decrease in WP Diseased (4.77±1.8) compared with Healthy tissues (19.21±15.2). The ratio of bacteriophage to eukaryotic virus sequence similarities in the SWB sample also was 1.47-fold higher than in the SWBD library (Table 1).
Table 1. Mean percentage of reads with similarity to eukaryotic viruses and mean ratio of bacteriophage (phage) to eukaryotic (euk) virus similarities in metagenomes generated from corals of different health states or adjacent seawater.
| Healthy (N=2) | Bleached (N=5) | Bleached+ WP Diseased (N=7) | WP Diseased (N=7) | SW Bleached (N=1) | SW Bleached+ WP Diseased (N=1) | |
|---|---|---|---|---|---|---|
| Mean % euk viral similarities (mean±s.e.) | 11.11±8.30 | 34.47±17.00 | 32.61±18.79 | 26.82 ±16.53 | 17.37 | 12.49 |
| Mean ratio of phage: euk viral similarities (mean±s.e.) | 19.31±15.16 | 3.80±2.03 | 3.23±0.93 | 4.77±1.76 | 7.00 | 4.76 |
Abbreviations: SW, seawater; WP, white plague.
Values in bold are statistically different from healthy sample values.
SCSDV and satellite DNA similarities dominated diseased coral tissues
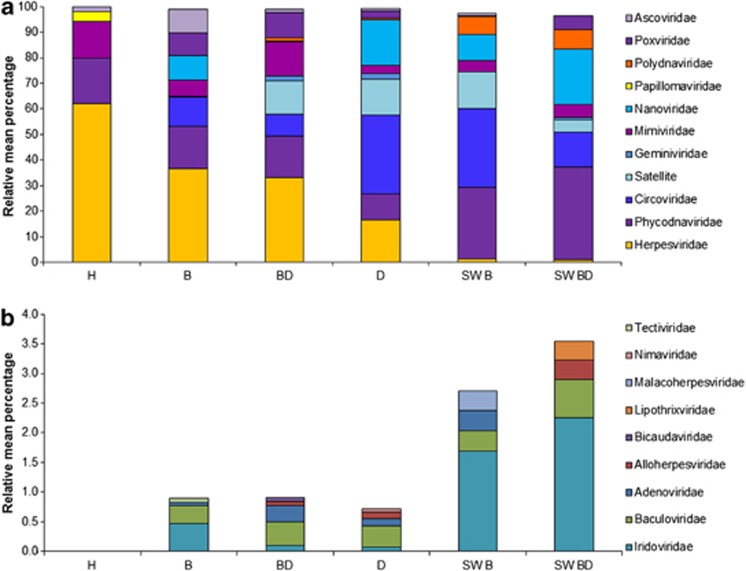
All viral types comprising ⩾4% of known eukaryotic virus similarities in at least one coral tissue or seawater sample were compared among the coral tissues health states and seawater viromes (Figure 2a). These comparisons indicated that significant differences (df=3) in viral consortia exist among coral tissue health states. For example, similarities to SCSDVs, including Circoviridae, Nanoviridae and satellite DNAs were each statistically more abundant in the WP Diseased libraries, relative to the Healthy libraries (P<0.05 for all tests). Collectively, SCSDV similarities comprised 64.5% of known eukaryotic viral similarities in WP Diseased tissues (Figure 3), whereas similarities to SCSDVs were not detected in Healthy coral viromes (Table 2). Furthermore, similarities to Nanoviridae were more abundant in WP Diseased than in Bleached+WP Diseased viromes (17.57±5.1% vs 0.35±0.4% P<0.03; Table 2). Satellite DNA similarities also were in higher relative abundance in WP Diseased (14.05±5.2%) than Bleached (0.00%) tissue libraries (P<0.01). Both seawater viromes also contained high abundances of SCSDV similarities (SWD=41.29% and SWB=55.59% Figure 2a).
Figure 2.
Mean percentage of similarities to viral families (and satellite DNAs) in metagenomes generated from different coral health states and seawater. Viral similarities that comprised ⩾4% of the total known eukaryotic viral similarities for a given sample type (a). The rare virome (b) or the viral similarities comprising <4% of the total known eukaryotic viral similarities detected for a given sample type.
Figure 3.

Viral ‘groups' among different coral health states. The mean relative percentage of similarities to each viral type was lumped into ‘groups' based on their common evolutionary history. ‘NCLDVs' are nucleocytoplasmic large double-stranded DNA (dsDNA) viruses, which include Ascoviridae, Phycodnaviridae, Poxviridae and Mimiviridae. ‘SCSDVs' are small circular ssDNA viruses and their associated satellites and include: Nanoviridae, Circoviridae and Gemniviridae, and satellite DNAs. The ‘other' category includes similarities to all viral types not fitting into the two previous groups (that is, in this study: Herpesviridae, Papillomaviridae and Polydnaviridae).
Table 2. Univariate comparisons of the relative abundance of the sequence similarities to eukaryotic viral families and satellite DNAs comprising ⩾4% of coral tissue metagenomes.
| Viral type | Significant pairwise differences | Kruskall–Wallis statistic | P-value |
|---|---|---|---|
| Circoviridae | H-D | 8.42 | ⩽0.04 |
| Herpesviridae | H-D | 9.20 | ⩽0.03 |
| Nanoviridae | H-D, BD-D | 9.57 | ⩽0.02 |
| Satellite DNAs | H-D, B-D | 10.92 | ⩽0.01 |
| Papillomaviridae | H-B, H-BD, H-D | 9.50 | ⩽0.02 |
| Ascoviridae, | — | 2.33 | >0.05 |
| Gemniviridae | — | 4.22 | >0.05 |
| Mimiviridae | — | 1.45 | >0.05 |
| Phycodnaviridae | — | 1.78 | >0.05 |
| Polydnaviridae | — | 1.01 | >0.05 |
| Poxviridae | — | 4.53 | >0.05 |
Abbreviations: B, Bleached; B, Bleached+WP Diseased; D, WP Diseased; H, Healthy; WP, white plague.
Bold value indicates significant difference (P<0.05).
Herpesviridae similarities were identified in all coral tissue types but at a higher relative abundance in Healthy than in WP Diseased tissue (62.18±4.5% vs 16.47±3.4% P<0.03; Table 2). Although common in coral tissues, these herpesvirus similarities were rare in adjacent seawater and comprised<1.5% of the similarities in either seawater virome (Figure 2a). Sequence similarities to at least some members of the NCLDV group also were present in all six virome types including similarities to members of the Phycodnaviridae, Mimiviridae and Ascoviridae. A higher relative proportion (>40%) of these viral sequences were identified in both bleached (B and BD) tissue types compared with Healthy or WP Diseased tissues (33% and 18%, respectively; Figures 2a and 3). Viral sequence similarities that constituted a minority (that is, <4% of eukaryotic viruses similarities) of the best annotations for each read also were examined for apparent patterns (Figure 2b). With the exception of the Healthy viromes, Baculoviridae and Iridoviridae similarities were present in viromes generated from all coral health states and seawater samples.
Diseased coral tissues contain a unique consortium of viral sequence similarities
To determine whether viral consortia differed among viromes generated from different coral health states as well as from adjacent seawater, multivariate analyses were performed. The WP Diseased viromes grouped (Supplementary Figure S1, red stars) in this analysis, and analysis of similarity confirmed that these viromes were statistically different from the other three tissue type viromes (Global R=0.238 and P-value=0.012; Table 3). The two Healthy samples (green hexagons) were located furthest from the WP Diseased tissues, depicting the minimal similarity between their viral consortia (Supplementary Figure S1). Seawater viromes (dark and light blue circles) also grouped closer to each other than to viromes generated from any coral tissue types (Supplementary Figure S1). Cluster analysis depicted relatedness between paired viromes generated from Bleached+WP Diseased and WP Diseased tissue of an individual colony; viral consortia from the same individual colonies clustered on different branches, with up to 55% dissimilarity (Supplementary Figure S2).
Table 3. ANOSIM results comparing sequence similarities in viral consortia from different health states and adjacent seawater.
| Sample type | H | B | BD | D | SWB | SWBD |
|---|---|---|---|---|---|---|
| H (n=2) | ||||||
| B (n=5) | R=−0.16 P=0.52 | |||||
| BD (n=7) | R=−0.18 P=0.69 | R=−0.08 P=0.76 | ||||
| D (n=7) | R=0.83 P=0.03 | R=0.36 P=0.03 | R=0.37 P=0.01 | |||
| SWB (n=1) | R=1.00 P=0.33 | R=0.24 P=0.33 | R=0.36 P=0.25 | R=−0.03 P=0.50 | ||
| SWBD (n=1) | R=1.00 P=0.33 | R=0.20 P=0.33 | R=0.43 P=0.25 | R=0.33 P=0.25 | NA | NA |
Abbreviations: ANOSIM, analysis of similarity; B, Bleached tissue; BD, Bleached+WP Diseased tissue; D, Diseased tissue; H, Healthy tissue; NA, not applicable; SWB, Seawater Bleached; SWBD, Seawater Bleached+WP Diseased.
Bold text indicates a significant difference between sample types.
Similarity of percentage analysis determined which viral families contributed to virome dissimilarity (Supplementary Table S2). Although similarities to Herpesviridae were present in viromes generated from all tissue health states, these sequences contributed to >31% of the dissimilarity between Healthy and WP Diseased tissue viromes, with herpesvirus similarities being the most abundant in Healthy tissues (Supplementary Table S2). Sequences similar to members of the SCSDVs contributed from 9% to 20% of the dissimilarity between WP Diseased and other coral virome types, and are strongly responsible for the uniqueness of the WP Diseased viral consortia (Supplementary Table S3). Poxviridae similarities contributed to dissimilarities among the Bleached+WP Diseased and Bleached viromes compared with the Healthy viromes (9.78% and 9.46%, respectively; Supplementary Table S2). Out of all health states, similarity of percentage analysis showed that the Healthy viromes were most similar to each other (70.94% similarity). Both Bleached+WP Diseased viromes and Bleached viromes each had approximately 43% similarity, whereas WP Diseased tissues viromes had intermediate similarity at 55.90% (Supplementary Table S2).
Diseased viromes contain higher eukaryotic viral diversity than healthy corals
To determine whether there were differences in viral diversity among coral tissue health states, Shannon's diversity index was calculated (Supplementary Table S3). Eukaryotic viral diversity was statistically variable among coral tissue types (P<0.05), with WP Diseased viromes having higher diversity than Healthy viromes (1.96±0.08 vs 0.98±0.19). Diversity between different types of bleached tissues (B and BD) was similar (1.30±0.10 and 1.35±0.11; Supplementary Table S3).
Diseased coral SCSDVs are distinct from Seawater SCSDVs
Phylogenetic trees were created using the viral Rep protein of SCSDVs identified within the seven WP Diseased and two Seawater viromes. The tree contained over 50 sequences, with all of the WP Diseased and Seawater viromes contributing at least one Rep sequence similarity (Supplementary Figure S3). Rep sequences from both Seawater viromes were generally located on separate branches (>60% support) of the tree than Rep sequences from WP Diseased coral viromes (Supplementary Figure S3). For example, two main coral SCSDV clades (stars) were distinct (72.4% and 95.0% bootstrap support) from a well-supported Seawater clade (89.9% bootstrap support; triangle; Supplementary Figure S3).
To confirm the origin of SCSDVs in WP Diseased libraries, representative SCSDV contigs were aligned to completely assembled and annotated genomes of circoviruses, nanoviruses and satellite DNAs. It was determined that putative WP Diseased contigs had broad similarities to fully sequenced and annotated circo- and nanoviruses, and not just within the Rep gene (Supplementary Figure S4). Further, average nucleotide identity of putative circoviruses were more similar to known circoviruses than other viral types (Supplementary Figure S4; df=11, P<0.01) when all pairwise comparisons were evaluated. Surprisingly, the putative nanoviruses from WP diseased libraries also were more similar to known circoviruses and our own putative circoviruses than to known and annotated nanoviruses (df=11, P<0.01).
Identification of intracellular VLPs corroborates genomic-based findings
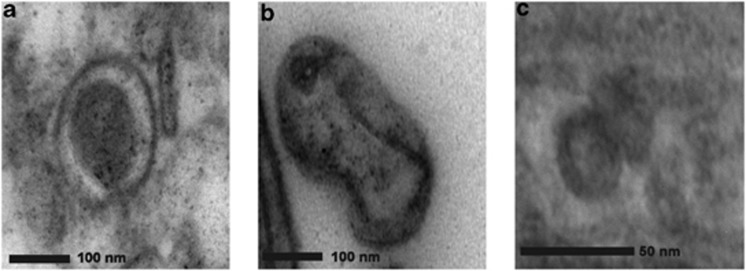
Electron micrographs were used to identify potential active viral infections within corals, and to corroborate the sequence-based findings that herpes-like viruses, SCSDVs and NCLDVs, were present in coral tissue. However, quantitative TEM analysis could not be performed because of the extensive variation in the quality of the tissues among different health states. For example, tissues were degraded in the WP Diseased samples and cell layers were difficult to discern, whereas Bleached tissues appeared more intact than either WP Diseased or Bleached+WP Diseased tissues. Healthy tissues exhibited intact Symbiodinium-host cell attachment and defined tissue layers, whereas signs of degradation were evident in Bleached tissue types (that is, Symbiodinium detachment; data not shown). Herpesvirus-like particles with a characteristic envelope that were ∼180 nm in diameter were observed in Bleached+WP Diseased and WP Diseased tissues (Figure 4a). In addition, distinctive figure 8-shaped poxvirus-like particles were visualized in WP Diseased tissues (Figure 4b). Finally, SCSDV-like particles were observed in WP Diseased tissues (Figure 4c; twinned gemini-like particle), however, TEM resolution was low, ∼20 nm (the size of a typical SCSDV particle), so the features of these VLPs are not well defined. No bacterial cells were observed within the degraded WP Diseased tissue, while in Healthy tissues small cell-like structures (<0.22 um) were commonly identified (data not shown).
Figure 4.
Transmission electron micrograph of viral particles detected in diseased M. annularis tissues. A herpes-like viral particle approximately 180 nm in width (a). A pox-like viral particle, approximately 200 nm in length (b). A gemini-like particle approximately 40 nm length (c).
Discussion
SCSDVs are associated with WP
Combining TEM and next-generation sequencing we have shown here that viruses are variably associated with different M. annularis coral health states. To examine the local oceanic viral consortia and to give insight into transmission mechanisms, we also generated viromes from seawater above Bleached and WP diseased corals. Our robust sampling, which in some cases generated up to seven replicate viromes per health state, allowed us to determine that a unique viral group, the coral small circular ssDNA viruses (SCSDVs), was associated with a WP disease outbreak in the Caribbean (Figures 2 and 3; Table 2).
SCSDVs are common pathogens of plants and animals and include the families: Circoviridae, Nanoviridae, Geminiviridae, but their prevalence and distribution in the environment was unknown until recently. Based on metagenomic analyses of animal hosts (for example, human, bat, rodent, pig and chimpanzee) and water samples, SCSDVs are now thought to be more common than previously thought (for review, Rosario et al., 2012).
Interestingly, SCSDVs have been found to be abundant in reclaimed waters and sewage, suggesting that SCSDVs in the diseased corals examined here could be linked to environment degradation (Blinkova et al., 2009; Rosario et al., 2009). Previously it has been shown that environmental viruses such as human enteroviruses and adenoviruses are present on coral mucus, suggesting that these associations were the result of anthropogenic pollution (for example, runoff; Lipp et al., 2002; Futch et al., 2010). Serratia marcescens, a human bacterial pathogen, also found in sewage was demonstrated to be the cause of white pox disease in acroporids; it is likely that other pathogens including viruses originate from sewage and negatively affect corals (Patterson et al., 2002; Sutherland et al., 2011). However, our analysis of the Rep proteins and genomes of coral and seawater SCSDVs sequenced in this study suggested that the coral SCSDVs are genetically distinct from those found in the surrounding seawater samples and are likely a unique component of the coral virome. An alternate hypothesis is that SCSDVs are present in healthy corals but are undetectable because they are either below the detection threshold of the methods used or present in a quiescent stage.
Nevertheless, it is evident from our analysis of both the conserved SCSDV Rep (Supplementary Figure S2 red lettering) sequences and the genome–genome alignments (Supplementary Figure S4) of our putative SCSDVs and known circo-, nano- and satellite viruses that these WP-associated SCSDV types are novel.
The host for the coral-associated SCSDVs presented here is yet to be determined, but may include the algal endosymbiont (Symbiodinium spp., because nanovirus/geminiviruses infect plants), the coral host itself (because circoviruses infect animals), or another member of the coral holobiont (Yu et al., 2010; Delwart and Li, 2012; Rosario et al., 2012;). However, a previous study examining viruses associated with Montastraea cavernosa coral colonies searched Symbiodinium EST libraries and found few similarities to SCSDVs (Correa et al., 2013) suggesting that the SCSDVs associated with diseased corals in this study are infecting the coral animal. In addition, WP disease signs include rapid tissue loss that suggests a coral (and not algal) pathogen is causing tissue necrosis. Furthermore if algal symbionts were affected, bleaching also would have been expected in the area of lesions; this was not observed (Figure 1).
NCLDVs and bleaching
Sequence similarities to NCLDVs, including similarities to members of the Phycodnaviridae, Poxviridae, Mimivirdae and Ascoviridae, were relatively more abundant in bleached coral tissue types than in non-bleached tissue types (Figure 3). Previously, poxvirus sequences were found to be more abundant in temperature-stressed corals (Vega Thurber et al., 2008), and a recent study on the scleractinian coral, Acropora millerpora, which is both geographically and evolutionary distinct from the Caribbean M. annularis, also contained pox-like virus sequences (Littman et al., 2011). This study also corroborates findings that have implicated NCLDVs in the infection of the algal symbiont Symbiodinium and provides more evidence that bleached host tissue contains viral types different from those found in healthy coral tissues, suggesting viruses may be involved in some thermally induced bleaching responses (Marhaver et al., 2008; Correa et al., 2013).
Herpesviruses dominate healthy tissues
Previous studies have shown that herpes-like viruses are a commonly observed in cnidarian viromes (Vega Thurber et al., 2008). This new work provides physical evidence that herpes-like particles are produced in coral tissues (Figure 4a). Given their cosmopolitan presence in healthy individuals of every coral genus tested (for example, Porites, Acropora, Montastraea and Diploria), we hypothesize that herpes-like viruses establish long-term non-fatal infections in corals, in a manner similar to their infections of vertebrate hosts (Knipe and Cliffe, 2008).
Viral diversity is altered in diseased corals
WP Diseased corals hosted the highest diversity of eukaryotic viruses, bleached corals had an intermediate viral diversity, and healthy corals exhibited the lowest viral diversity. As both the WP Diseased and Healthy tissues contained high abundances of Symbiodinium (based on qualitative analysis of TEM images), we suggest that the higher viral diversity in WP Diseased viromes (relative to Healthy viromes) was not related to the relative abundance of Symbiodinium in specimens of the two health states. Rather, the higher diversity of viral types found in WP Diseased corals may be a consequence of secondary infections resulting from a coral weakened from other viral infections. On the other hand, a higher viral diversity in WP Diseased corals may be the direct cause of disease signs. In either case, higher viral diversity may be an indicator of coral stress and may be used to diagnostically characterize corals in a disease state.
Caveats
MDA has been shown to bias metagenomic libraries toward ssDNA sequences and genomes because of the rolling circular amplification method that the Phi29 polymerase uses (Kim et al., 2008; Kim and Bae, 2011). Although it is unclear whether MDA bias is stochastic (Abulencia et al., 2006) or linear (Yilmaz et al., 2010), we calculated that the probability that all WP Diseased (n=7) but no Healthy (n=2) samples contained SCSDV-like sequences was <0.0079 (assuming MDA bias toward ssDNA is 50% or less). Even with a 95% bias of MDA toward SCSDVs, we calculated that there is still <0.7% chance of obtaining these results. Further, much of the debate about the utility of MDA for viral metagenomics comes from studies where samples were processed in different ways. For example, some studies compared the results of MDA amplified versus unamplified libraries, or to data sets generated using amplification methods that do not amplify ssDNA viruses (such as LASLs), or even after the elimination of a denaturing step that would enrich for ssDNA viruses (Kim et al.; 2008; Kim and Bae, 2011). As all of our samples were processed in an identical, scientifically rigorous manner, any bias toward ssDNA sequences should be expressed uniformly in each of the sample libraries. As only the WP Diseased and seawater viromes, but not the bleached and healthy viromes, contained high amounts of these viral types, it is evident that the changes detected in the relative abundance of similarities to these viral types are not the result of MDA. Finally, a previous comparative study on stressed Pacific coral species (not infected with WP) also used MDA but detected SCSDV sequences only in 2 viromes out of 6 (Vega Thurber et al., 2008), while another study on another Pacific coral species using MDA amplification identified <5% nano/circo viruses with no similarities to satellite DNAs or geminiviruses (Wegley et al., 2007).
These conclusions are based on correlated data, yet, ultimately only direct infection studies, such as exposing healthy corals to viruses isolated from diseased corals in a controlled laboratory setting, can determine whether these suspected SCSDV pathogens cause WP-like disease or are alternatively secondary infections resulting from altered coral physiology. Until a proper infection model is developed, correlation with disease prevalence and incidence will be the best evidence as to causes of coral diseases. In addition, this study only examined DNA viruses; it is possible that RNA viruses are involved in WP and future experiments should aim to evaluate viruses with RNA genomes.
Finally, a majority of our viral sequences (78.33%) were not similar to a known virus or group of viruses. Therefore, although we find significant differences in these libraries, they are based on a fraction of the total data. Yet to pointedly assess the prevalence of already known virus, described as disease agents across these healthy and disease states, we narrowed our focus to taxonomically described viruses.
Conclusions
This study aimed to determine the viruses associated with WP-infected corals. Our microscopy data found no evidence of foreign microbial cells present in WP Diseased corals, strongly suggesting that bacterial or small eukaryotic pathogen infection are not the causes of the examined disease. We also found heterogeneity in the viral consortia among tissue types isolated from the same coral colonies. Bleached+WP Diseased tissues were more similar to Bleached tissues than the WP-Diseased area of the same colony. We thus hypothesize that WP infection is only localized to the disease front, and that bleached and WP Diseased tissues are distinct in their viral composition. Bacteria associated with coral mucus have been shown to be spatially heterogeneous (Daniels et al., 2011), and here we demonstrate viral heterogeneity in a coral colony. Ultimately, we have shown that healthy corals have more abundant viral similarities to Herpesviridae, bleached corals possess more viral similarities to NCLDVs, such as Phycodnaviridae and Poxviridae, and diseased tissues contain an abundance of unique SCSDVs including members similar to Circoviridae and Nanoviridae, as well as their associated satellites; this novel SCSD viral group therefore may be responsible for WP infections in M. annularis.
Acknowledgments
We thank the USVI Department of Planning and Natural Resources for allowing us to perform coral collections (permit #STT-050-10). We thank the faculty at the University of the Virgin Islands St Thomas campus for their hospitality. We also thank Dr Pat Blackwelder and Husain Al-Sayegh from University of Miami Center for Advanced Microscopy for their assistance in sample preparation and viewing. We thank four anonymous reviewers for their efforts and input. This work was funded by the National Science Foundation OCE Grant/Award ID 0960937 to RVT and the National Graduate Research Fellowship to NS (1000036136). We also thank the National Science Foundation, through the VI Experimental Program to Stimulate Competitive Research (VI-EPSCoR), which contributed valuable infrastructure support and research equipment.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abulencia CB, Wyborski DL, Garcia JA, Podar M, Chen W, Chang SH. Environmental whole-genome amplification to access microbial populations in contaminated sediments. Appl Environ Microbiol. 2006;72:3291–3301. doi: 10.1128/AEM.72.5.3291-3301.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Atad I, Zvuloni A, Loya Y, Rosenberg E. Phage therapy of the white plague-like disease of Favia favus in the Red Sea. Coral reefs. 1990;31:665–670. [Google Scholar]
- Bermingham A, Chand M, Brown C, Aarons E, Tong C, Langrish C, et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Eur Comm Dis Bull. 2012;17:1–5. [PubMed] [Google Scholar]
- Blinkova O, Rosario K, Li L, Kapoor A, Slikas B, Bernardin F, et al. Frequent detection of highly diverse variants of cardiovirus, cosavirus, bocavirus, and circovirus in sewage samples collected in the United States. J of clin Microbio. 2009;l47:3507–3513. doi: 10.1128/JCM.01062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomström AL, Widén F, Hammer AS, Belák S, Berg M. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J Clin Microbiol. 2010;48:4392–4396. doi: 10.1128/JCM.01040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne DG, Garren M, Wor TM, Rosenberg E, Smith GW, Harvell CD. Microbial disease and the coral holobiont. Trends Microbiol. 2009;17:554–562. doi: 10.1016/j.tim.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Brandt ME, Smith TB, AMS Correa, Vega-Thurber R. Disturbance driven colony fragmentation as a driver of a coral disease outbreak. PLoS One. 2013;8:e57164. doi: 10.1371/journal.pone.0057164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bythell JC, Pantos O, Richardson L.2004White plague, white band and other ‘white' diseasesIn Eugene Rosenberg, Loya Yossi, (eds)Coral Health and Disease 351–365.Springer-Verlag: New York, NY, USA [Google Scholar]
- Cárdenas A, Rodriguez-R LM, Pizarro V, Cadavid LF, Arévalo-Ferro C. Shifts in bacterial communities of two Caribbean reef-building coral species affected by white plague disease. ISME J. 2012;6:502–512. doi: 10.1038/ismej.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa AMS, Welsh RM, Vega Thurber RL. Unique nucleocytoplasmic dsDNA and +ssRNA viruses are associated with the dinoflagellate endosymbionts of corals. ISME J. 2013;7:13–27. doi: 10.1038/ismej.2012.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KR, Gorley RN. PRIMER v6: User Manual/Tutorial. PRIMER-E: Plymouth; 2006. [Google Scholar]
- Clarke KR, Warwick RM.2001Change in Marine Communities: An Approach to Statistical Analysis and Interpretation2nd ednPRIMER-E Plymouth, MA, USA [Google Scholar]
- Daniels C, Zeifman A, Heym K, Ritchie K, Watson C, Berzins I, et al. Spatial heterogeneity of bacterial communities in the mucus of Montastraea annularis. MEPS. 2011;426:29–40. [Google Scholar]
- Davy JE, Patten NL. Morphological diversity of virus-like particles within the surface microlayer of scleractinian corals. Aquat Microb Ecol. 2007;47:37–44. [Google Scholar]
- Delwart E, Li L. Rapidly expanding genetic diversity and host range of the Circoviridae viral family and other Rep encoding small circular ssDNA genomes. Virus Res. 2012;164:1–2. doi: 10.1016/j.virusres.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denner EBM, Smith GW, Busse HJ, Schumann P, Narzt T, Polson SW, et al. Aurantimonas coralicida gen. nov., sp. nov., the causative agent of white plague type II on Caribbean scleractinian corals. Int J Syst Evol Microbiol. 2003;53:1115–1122. doi: 10.1099/ijs.0.02359-0. [DOI] [PubMed] [Google Scholar]
- Dustan P. Vitality of reef coral populations off Key Largo, Florida: recruitment and mortality. Environ Geol. 1977;2:51–58. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efrony R, Atad I, Rosenberg E. Phage therapy of coral white plague disease: properties of phage BA3. Curr Microbiol. 2009;58:139–145. doi: 10.1007/s00284-008-9290-x. [DOI] [PubMed] [Google Scholar]
- Futch JC, Griffin DW, Lipp EK. Human enteric viruses in groundwater indicate offshore transport of human sewage to coral reefs of the Upper Florida Keys. Environ Microbial. 2010;12:964–974. doi: 10.1111/j.1462-2920.2010.02141.x. [DOI] [PubMed] [Google Scholar]
- Gardner TA, Côté IM, Gill JA, Grant A, Watkinson AR. Long-term region-wide declines in Caribbean corals. Science. 2003;301:958–960. doi: 10.1126/science.1086050. [DOI] [PubMed] [Google Scholar]
- Gibbs MJ, Weiller GF. Evidence that a plant virus switched hosts to infect a vertebrate and then recombined with a vertebrate-infecting virus. Proc Natl Acad Sci USA. 1999;96:8022–8027. doi: 10.1073/pnas.96.14.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, The Galaxy Team Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EP, Bruckner AW. The significance of coral disease epizootiology for coral reef conservation. Biol Cons. 2000;96:347–361. [Google Scholar]
- Harvell CD, Kim K, Burkholder JM, Colwell RR, Epstein PR, Grimes DJ, et al. Emerging marine diseases-climate links and anthropogenic factors. Science. 2004;285:1505–1510. doi: 10.1126/science.285.5433.1505. [DOI] [PubMed] [Google Scholar]
- Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Bae JW. Amplification methods bias metagenomics libraries of uncultured single-stranded and double-stranded DNA viruses. Appl Environ Microbiol. 2011;77:7663–7668. doi: 10.1128/AEM.00289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Chang HW, Nam YD, Roh SW, Kim MS, Sung Y. Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl Environ Microbiol. 2008;74:5975–5985. doi: 10.1128/AEM.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe DM, Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol. 2008;6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- Lipp EK, Jarrell JL, Griffin DW, Lukasik J, Jacukiewicz J, Rose JB. Preliminary evidence for human fecal contamination in corals of the Florida Keys, USA. Mar Pol Bull. 2002;44:666–670. doi: 10.1016/s0025-326x(01)00332-0. [DOI] [PubMed] [Google Scholar]
- Lipp EK, Futch JC, Griffin DW. Analysis of multiple enteric viral targets as sewage markers in coral reefs. Mar Poll Bull. 2007;54:1897–1902. doi: 10.1016/j.marpolbul.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Littman R, Willis BL, Bourne DG. Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ Microbiol. 2011;3:651–660. doi: 10.1111/j.1758-2229.2010.00234.x. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhaver KL, Edwards RA, Rohwer F. Viral communities associated with healthy and bleaching corals. Environ Microbiol. 2008;10:2277–2286. doi: 10.1111/j.1462-2920.2008.01652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DP, Biagini P, Lefeuvre P, Golden M, Roumagnac P, Varsani A. Recombination in eukaryotic single stranded DNA viruses. Viruses. 2011;3:1699–1738. doi: 10.3390/v3091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AW, Blackwelder P, Al-Sayegh H, Richardson LL. Fine-structural analysis of black band disease-infected coral reveals boring cyanobacteria and novel bacteria. Dis Aqua Org. 2011;93:179–190. doi: 10.3354/dao02305. [DOI] [PubMed] [Google Scholar]
- Miller J, Waara R, Muller E, Rogers C. Coral bleaching and disease combine to cause extensive mortality on reefs in US Virgin Islands. Coral Reefs. 2006;25:418. [Google Scholar]
- Mokili JL, Rohwer F, Dutilh BE. Metagenomics and future perspectives in virus discovery. Curr Opin Virol. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TF, Suedmeyer WK, Wheeler E, Gulland F, Breitbart M. Novel anellovirus discovered from a mortality event of captive California sea lions. J Gen Virol. 2009;90:1256–1261. doi: 10.1099/vir.0.008987-0. [DOI] [PubMed] [Google Scholar]
- Pantos O, Cooney RP, MDAL Tissier, Barer MR, Donnell AGO, Bythell JC. The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environ Microbiol. 2003;5:370–382. doi: 10.1046/j.1462-2920.2003.00427.x. [DOI] [PubMed] [Google Scholar]
- Patten NL, Harrison PL, Mitchell JG. Prevalence of virus-like particles within a staghorn scleractinian coral (Acropora muricata) from the Great Barrier Reef. Coral Reefs. 2008;27:569–580. [Google Scholar]
- Patterson KL, Porter JW, Ritchie KB, Polson SW, Mueller E, Peters EC, et al. The etiology of white pox, a lethal disease of the Caribbean elkhorn coral, Acropora palmata. Proc Natl Acad Sci USA. 2002;99:8725–8730. doi: 10.1073/pnas.092260099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock FJ, Morris PJ, Willis BL, Bourne DG. The urgent need for robust coral disease diagnostics. PLoS Path. 2011;7:1–10. doi: 10.1371/journal.ppat.1002183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson LL, Goldberg W, Kuta K. Florida's mystery coral-killer identified. Nature. 1998;392:557–558. [Google Scholar]
- Richardson LL, Smith GW, Ritchie KB, Carlton RG. Integrating microbiological, microsensor, molecular, and physiologic techniques in the study of coral disease pathogenesis. Hydrobiologia. 2001;460:71–89. [Google Scholar]
- Rogers C. Coral bleaching and disease should not be underestimated as causes of Caribbean coral reef decline. Proc R Soc B. 2009;276:197–198. doi: 10.1098/rspb.2008.0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K, Duffy S, Breitbart M. A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch Virol. 2012;157:1851–1871. doi: 10.1007/s00705-012-1391-y. [DOI] [PubMed] [Google Scholar]
- Rosario K, Nilsson C, Lim YW, Ruan Y, Breitbart M. Metagenomic analysis of viruses in reclaimed water. Environ Microbiol. 2009;11:2806–2820. doi: 10.1111/j.1462-2920.2009.01964.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol. 2007;5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- Roux S, Faubladier M, Mahul A, Paulhe N, Bernard A, Debroas D, et al. Metavir: a web server dedicated to virome analysis. Bioinformatics. 2011;27:3074–3075. doi: 10.1093/bioinformatics/btr519. [DOI] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS One. 2011;6:e17288. doi: 10.1371/journal.pone.0017288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolow S. Effects of a changing climate on the dynamics of coral infectious disease: a review of the evidence. Dis Aquat Org. 2009;87:5–18. doi: 10.3354/dao02099. [DOI] [PubMed] [Google Scholar]
- Sun S, Chen J, Li W, Altinatas I, Lin A, Peltier S, et al. Community cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: the CAMERA resource. Nucleic Acids Res. 2011;39:546–551. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, Desantis TZ, Piceno YM, Brodie EL, Desalvo MK, Voolstra CR, et al. Bacterial diversity and white plague disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 2009;131:512–521. doi: 10.1038/ismej.2008.131. [DOI] [PubMed] [Google Scholar]
- Sutherland K, Porter JW, Cecilia T. Disease and immunity in Caribbean and Indo-Pacific zooxanthellate corals. Marine Ecol Prog Ser. 2004;266:273–302. [Google Scholar]
- Sutherland KP, Shaban S, Joyner JL, Porter JW, Lipp EK. Human pathogen shown to cause disease in the threatened Eklhorn coral Acropora palmata. PLoS One. 2011;6:e23468. doi: 10.1371/journal.pone.0023468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson FL, Barash Y, Sawabe T, Sharon G, Swings J, Rosenberg E. Thalassomonas loyana sp. nov., a causative agent of the white plague-like disease of corals on the Eilat coral reef. Int J Syst Evol Microbiol. 2006;56:365–368. doi: 10.1099/ijs.0.63800-0. [DOI] [PubMed] [Google Scholar]
- Vega Thurber RL, Barott KL, Hall D, Liu H, Rodriguez-Mueller B, Desnues C, et al. Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc Natl Acad Sci USA. 2008;105:18413–18418. doi: 10.1073/pnas.0808985105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega Thurber RL, Correa AMS. Viruses of reef-building scleractinian corals. JEMBE. 2011;408:102–113. [Google Scholar]
- Vega Thurber RL, Willner-hall D, Rodriguez-mueller B, Desnues C, Edwards RA, Angly F, et al. Metagenomic analysis of stressed coral holobionts. Environ Microbiol. 2009;11:2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- Ward JR, Lafferty KD. The elusive baseline of marine disease: are diseases in ocean ecosystems increasing. PLoS Biology. 2004;2:0543–0547. doi: 10.1371/journal.pbio.0020120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol. 2007;9:2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- Weil E, Smith G, Gil-agudelo DL. Status and progress in coral reef disease research. Dis Aqua Organ. 2006;69:1–7. doi: 10.3354/dao069001. [DOI] [PubMed] [Google Scholar]
- Wilson WH, Dale AL, Davy JE, Davy SK. An enemy within? Observations of virus-like particles in reef corals. Coral Reefs. 2004;24:145–148. [Google Scholar]
- Wilson WH, Van Etten JL, Allen MJ. The Phycodnaviridae: the story of how tiny giants rule the world. Curr Top Microbiol Immunol. 2009;328:1–42. doi: 10.1007/978-3-540-68618-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz S, Allgaier M, Hugenholtz P. Multiple displacement amplification compromises quantitative analysis of metagenomes. Nat Methods. 2010;7:943–944. doi: 10.1038/nmeth1210-943. [DOI] [PubMed] [Google Scholar]
- Yu X, Li B, Fu YP, Jiang DH, Ghabrial SA, Li GQ, et al. A Gemini virus related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci USA. 2010;107:8387–8392. doi: 10.1073/pnas.0913535107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.