Abstract
Marine Group A (MGA) is a deeply branching and uncultivated phylum of bacteria. Although their functional roles remain elusive, MGA subgroups are particularly abundant and diverse in oxygen minimum zones and permanent or seasonally stratified anoxic basins, suggesting metabolic adaptation to oxygen-deficiency. Here, we expand a previous survey of MGA diversity in O2-deficient waters of the Northeast subarctic Pacific Ocean (NESAP) to include Saanich Inlet (SI), an anoxic fjord with seasonal O2 gradients and periodic sulfide accumulation. Phylogenetic analysis of small subunit ribosomal RNA (16S rRNA) gene clone libraries recovered five previously described MGA subgroups and defined three novel subgroups (SHBH1141, SHBH391, and SHAN400) in SI. To discern the functional properties of MGA residing along gradients of O2 in the NESAP and SI, we identified and sequenced to completion 14 fosmids harboring MGA-associated 16S RNA genes from a collection of 46 fosmid libraries sourced from NESAP and SI waters. Comparative analysis of these fosmids, in addition to four publicly available MGA-associated large-insert DNA fragments from Hawaii Ocean Time-series and Monterey Bay, revealed widespread genomic differentiation proximal to the ribosomal RNA operon that did not consistently reflect subgroup partitioning patterns observed in 16S rRNA gene clone libraries. Predicted protein-coding genes associated with adaptation to O2-deficiency and sulfur-based energy metabolism were detected on multiple fosmids, including polysulfide reductase (psrABC), implicated in dissimilatory polysulfide reduction to hydrogen sulfide and dissimilatory sulfur oxidation. These results posit a potential role for specific MGA subgroups in the marine sulfur cycle.
Keywords: marine, candidate phylum, sulfur cycle, marine group A, oxygen minimum zone, polysulfide
Introduction
Marine Group A (MGA) bacteria were first identified in small subunit ribosomal RNA (16S rRNA) gene clone libraries generated from surface waters of the Atlantic and Pacific Oceans (Fuhrman et al., 1993; Gordon and Giovannoni, 1996; Fuhrman and Davis, 1997). MGA, originally referred to as the ‘SAR406 gene lineage', represents a deeply branching lineage of bacteria related to the genus Fibrobacter and the green sulfur bacterial (GSB) phylum, which includes the genus Chlorobium (Gordon and Giovannoni, 1996). To date, MGA remains a candidate phylum with no cultured representatives. Modern phylogenetic analyses indicate that the closest cultivated relatives of MGA are Caldithrix abyssi and Caldithrix palaeochoryensis, both belonging to the phylum Caldithrix. These isolates are anaerobic, mixotrophic, thermophiles obtained from hydrothermal vent and sediment environments, respectively (Miroshnichenko et al., 2003, 2010). Although ubiquitous in the dark ocean, MGA are most prevalent and diverse in interior regions of the ocean with distinct oxyclines, such as in oxygen minimum zones (OMZs) and permanent or seasonally stratified anoxic basins (Madrid et al., 2001; Fuchs et al., 2005; Stevens and Ulloa, 2008; Schattenhofer et al., 2009; Zaikova et al., 2010; Allers et al., 2012; Wright et al., 2012). At present, the metabolic capacity and ecological roles of MGA in OMZs or in the ocean at large remain entirely unknown. Given that OMZs are expanding and intensifying (Emerson et al., 2004; Whitney et al., 2007; Bograd et al., 2008; Stramma et al., 2008; Helm et al., 2011), primarily as a result of global climate change (Keeling et al., 2010), it is of increasing importance to define the metabolic diversity and ecosystem function of dominant microorganisms within these systems in order to predict the systemic impacts of OMZ expansion on ocean ecology and biogeochemistry.
The distribution of certain MGA subgroups was reported as being negatively correlated with the concentration of dissolved oxygen (O2) in the OMZ of the Northeast subarctic Pacific Ocean (NESAP), suggesting a potential role for O2 as a driver of MGA habitat selection and metabolic adaption in these waters (Allers et al., 2012). Determining the extent to which 16S rRNA-based patterns of MGA distribution represent ecological types (ecotypes) differentiating in response to selective environmental pressures such as O2 deficiency requires genome-scale sequence data associated with multiple MGA subgroups to query for changes in genome composition that might promote differential fitness across the oxycline. Here, we explore potential niche partitioning among and within MGA subgroups along oxic to anoxic-sulfidic gradients of dissolved O2 in the North Pacific Ocean. In the absence of reference genomes representative of the MGA candidate phylum, we use phylogenetic anchor screening to identify 18 large-insert DNA fragments affiliated with various MGA subgroups as a direct route to studying MGA function. We describe and compare the genetic content and organization of these large-insert DNA fragments to gain preliminary insights into MGA metabolism.
Materials and methods
Sample collection and processing in the NESAP
Sampling in the NESAP was conducted via multiple hydrocasts using a Conductivity, Temperature, Depth (CTD) rosette water sampler aboard the CCGS John P. Tully during Line P cruises 2009-09 June 2009 (major stations: P4 (48°39.0N, 126°4.0W) - 7 June, P12 (48°58.2N, 130°40.0W) - 9 June and P26 (50°N, 145°W) - 14 June; 2009-10 August 2009 (major stations P4 - 21 August, P12 - 23 August, P26 - 27 August; and 2010-01 February 2010 (major stations: P4 - 4 February, P12 - 11 February) (Supplementary Figure S1). At these stations, large volume (20 l) samples for DNA isolation were collected from the surface (10 m), whereas 120 l samples were taken from three depths spanning the OMZ core and upper and deep oxyclines (500 m, 1000 m and 1300 m at station P4; 500 m, 1000 m and 2000 m at station P12). Sampling at Saanich Inlet (SI) station S3 (48°35.30N, 123°30.22W) was performed as previously described (Zaikova et al., 2010) as part of a monthly monitoring program aboard the MSV John Strickland. Sample collection and filtration protocols can be viewed as visualized experiments at http://www.jove.com/video/1159/ (Zaikova et al., 2009) and http://www.jove.com/video/1161/ (Walsh et al., 2009), respectively.
Environmental DNA extraction
DNA was extracted from sterivex filters as described by Zaikova and colleagues (Zaikova et al., 2010) and DeLong and colleagues (DeLong et al., 2006). The DNA extraction protocol can be viewed as a visualized experiment at http://www.jove.com/video/1352/ (Wright et al., 2009).
Phylogenetic analysis and tree construction using MGA 16S rRNA gene sequences
Phylogenetic analysis and tree construction using full-length 16S rRNA gene clone sequences from the NESAP and SI and 16S rRNA gene sequences identified on large-insert DNA fragments was performed as reported previously (Allers et al., 2012); see Supplementary Methods for details.
Fosmid library construction and end sequencing
Thirty fosmid libraries (∼7680 clones/libraries) were constructed from DNA samples collected from Line P stations P4, P12, and P26 in June and August of 2009, and stations P4 and P12 during February 2010 (Supplementary Table S1, Supplementary Figure S1). An additional 16 fosmid libraries were constructed from DNA samples collected from SI station S3 during the 2006-2007 seasonal stratification and deep-water renewal cycle (Supplementary Table S1, Supplementary Figure S1) (Walsh et al., 2009). Further details on fosmid library construction and sequencing can be found in Supplementary Methods.
Fosmid library screening, preparation, and full-length sequencing
Twenty three of the 46 fosmid end sequenced libraries described above including 7 from Line P and 16 from SI were screened for the presence of 16S rRNA genes using the NAST aligner (DeSantis et al., 2006a) and BLAST using default parameters against the 2008 Greengenes database (DeSantis et al., 2006) (Supplementary Figure S1). After preliminary phylogenetic analyses, 14 fosmid clones containing MGA-affiliated 16S rRNA genes were selected for complete sequencing (8 fosmids from Line P libraries and 6 fosmids from SI libraries; Table 1, Supplementary Figure S1). For sequencing protocols, see Supplementary Methods.
Table 1. Characterization of large-insert DNA fragments containing MGA 16S rRNA genes.
| Fosmid insert size (bp) | GC content (%) | No. of ORFS | No. of ORFs encoding hypotheticals | Accession number | Sampling location | Sampling coordinates | Collection date | Depth (m) | [O2] (μmol kg−1) | Phylogenetic Identity | Syntenic group | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Saanich Inlet | |||||||||||||
| FPPP_13C3 | 40 370 | 32.8 | 36 | 19 | KF170421 | Station S3 | 48°58′N, 123°50′W | Nov-06 | 10 | 249.0 | Arctic96B-7 | II | This study |
| FPPP_33K14 | 27 446 | 37.4 | 17 | 15 | KF170417 | Station S3 | 48°58′N, 123°50′W | Nov-06 | 10 | 249.0 | A714018 | IV | This study |
| FGYC_13M19 | 32 678 | 43.6 | 16 | 11 | KF170416 | Station S3 | 48°58′N, 123°50′W | Feb-06 | 125 | 5.0 | SHBH391 | I | This study |
| FPPS_57A9 | 32 217 | 37.9 | 21 | 13 | KF170418 | Station S3 | 48°58′N, 123°50′W | Nov-06 | 100 | 15.4 | SHAN400 | I | This study |
| FPPU_33B15 | 32 230 | 37.0 | 14 | 12 | KF170419 | Station S3 | 48°58′N, 123°50′W | Nov-06 | 200 | 54.0 | SAR406 | IV | This study |
| FPPZ_5C6a | 41 899 | 38.2 | 36 | 26 | KF170420 | Station S3 | 48°58′N, 123°50′W | Apr-07 | 200 | 1.1 | Arctic96B-7 | I | This study |
| Monterey Bay | |||||||||||||
| EBAC750-03B02 | 34 714 | 42.0 | 21 | 15 | AY458631 | Monterey Bay | 36°41′N, 122°02′W | Apr-00 | 750 | 16.5 | Arctic95A-2 | III | Suzuki et al., 2004 |
| Hawaii Ocean Time-series | |||||||||||||
| HF0010_18O13 | 37 088 | 33.0 | 33 | 17 | GU474850 | Station ALOHA | 22°45′'N, 158°00'W | Oct-02 | 10 | 204.6 | ZA3312c | I | DeLong et al., 2006 |
| HF0500_01L02 | 32 792 | 37.7 | 25 | 19 | GU474916 | Station ALOHA | 22°45′'N, 158°00'W | Oct-02 | 500 | 118.0 | Arctic96B-7 | I | DeLong et al., 2006 |
| HF4000_22B16 | 31 597 | 43.0 | 17 | 15 | GU474892 | Station ALOHA | 22°45′'N, 158°00'W | Dec-03 | 4000 | 147.8 | P262000N21 | IV | DeLong et al., 2006 |
| NESAP | |||||||||||||
| 405006-B04 | 43 543 | 42.8 | 46 | 32 | KF170424 | Station P4 | 48°39′N, 126°40′W | Feb-10 | 500 | 40.4 | SAR406 | I | This study |
| 4050020-J15 | 43 860 | 39.3 | 49 | 39 | KF170415 | Station P4 | 22°45′'N, 158°00'W | Feb-10 | 500 | 40.4 | Deferribacteres-like | V | This study |
| 413004-H17 | 35 813 | 39.8 | 45 | 29 | KF170425 | Station P4 | 22°45′'N, 158°00'W | Feb-10 | 1300 | 22.7 | Arctic96B-7 | I | This study |
| 4130011-I07 | 34 533 | 41.5 | 39 | 27 | KF170426 | Station P4 | 22°45′'N, 158°00'W | Feb-10 | 1300 | 22.7 | Arctic95A-2 | III | This study |
| 413009-K18 | 35 438 | 41.4 | 25 | 23 | KF170413 | Station P4 | 22°45′'N, 158°00'W | Feb-10 | 1300 | 22.7 | SAR406 | IV | This study |
| 125003-E23 | 34 144 | 37.3 | 41 | 28 | KF170423 | Station P12 | 48°58′N, 130°40′W | Feb-10 | 500 | 32.2 | Arctic96B-7 | I | This study |
| 1250012-L08 | 34 444 | 40.6 | 35 | 26 | KF170414 | Station P12 | 22°45′'N, 158°00'W | Feb-10 | 500 | 32.2 | Arctic95A-2 | III | This study |
| 122006-I05 | 35 932 | 47.7 | 45 | 26 | KF170422 | Station P12 | 22°45′'N, 158°00'W | Feb-10 | 2000 | 59.5 | P262000D03 | II | This study |
Abbreviations: MGA, Marine Group A; ORF, open reading frames.
This fosmid was derived from an H2S-containing sample.
GC content and oligonucleotide frequency analysis
GC content of large-insert DNA fragments (14 fosmids from NESAP and SI in addition to four large-insert DNA fragments from other North Pacific Ocean environments; Table 1) was calculated using gccontent.pl with default parameters, available for download at https://github.com/hallamlab/utilities. Tetranucleotide frequencies were calculated as normalized Z-scores using TETRA (Teeling et al., 2004a, 2004b; http://www.megx.net/tetra). Principal component analysis was performed on normalized Z-score profiles for each insert using PRIMER v6.1.13 (Clarke, 1993; Clarke and Gorley, 2006). Principal component analysis was overlaid with clusters determined by Hierarchical Cluster Analysis of normalized Z-scores using a Euclidean distance matrix (also performed in PRIMER).
Global nucleotide similarity analysis
Global nucleotide similarity in large-insert DNA fragments was determined by performing pairwise blastn comparisons between all fragments using onecircos.pl with default settings for all parameters except percent_identity (-p), which was calculated at 50%, 80%, 90% and 95% in separate analyses. onecircos.pl is available for download at: https://github.com/hallamlab/utilities and is based on Circos (http://circos.ca/; Krzywinski et al., 2009).
Open reading frame prediction and gene annotation
Open reading frames (ORFS) were predicted and annotated using the in-house MetaPathways pipeline (Konwar et al., 2013), available for download at: http://hallam.microbiology.ubc.ca/MetaPathways/. Briefly, primary nucleotide sequences from large-insert DNA fragments were quality controlled for ambiguous bases and file-format errors. ORFs were predicted using Prodigal (Hyatt et al., 2010). ORFs shorter than 60 amino acids in length were removed and were annotated using Protein BLAST (Altschul et al., 1990) (bit-score ratio >0.4 (Rasko et al., 2005), e-value=1e−5) against the RefSeq (Pruitt and Maglott, 2001), KEGG (Kanehisa and Goto, 1999), COG (Tatusov et al., 2001 ) and MetaCyc (Karp et al., 2000) databases. Annotations were assigned to predicted ORFs based on the following four criteria: (i) the BLAST hit with top e-value was selected from each database; (ii) each BLAST hit was assigned an ‘information score' based on the sum of distinct and shared enzymatic words (prepositions, articles and auxiliary verbs were removed) with a preference for Enzyme Commission numbers (+10 score); (iii) the annotation with the highest score was selected and assigned to the respective ORF; (iv) ORFs with no hits were assigned the annotation ‘hypothetical protein'.
Amino acid similarity analysis
Predicted amino acid similarity of large-insert DNA fragments was plotted in Trebol (available for download at: http://bioinf.udec.cl/trebol) using tblastx with a minimum bit-score cutoff of 50. COG categories present on large-insert fragments were plotted using tblastn of COG proteins against large-insert DNA fragments with a minimum e-value cutoff of 1e-4.
Fragment recruitment of fosmid end sequences
Coverage plots relating fosmid end sequences from individual NESAP and SI fosmid end libraries to large-insert DNA fragments were generated using the Nucmer program implemented in MUMmer 3.23 (Kurtz et al., 2004) as cited in (Hallam et al., 2006). Further details on fragment recruitment can be found in Supplementary Methods.
Phylogenetic analysis of PsrABC
Protein sequences (including predicted protein sequences for PsrA, PsrB, and PsrC identified on fosmids FPPP_13C3 and 122006-I05) were aligned using MUSCLE v3.6 with default parameters (Edgar, 2004). For the purposes of this analysis, the PsrBC fusion proteins encoded by psrBC on fosmid FPPP_13C3 and on certain reference sequences were divided into PsrB and PsrC subunits and analyzed in separate trees. Phylogenetic analyses were performed using PHYML (Guindon et al., 2005) using a WAG model of amino-acid substitution, where the parameter of the G distribution and the proportion of invariable sites were estimated for each data set. The confidence of each node was determined by assembling a consensus tree of 100 bootstrap replicates. The presence of TAT signal sequences on PsrA proteins was predicted using TatP 1.0 (Bendtsen et al., 2005), available at: http://www.cbs.dtu.dk/services/TatP/.
Results
Physiochemical characteristics of the NESAP and SI
This study was conducted along the Line P transect of the NESAP (Supplementary Figure S1), beginning in SI, Vancouver Island, British Columbia (SI, Station S3: 48°58′N, 123°50′W) and ending at Ocean Station Papa (also referred to as station P26: 50°N, 145°W) (Freeland, 2007). Owing to strong stratification and sluggish circulation of the interior NESAP waters, a large region of O2-deficient (<90 μmol kg−1) water containing dysoxic (20–90 μmol kg−1) and suboxic (1–20 μmol kg−1) compartments spans from ∼400 m to 2000 m in depth resulting in a persistent OMZ (O2 <20 μmol kg−1). The OMZ is centered at 1000 m, wherein dissolved O2 concentrations typically drop to ∼9 μmol kg−1 (Whitney et al., 2007). During the past 50 years of oceanographic observation, O2 concentrations in the OMZ of coastal to open-ocean regions of the NESAP have not been observed to reach anoxic (<1 μmol kg−1) levels. However, interior and basin waters of SI typically experience seasonal periods of anoxia and sulfide accumulation on an annually recurring basis (Anderson and Devol, 1973; Lilley et al., 1982; Ward et al., 1989). Physicochemical data from basin (S3), coastal (P4), transition (P12) and open-ocean (P26) stations measured along the Line P transect relevant to the present study are provided in Table 1 and Supplementary Table S1.
Taxonomic diversity of MGA in the NESAP and SI
To identify 16S rRNA genes affiliated with MGA inhabiting SI waters, we screened 19 previously published bacterial 16S rRNA gene clone libraries (containing a total of 6645 sequences) generated from samples traversing the water column during the 2006-2007 seasonal stratification and deep-water renewal cycle and during the spring stratification in 2008 at Station S3 (Supplementary Table S1; Walsh and Hallam, 2011). A total of 415 16S rRNA gene sequences affiliated with MGA were recovered from SI clone libraries. These sequences were added to a data set containing 290 MGA 16S rRNA sequences previously reported from Line P stations P4, P12, and P26 (Allers et al., 2012) and clustered at 97% identity, forming 156 distinct operational taxonomic units (OTUs), 120 of which contained only singletons. Representative sequences were obtained for each non-singleton OTU and placed in phylogenetic context with relevant reference sequences from other locations (Supplementary Figure S2). Five out of 10 previously defined MGA subgroups were recovered in SI clone libraries (ZA3648c and ZA3312c (Fuchs, unpublished); Arctic96B-7 (Bano and Hollibaugh, 2002); SAR406 (Gordon and Giovannoni, 1996); and A714018 (Allers et al., 2012), and three novel subgroups were identified (SHBH1141, SHBH391, and SHAN400) (Supplementary Figure S2). These novel subgroups were found exclusively in SI and contained the most abundant OTUs identified in this location (Supplementary Figures S2, S3).
As described by Allers and colleagues (Allers et al., 2012), MGA sequences identified in coastal and open ocean waters of the NESAP comprised 0.7±0.84% of 10 m clone libraries and 11.2±3.9% of clone libraries from O2-deficient waters, with a maximum of 16.4% at P26 1000 m. The most abundant MGA OTUs present in these locations comprised between 1% and 4% of clone libraries and belonged to subgroups Arctic95A-2, ZA3312c, Arctic96B-7, SAR406, and HF770D10, in order of decreasing OTU abundance (Supplementary Figures S2, S3). In comparison, MGA OTUs identified in SI comprised 1.6±0.81% of 10 m clone libraries and 7.1±3.6% of clone libraries from O2-deficient waters. The most abundant OTUs present in SI comprised between 1% and 5% of clone libraries, and belonged to subgroups SHBH391, SHAN400, SHBH1141, ZA3312c, SAR406 and Arctic96B-7, in order of decreasing OTU abundance (Supplementary Figures S2, S3).
Characterization and phylogenetic assignment of large-insert DNA fragments
To connect 16S rRNA-based patterns of distribution across the oxycline in the NESAP and SI to genomic information associated with specific MGA subgroups, we screened 23 end sequenced fosmid libraries for the presence of clones containing 16S rRNA gene sequences (Supplementary Figure S1). Collectively, fosmid end libraries contained a total of 164 736 genomic clones representing 255.3 Mb of environmental genomic DNA (Supplementary Table S1). Screening of fosmid end sequences for 16S rRNA genes uncovered 14 fosmid inserts containing partial or full-length 16S rRNA gene sequences affiliated with MGA (Table 1; Supplementary Figure S1). These 14 fosmid inserts were fully sequenced (Materials and methods) for downstream analyses, generating ∼540 kb of DNA sequence linked to MGA. In addition, four large-insert DNA fragments from Hawaii Ocean Time-series Station ALOHA (DeLong et al., 2006; Rich et al., 2011) and Monterey Bay (Suzuki et al., 2004) harboring MGA 16S rRNA gene sequences were identified in public databases and used in comparative analyses (Table 1; Supplementary Figure S1).
To identify subgroup affiliations, all 18 MGA 16S rRNA gene sequences identified on large-insert fragments from North Pacific Ocean environments were placed into the MGA reference tree described above (Supplementary Figure S2). Seventeen out of 18 16S rRNA gene sequences identified on large-inserts grouped with 10 defined MGA subgroups. The remaining 16S rRNA gene (on fosmid 4050020-J15) appeared to group outside of MGA and was most closely affiliated with sequences in the phylum Deferribacteres. We chose to include this fosmid in downstream analyses to represent a close relative of MGA.
Genomic content and organization of large-insert DNA fragments derived from MGA
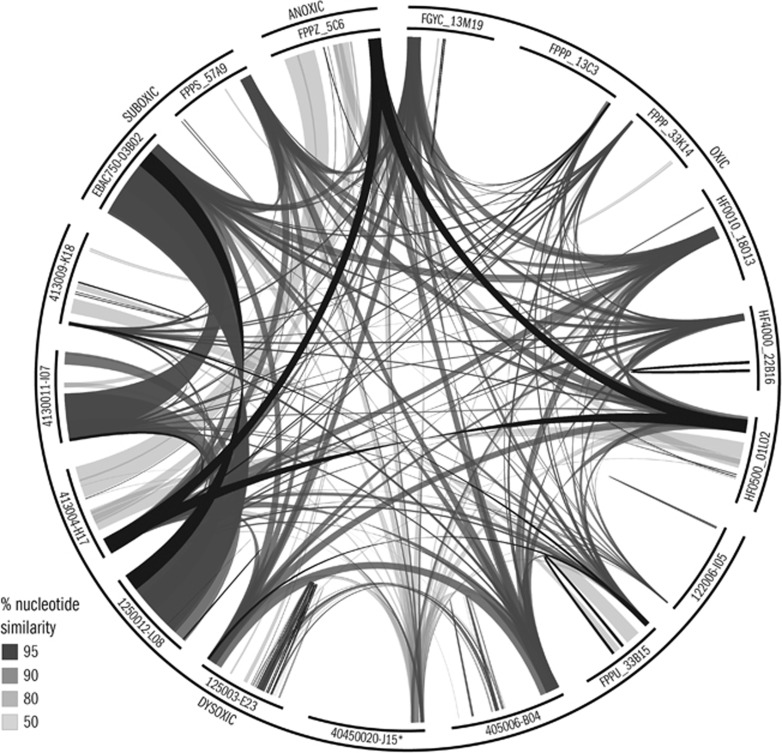
Four criteria were used to determine the extent to which large-insert DNA fragments partitioned into groups consistent with shared environmental context or phylogenetic association, including GC content, tetranucleotide frequency, global nucleotide similarity, and amino acid similarity of predicted ORFs. The size of the large-insert fragments containing MGA 16S rRNA genes ranged from 27.4 kb to 43.5 kb with a GC content ranging from 32.8% to 47.7% (Table 1). Large-insert fragments did not differentiate into discrete groups based on similar GC content (Table 1) or tetranucleotide frequency (Supplementary Figure S4). To further investigate potential similarities among fragments associated with nucleotide arrangement, pairwise blastn analyses were performed between all fragments (Figure 1). Bit-scores for pairwise blastn analyses ranged between 0 and 4.5 × 104 for nonidentical fragments. Large-insert fragments from Monterey Bay (EBAC750-03B02) and the NESAP (1250012-L08 and 4130011-I07), affiliated with subgroup Arctic95A-2, were most similar to one another and formed a distinct group based on global nucleotide similarity (Figure 1). The remaining inserts did not form distinct groups based on global nucleotide similarity, but displayed a gradient of similarity, with bit scores for pairwise blastn analyses averaging (2.2±1.5) × 103. Fosmid 122006-I05, affiliated with subgroup P262000D03, was most unique at the nucleotide level.
Figure 1.
Global nucleotide similarity among 17 MGA-affiliated and 1 Deferribacteres-affiliated* large-insert DNA fragments at 50%, 80%, 90%, and 95%.
To investigate potential similarities among large-insert fragments at the protein-coding level, ORFs were predicted and annotated (Materials and methods). The number of predicted ORFs per insert ranged from 14 to 49, and the number of ORFs on each fragment annotated as ‘hypothetical protein' ranged from 11 to 39 (51–92% of ORFs per insert) (Table 1). Four groups with shared but not identical amino-acid sequences of predicted ORFs surrounding the 16S rRNA gene were identified (groups I–IV), whereas the Deferribacteres-affiliated fosmid (4050020-J15) did not show significant similarity to any other fragments at the protein-coding level and was placed in its own group (group V) (Figure 2). These groups did not uniformly correlate with shared environmental origin or 16S rRNA sequence identity at the level of defined subgroups (Table 1, Supplementary Figure S2). In some cases it was clear that fosmid groups represented different flanking regions of the rRNA operon (that is, groups I and II; Figure 2, Supplementary Figure S5).
Figure 2.
Genes and similarity comparison of large-insert DNA fragments containing MGA 16S rRNA genes representative of syntenic groups I–V. COG categories detected on large-insert fragments are shown in color. 5S, 5S rRNA; 16S: 16S rRNA; 23S, 23S rRNA; ABC, ABC-type multidrug transport system; ACA, acetyl-CoA carboxylase carboxyl transferase; ACP, ATP-dependent CLP protease; GF6P, glucosamine-fructose-6-phosphate aminotransferase; GMP, GMP synthase; MCB, molybdenum cofactor biosynthesis; MS, molybdopterin synthase; NQO, NADH quinone oxidoreductase; PPP, pentose phosphate pathway enzymes; PSRA, polysulfide reductase subunit A; PSRB, polysulfide reductase subunit B; PSRC, polysulfide reductase subunit C; PSRBC, polysulfide reductase subunit BC gene fusion; RRR, response regulator receiver protein; SDD, succinyl-diaminopimelate desuccinylase; SPS, stationary-phase survival protein; TONB, TonB-dependent receptor; and tRNA, transfer RNA.
Four out of eight fosmids in group I were affiliated with the Arctic96B-7 subgroup, whereas the remaining four fosmids were affiliated with ZA3312c, SHBH391, SAR406 and SHAN400. Fosmids in group I contained a conserved gene cluster with genes encoding glucosamine-fructose-6-phosphate aminotransferase (involved in glucosamine biosynthesis), GMP synthase (involved in purine nucleotide biosynthesis) and acetyl-coenzyme A carboxylase carboxyl transferase subunits alpha and beta (potentially involved in fatty acid biosynthesis or CO2 fixation) (Figure 2, Supplementary Figure S5). SI fosmid FPPZ_5C6 also contained a gene encoding RNA polymerase sigma-70 factor (rpoE), known to have a role in high temperature and oxidative stress response (Hild et al., 2000). Fosmid HF0010_18O13 contained the conserved cluster of genes found in group I fosmids as well as a cluster of cytochrome c oxidase subunit genes present in Fe(II) oxidation and three pentose phosphate pathway genes also found in group III fosmids (ribulose-phosphate 3-epimerase, ribose-5-phosphate isomerase b and transketolase).
Group II fosmids were affiliated with Arctic96B-7 and P262000D03. Both fosmids in this group (FPPP_13C3 and 122006-I05) contained a cluster of genes encoding enzymes involved in the pentose phosphate pathway of carbon metabolism, including ribulose-phosphate 3-epimerase, ribose-5-phosphate isomerase b, and in one case, transketolase. Both fosmids also contained an operon encoding an enzyme complex related to polysulfide reductase (Psr). The operon on fosmid 122006-I05 contained three genes encoding homologs of the three Psr subunits: PsrA, a molybdopterin oxidoreductase; PsrB, a [4Fe-4S]-binding subunit; and PsrC, a membrane anchor subunit carrying the site of quinol oxidation, whereas the operon on fosmid FPPP_13C3 contained two genes encoding PsrA and a PsrBC fusion protein. Fosmid FPPP_13C3 contained additional neighboring genes encoding molybdenum cofactor and molybdopterin biosynthesis proteins potentially associated with the assembly of the molybdenum and molybdopterin guanine dinucleotide-containing subunit PsrA. Fosmid FPPP_13C3 also contained a gene for glutamate synthase, often involved in nitrogen assimilation (Vanoni and Curti, 2008), and a gene for rubrerythrin, involved in oxidative stress protection in some anaerobic bacteria and archaea (deMaré et al., 1996; Sztukowska et al., 2002). Fosmid 122006-I05 contained a gene encoding a rhodanese-like protein, belonging to a superfamily of sulfur transferases (Cipollone et al., 2007), upstream of the Psr operon.
All three genomic inserts belonging to group III were affiliated with subgroup Arctic95A-2 and were derived from Monterey Bay and the NESAP (Table 1, Figure 2, Supplementary Figure S6). These three fosmids also formed a discrete group based on global nucleotide similarity analysis (Figure 1). The main organizational feature shared by these inserts was a set of genes encoding transporters, including an ABC-type multidrug transporter, ATPase component, ABC-2 permease and a Tonb-dependent receptor. Group III inserts also contained genes encoding succinyl-diaminopimelate desuccinylase, involved in lysine biosynthesis. Monterey Bay insert EBAC750-03B02 contained a gene affiliated with methionine sulfoxide reductase (msrB). In Escherichia coli, MsrB has been shown to have sulfoxide and dimethyl sulfoxide reductase activity (Grimaud et al., 2001). This insert also contained a gene encoding a rhodanese-like protein.
Fosmids in group IV were affiliated with subgroups P262000N21, SAR406, and A714018, and primarily contained genes encoding hypothetical proteins except for two conserved genes encoding an ATP-dependent protease Clp ATPase subunit and protease subunit. Group IV fosmid HF4000_22B16 was assembled as two unordered pieces, as such, it contained a break point within the 23S rRNA gene (Supplementary Figure S6).
The only fosmid in group V (4050020-J15; most closely related at the 16S rRNA gene sequence level to members of the phylum Deferribacteres) did not exhibit much protein similarity to any of the MGA-affiliated fosmids. This fosmid contained genes for NADH-ubiquinone and quinone oxidoreductase involved in energy metabolism, a major facilitator superfamily transporter, a dihydroorotate dehydrogenase, a cell wall associated hydrolase, and a tRNA nucleotidyltransferase, in addition to genes encoding a number of hypothetical proteins.
Population structure of MGA syntenic groups
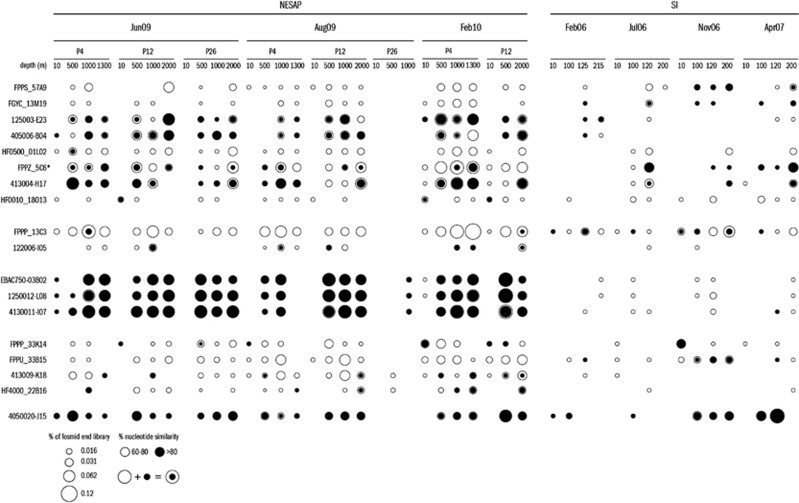
To determine the prevalence and distribution of MGA subgroups represented by large-insert DNA fragments detected in this study, the proportion of fosmid end sequences from each NESAP and SI library recruiting to large-insert fragments was determined (Figure 3). The largest proportions of sequences recruiting to large-insert fragments were derived from depths ⩾500 m in the NESAP and ⩾100 m in the SI. A very small proportion of end sequences were recruited from Aug-09 P26 libraries, which could be due to the relatively small size of these libraries (Supplementary Table S1). End sequences from NESAP libraries generally recruited to large-insert fragments in larger numbers and with a higher degree of nucleotide similarity than end sequences from SI libraries, even for large-insert fragments derived from SI (Figure 3). End sequences similar to group III fragments were most highly and consistently represented in NESAP fosmid end libraries, followed by end sequences similar to several group I fragments. End sequences similar to the Deferribacteres-like fosmid 4050020-J15 were also well represented and very similar to sequences derived from oxic through suboxic (but not anoxic) NESAP and SI libraries.
Figure 3.
Dot plot showing the proportion of fosmid end sequenced libraries recruiting to MGA large-insert DNA fragments at various sample locations and depths in the NESAP (at stations P4, P12, and P26) and SI (station S3). Hollow circles represent proportion of fosmid end sequenced libraries recruiting to large-insert fragments with nucleotide similarity 60–80% solid circles >80%.
Phylogenetic analysis and distribution of Psr
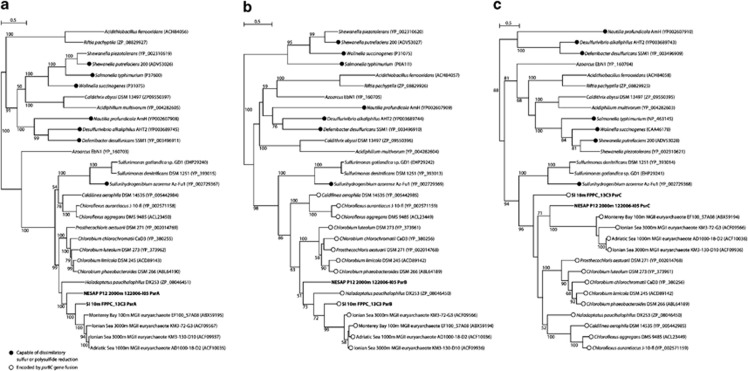
To gain insight into the evolutionary history of psrA, psrB, and psrC genes detected on MGA fosmids, phylogenetic trees of their predicted protein products were constructed. Phylogenetic analysis of the catalytic subunit, PsrA, confirmed that predicted PsrA homologs detected on MGA fosmids were most closely related to Psr and thiosulfate reductase (Phs) of the dimethyl sulfoxide reductase family of molybdenum-containing enzymes (Supplementary Figure S7). Predicted PsrA homologs from MGA fosmids were ∼63% similar to one another, and most closely related to proteins encoded on fosmids from the Mediterranean Sea and Monterey Bay derived from Marine Group II euryarchaeota (Figure 4a). Predicted MGA proteins were less similar to canonical PsrA proteins originally characterized in Wolinella succinogenes (Krafft et al., 1995). Phylogenetic trees of predicted PsrB with PsrB-like respiratory proteins containing [4Fe-4S]-binding-subunits and of predicted PsrC with PsrC-like membrane anchor subunits indicated similar phylogenetic relationships (Figures 4b and c). Predicted PsrA proteins encoded on MGA fosmids did not contain any obvious signal sequences (for example, twin-arginine translocation (TAT) signal sequences) suggesting that these proteins are located in the cytoplasm, similar to PsrA proteins detected in most green sulfur bacteria (GSB) (Frigaard and Bryant, 2008). PsrA encoded by W. succinogenes, by comparison, encodes a TAT signal sequence and is translocated into the periplasm (Krafft et al., 1995).
Figure 4.
Unrooted phylogenetic trees based on protein sequences with homology to (a) predicted polysulfide reductase molybdopterin-containing subunit (PsrA); (b) predicted [4Fe-4S]-binding subunit (PsrB); and (c) membrane anchor subunit (PsrC) identified on fosmids FPPP_13C3 and 122006-I05. The trees were inferred using maximum likelihood implemented in PhyML. Solid circle indicates proteins derived from organisms that have been demonstrated to grow by reducing elemental sulfur or polysulfide with concomitant H2S production; hollow circle indicates presence of a psrBC gene fusion. The scale bar represents estimated number of amino-acid substitutions per site. Bootstrap values below 50% are not shown.
In contrast to the organization of the psrABC operon originally described in W. succinogenes (Krafft et al., 1995), the ORFs encoding PsrB and PsrC homologs on both MGA fosmids were located upstream of ORFs encoding PsrA homologs (Figure 2). Also in contrast to the W. succinogenes psrABC operon, the genes encoding PsrB and PsrC on fosmid FPPP_13C3 appeared to form a gene fusion (psrBC), a feature also detected in several PSR-containing GSB (Frigaard and Bryant, 2008) and other PSR-containing bacteria and archaea. The psrBCA format of operon organization detected on MGA fosmids was also detected on Marine Group II fosmids and several PSR-containing GSB in addition to Sulfurimonas denitrificans DSM 1251, Caldilinea aerophila DSM 14535, Chloroflexus aggregans DSM 9485, and Haladaptatus paucihalophilus DX253 (Figures 4b and c). A third format of operon organization (psrACB) was detected in Sulfurimonas gotlandica GD1 and Sulfurihydrogenibium azorense Az-Fu1.
To determine the prevalence of predicted MGA psr genes in NESAP and SI fosmid end sequenced libraries, the proportion of fosmid end sequences that recruited to the psrBCA operon on fosmids FPPP_13C3 and 122006-I05 was calculated for each end sequenced library (Supplementary Figure S8). The majority of end sequences recruiting to psr genes were derived from ⩾500 m depth in the NESAP and ⩾100 m depth in SI, and psr homologs were most consistently present throughout O2-deficient waters of the NESAP in August 2009 at station P4.
Discussion
The 17 large-insert DNA fragments containing MGA 16S rRNA genes derived from North Pacific Ocean metagenomic libraries were affiliated with seven previously defined and two novel MGA subgroups, whereas the 16S rRNA gene on an 18th insert was more closely related to the phylum Deferribacteres (Supplementary Figure S2). Although large-insert DNA fragments were obtained from multiple environments manifesting distinct oxyclines, fragments did not coalesce into coherent groups based on GC content, tetranucleotide frequency, or global nucleotide similarity. However, fragments did coalesce into five syntenic groups based on shared amino acid similarity of predicted ORFs. Group membership was not generally consistent with shared environmental origin, O2 concentration, or 16S rRNA gene sequence identity (Table 1). These observations could be explained in several ways. MGA subgroups may contain multiple unlinked copies of the 16S rRNA operon (Acinas et al., 2004). Alternatively, large-insert fragments may be derived from flanking regions of the same 16S rRNA operon, as observed for syntenic groups I and II. It is also possible that subgroups ZA3312c through A714018 actually represent one subgroup of MGA, evidenced by a lack of bootstrap support for nodes encompassing these subgroups within the MGA 16S rRNA gene tree (Supplementary Figure S2).
Recruitment of fosmid end sequences from Line P and SI libraries to large-insert DNA fragments reflected 16S rRNA-based patterns of MGA distribution in that the proportion of MGA sequences was maximal in waters ⩾500 m depth in the NESAP and ⩾100 m depth in SI (Figure 3). MGA sequences comprised a much larger proportion of NESAP (open ocean) than SI (coastal basin) end libraries, a pattern also reflected in MGA 16S rRNA distribution and representative of the overall higher proportion of MGA detected in the NESAP than in SI microbial communities (Zaikova et al., 2010; Allers et al., 2012). The proportion of SI end sequences recruiting to large-insert fragments was maximal in dysoxic and suboxic samples from Nov 2006 and April 2007, supporting the hypothesis that dominant MGA subgroups are adapted to O2-deficiency in this location. The largest proportion of end sequences from NESAP libraries recruited to group III fragments affiliated with subgroup Arctic95A-2 supporting 16S rRNA-based observations that Arctic95A-2 is a dominant subgroup in the NESAP open-ocean. Group I fragments affiliated with subgroups Arctic96B-7 and SAR406 also recruited a relatively large proportion of NESAP end sequences. A reasonable proportion of end sequences from the NESAP and SI libraries also recruited to Deferribacteres-like fosmid 4050020-J15, with a pattern of distribution suggesting adaptation to suboxic and dysoxic, but not anoxic, conditions.
Although large-insert fragments did not clearly partition into ecologically distinct groups based on O2 concentration, predicted protein-coding genes associated with adaptation to O2-deficiency and sulfur-based energy metabolism were detected on multiple fosmids. With respect to adaptation to O2-deficiency, a gene encoding rpoE RNA polymerase sigma-70 factor, known to have a role in oxidative stress response, was detected on SI fosmid FPPZ_5C6, obtained from an anoxic-sulfidic 200 m sample. A gene encoding rubrerythrin, also involved in oxidative stress response in some anaerobic prokaryotes, was detected on SI fosmid FPPP_13C3, obtained from an oxic 10 m sample. With respect to sulfur-based energy metabolism, a gene encoding methionine sulfoxide reductase (MsrB) was detected on Monterey Bay insert EBAC750-03B02, which in E. coli has been shown to have sulfoxide and dimethyl sulfoxide reductase activity (Grimaud et al., 2001). In addition, four fosmids encoded rhodanese-like proteins, affiliated with a superfamily of sulfur transferases. Perhaps most interestingly, a psr operon was detected on SI fosmid FPPP_13C3 and on NESAP fosmid 122006-I05, obtained from a dysoxic 2000 m sample. Sequences similar to psr genes encoded on these fosmids were also detected in a number of fosmid end sequenced libraries derived from ⩾500 m depth in the NESAP and ⩾100 m depth in SI, suggesting that these genes are associated with O2-deficient environments (Supplementary Figure S8). In the anaerobic epsilonproteobacterium W. succinogenes, PSR and hydrogenase or formate dehydrogenase allows respiration on polysulfide (Sn) using H2 or formate as an electron donor, with concomitant production of H2S (Jankielewicz et al., 1995). The PSR complex isolated from W. succinogenes has also been documented to catalyze sulfide oxidation to polysulfide by dimethylnaphthoquinone, however, with much lower efficiency (Hedderich et al., 1999). The identification of proteins homologous to PSR on two fosmids suggests that specific MGA subgroups may have the capacity to generate energy via dissimilatory polysulfide reduction to hydrogen sulfide (H2S) or via dissimilatory H2S oxidation (Schröder et al., 1988; Klimmek et al., 1991; Krafft et al., 1992, 1995; Jormakka et al., 2008).
The PSR complex of W. succinogenes is encoded by the psrABC genes and consists of two periplasmic subunits (a catalytic molybdopterin-containing PsrA subunit and a [4Fe-4S]-binding PsrB subunit) and a membrane-anchoring PsrC subunit (Krafft et al., 1992). Predicted PsrA proteins detected on MGA fosmids were only distantly related to isolated PsrA from W. succinogenes but more closely related to PsrA homologs encoded on Marine Group II euryarchaeotal fosmids derived from the Mediterranean Sea and Monterey Bay. PsrA proteins detected on MGA fosmids were also similar to PsrA homologs found in the GSB Prostheticochloris aestuarii DSM 271, Chlorobium chlorochromatii CaD3, Chlorobium luteolum DSM273, Chlorobium limicola DSM 245, and Chlorobium phaeobacteroides DSM 266; the halophilic euryarchaeon Haladaptatus paucihalophilus DX253; the thermophilic Chloroflexi strain Caldilinea aerophila DSM 14535; the thermophilic Aquificales strain Sulfurihydrogenibium azorense Az-Fu1; and the sulfur-oxidizing Epsilonproteobacteria Sulfurimonas gotlandica GD1 and Sulfurimonas denitrificans DSM 1251. Interestingly, in GSB, the phylogeny of PsrA homologs is congruent with a number of phylogenetic anchor genes, suggesting that PSR was present in the last common ancestor of PSR-containing GSB (Gregersen et al., 2011). Given the proximal phylogenetic relationship of MGA and GSB based on 16S rRNA gene sequences (Supplementary Figure S2), it is possible that MGA inherited this operon from a common ancestor. The psrBC genes on MGA fosmid 122006-I05 were encoded by separate ORFs (psrB and psrC), whereas in fosmid FPPP_13C3, these genes were fused (psrBC). A psrBC gene fusion has been described previously in members of the PSR-containing GSB (including P. aestuarii, C. chlorochromatii, and C. luteolum; (Frigaard and Bryant, 2008)), and was detected in Marine Group II fosmids from the Mediterranean Sea and Monterey Bay in addition to H. paucihalophilus and C. aerophila. The broad phylogenetic origins of psrABC genes similar to those detected on MGA fosmids are consistent with multiple lateral transfer events across phyla and domains.
Although direct evidence for the role of PSR in sulfur-based energy metabolism has only been obtained from W. succinogenes, many cultivated reference strains encoding PSR are capable of generating energy using sulfur compounds. The PSR sequences derived from several such reference strains, including S. azorense Az-Fu1 and the GSB, branched with predicted PSR homologs detected on MGA fosmids. S. azorense Az-Fu1 is capable of growth by coupling reduction of elemental sulfur (S°) to hydrogen oxidation, although polysulfide was not directly tested as an electron acceptor (Aguiar et al., 2004). S. azorense Az-Fu1 has also been documented to oxidize S° and sulfite (SO23−) (Aguiar et al., 2004). Similarly, the cytoplasmic PSR complex found in many GSB (including P. aestuarii, C. chlorochromatii, C. luteolum, C. limicola and C. phaeobacteroides) has been proposed to oxidize sulfite produced by the dissimilatory sulfate reduction (Dsr) system (Gregersen et al., 2011). Although the actual substrate of PSR cannot be determined based on sequence similarity alone, the phylogenetic position of MGA PSR homologs provides a circumstantial link between MGA and sulfur cycling in the environment.
Oxygen-deficient marine systems, including OMZs and permanent or seasonally stratified anoxic basins, are known to harbor active sulfur cycles that have been linked to the activities of sulfur-oxidizing gamma and epsilonproteobacteria (Walsh et al., 2009; Canfield et al., 2010; Grote et al., 2012). Although this study provides only a glimpse into the metabolic diversity that is likely contained within the MGA candidate phylum, the presence of PSR homologs on MGA-affiliated genome fragments suggests a potential role for MGA in the cryptic sulfur cycle of O2-deficient marine systems, where the abundance of these bacteria is concentrated. Process rate measurements linking sulfur chemistry with MGA activity are required to support this hypothesis (Milucka et al., 2012). Given the lack of cultivated representatives of MGA, the application of single-cell genomics could aid in providing the genome-wide information needed to fully describe the metabolic capacity of defined MGA subgroups residing in distinct locations (Woyke et al., 2009; Swan et al., 2011; Stepanauskas, 2012). Such high-resolution genomic data may provide additional clues as to the evolutionary history and biogeochemical roles of these widely distributed marine bacteria.
Accession numbers
Fosmid end sequenced libraries reported in this study were deposited in the Genome Survey Sequences (GSS) Database with the accession numbers LIBGSS_039072–LIBGSS_039117 (individual sequences are also available in GenBank with accession numbers KG088956–KG619837). Fully sequenced fosmids reported in this study were deposited in GenBank with the accession numbers KF170413–KF170426.
Acknowledgments
We thank scientists and crew aboard the MSV John Strickland and the CCGS John P. Tully, as well as the Canadian Department of Fisheries and Oceans for logistical support. In particular, we thank Karl Schiffmacher and Marie Robert for assistance with data collection; Alyse Hawley, David Walsh, Olena Shevchuk and Charles Howes for assistance with data analysis; Martin Krzywinski and Salvador Ramirez for assistance with data visualization; and all members of the Hallam Lab for helpful comments along the way. We also thank the Joint Genome Institute, including Natasha Zvenigorodsky, Stephanie Malfatti, Phil Hugenholtz, Susan Yilmaz and Tijana Glavina del Rio for technical and project management assistance. This work was performed under the auspices of the US Department of Energy Joint Genome Institute (supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231), the Natural Sciences and Engineering Research Council (NSERC) of Canada, Canada Foundation for Innovation (CFI) and the Canadian Institute for Advanced Research (CIFAR) through grants awarded to SJH. JJW and KM were supported by NSERC, and KRM and KMK were supported by the Tula Foundation funded Centre for Microbial Diversity and Evolution.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, et al. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. 2004;430:551–554. doi: 10.1038/nature02649. [DOI] [PubMed] [Google Scholar]
- Aguiar P, Beveridge T, Reysenbach A. Sulfurihydrogenibium azorense, sp. nov., a thermophilic hydrogen-oxidizing microaerophile from terrestrial hot springs in the Azores. Int J Syst Evol Microbiol. 2004;54:33–39. doi: 10.1099/ijs.0.02790-0. [DOI] [PubMed] [Google Scholar]
- Allers E, Wright JJ, Konwar KM, Howes CG, Beneze E, Hallam SJ, et al. Diversity and population structure of Marine Group A bacteria in the Northeast subarctic Pacific Ocean. ISME J. 2012;7:256–268. doi: 10.1038/ismej.2012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson JJ, Devol AH. Deep water renewal in Saanich Inlet, an intermittently anoxic basin. Estuarine and Coastal Marine Science. 1973;1:1–10. [Google Scholar]
- Bano N, Hollibaugh JT. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl Environ Microbiol. 2002;68:505–518. doi: 10.1128/AEM.68.2.505-518.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. Prediction of twin-arginine signal peptides. BMC Bioinformatics. 2005;6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bograd SJ, Castro CG, Di Lorenzo E, Palacios DM, Bailey H, Gilly W, et al. Oxygen declines and the shoaling of the hypoxic boundary in the California Current. Geophys Res Lett. 2008;35:1–6. [Google Scholar]
- Canfield DE, Stewart FJ, Thamdrup B, De Brabandere L, Dalsgaard T, Delong EF, et al. A cryptic sulfur cycle in oxygen-minimum-zone waters off the Chilean coast. Science. 2010;330:1375–1378. doi: 10.1126/science.1196889. [DOI] [PubMed] [Google Scholar]
- Cipollone R, Ascenzi P, Visca P. Common themes and variations in the rhodanese superfamily. IUBMB Life. 2007;59:51–59. doi: 10.1080/15216540701206859. [DOI] [PubMed] [Google Scholar]
- Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–143. [Google Scholar]
- Clarke KR, Gorley RN. PRIMER v6. User manual/tutorial. Plymouth routine in mulitvariate ecological research. Plymouth Marine Laboratory: Plymouth, UK; 2006. [Google Scholar]
- DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- deMaré F, Kurtz DM, Nordlund P. The structure of Desulfovibrio vulgaris rubrerythrin reveals a unique combination of rubredoxin-like FeS4 and ferritin-like diiron domains. Nat Struct Mol Biol. 1996;3:539–546. doi: 10.1038/nsb0696-539. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZJ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006a;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S, Watanabe YW, Ono T, Mecking S. Temporal trends in apparent oxygen utilization in the upper pycnocline of the North Pacific: 1980-2000. J Oceanogr. 2004;60:139–147. [Google Scholar]
- Freeland H. A short history of ocean station papa and Line P. Prog Oceanogr. 2007;75:120–125. [Google Scholar]
- Frigaard NU, Bryant DA. Sulfur Metabolism in Phototrophic Organisms. Springer: New York; 2008. Genomic insights into the sulfur metabolism of phototrophic green sulfur bacteria; pp. pp 337–355. [Google Scholar]
- Fuchs BM, Woebken D, Zubkov MV, Burkill P, Amann R. Molecular identification of picoplankton populations in contrasting waters of the Arabian Sea. Aquat Microb Ecol. 2005;39:145–157. [Google Scholar]
- Fuhrman JA, Davis AA. Widespread Archaea and novel Bacteria from the deep sea as shown by 16S rRNA gene sequences. Marine Ecol Prog Series. 1997;150:275–285. [Google Scholar]
- Fuhrman JA, McCallum K, Davis AA. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DA, Giovannoni SJ. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific oceans. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen LH, Bryant DA, Frigaard NU. Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Front Microbiol. 2011;2:1–14. doi: 10.3389/fmicb.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud R, Ezraty B, Mitchell JK, Lafitte D, Briand C, Derrick PJ, et al. Repair of oxidized proteins: Identification of a new methionine sulfoxide reductase. J Biol Chem. 2001;276:48915–48920. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- Grote J, Schott T, Bruckner CG, Glöckner FO, Jost G, Teeling H, et al. Genome and physiology of a model Epsilonproteobacterium responsible for sulfide detoxification in marine oxygen depletion zones. Proc Natl Acad Sci USA. 2012;109:506–510. doi: 10.1073/pnas.1111262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedderich R, Klimmek O, Kröger A, Dirmeier R, Keller M, Stetter KO. Anaerobic respiration with elemental sulfur and with disulfides. Fems Microbiol Rev. 1999;22:353–381. [Google Scholar]
- Helm KP, Bindoff NL, Church JA. Observed decreases in oxygen content of the global ocean. Geophys Res Lett. 2011;38:1–6. [Google Scholar]
- Hild E, Takayama K, Olsson RM, Kjelleberg S. Evidence for a role of rpoE in stressed and unstressed cells of marine Vibrio angustum strain S14. J Bacteriol. 2000;182:6964–6974. doi: 10.1128/jb.182.24.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankielewicz A, Klimmek O, Kröger A. The electron transfer from hydrogenase and formate dehydrogenase to polysulfide reductase in the membrane of Wolinella succinogenes. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 1995;1231:157–162. [Google Scholar]
- Jormakka M, Yokoyama K, Yano T, Tamakoshi M, Akimoto S, Shimamura T, et al. Molecular mechanism of energy conservation in polysulfide respiration. Nat Struct Mol Biol. 2008;15:730–737. doi: 10.1038/nsmb.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S.1999KEGG: Kyoto encyclopedia of genes and genomes Nucleic Acids Res 2729–34.Downloaded 18 Jun 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Riley M, Saier M, Paulsen IT, Paley SM, Pellegrini-Toole A.2000The ecocyc and metacyc databases Nucleic Acids Res 2856–59.Downloaded 21 Oct 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling RF, Körtzinger A, Gruber N. Ocean deoxygenation in a warming world. Ann Rev Mar Sci. 2010;2:199–229. doi: 10.1146/annurev.marine.010908.163855. [DOI] [PubMed] [Google Scholar]
- Klimmek O, Kröger A, Steudel R, Holdt G. Growth of Wolinella succinogenes with polysulphide as terminal acceptor of phosphorylative electron transport. Arch Microbiol. 1991;155:177–182. [Google Scholar]
- Konwar KM, Hanson NW, Pagé AP, Hallam SJ. MetaPathways: a modular pipeline for constructing pathway/genome databases from environmental sequence information. BMC Bioinformatics. 2013;14:202. doi: 10.1186/1471-2105-14-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafft T, Bokranz M, Klimmek O, Schröder I, Fahrenholz F, Kojro E, et al. Cloning and nucleotide sequence of the pst A gene of Wolinella succinogenes polysulphide reductase. Eu J Biochem. 1992;206:503–510. doi: 10.1111/j.1432-1033.1992.tb16953.x. [DOI] [PubMed] [Google Scholar]
- Krafft T, Gross R, Kröger A. The function of Wolinella succinogenes psr genes in electron transport with polysulphide as the terminal electron acceptor. Eu J Biochem. 1995;230:601–606. doi: 10.1111/j.1432-1033.1995.0601h.x. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley MD, Baross JA, Gordon LI. Dissolved hydrogen and methane in Saanich Inlet, British Columbia. Deep Sea Res. 1982;29:1471–1484. [Google Scholar]
- Madrid VM, Taylor GT, Scranton MI, Chistoserdov AY. Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco Basin. Appl Environ Microb. 2001;67:1663–1674. doi: 10.1128/AEM.67.4.1663-1674.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milucka J, Ferdelman TG, Polerecky L, Franzke D, Wegener G, Schmid M, et al. Zero-valent sulphur is a key intermediate in marine methane oxidation. Nature. 2012;491:541–546. doi: 10.1038/nature11656. [DOI] [PubMed] [Google Scholar]
- Miroshnichenko ML, Kolganova TV, Spring S, Chernyh N, Bonch-Osmolovskaya EA. Caldithrix palaeochoryensis sp. nov., a thermophilic, anaerobic, chemo-organotrophic bacterium from a geothermally heated sediment, and emended description of the genus Caldithrix. Int J Syst Evol Microbiol. 2010;60:2120–2123. doi: 10.1099/ijs.0.016667-0. [DOI] [PubMed] [Google Scholar]
- Miroshnichenko ML, Kostrikina NA, Chernyh NA, Pimenov NV, Tourova TP, Antipov AN, et al. Caldithrix abyssi gen. nov., sp. nov., a nitrate-reducing, thermophilic, anaerobic bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent, represents a novel bacterial lineage. Int J Syst Evol Microbiol. 2003;53:323–329. doi: 10.1099/ijs.0.02390-0. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Maglott DR.2001RefSeq and LocusLink: NCBI gene-centered resources Nucleic Acids Res 29137–140.Downloaded 13 Nov 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko DA, Myers GS, Ravel J. Visualization of comparative genomic analyses by BLAST score ratio. Bmc Bioinformatics. 2005;6:2. doi: 10.1186/1471-2105-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich VI, Pham VD, Eppley J, Shi Y, DeLong EF. Time-series analyses of Monterey Bay coastal microbial picoplankton using a ‘genome proxy'microarray. Environ Microbiol. 2011;13:116–134. doi: 10.1111/j.1462-2920.2010.02314.x. [DOI] [PubMed] [Google Scholar]
- Schattenhofer M, Fuchs BM, Amann R, Zubkov MV, Tarran GA, Pernthaler J. Latitudinal distribution of prokaryotic picoplankton populations in the Atlantic Ocean. Environ Microbiol. 2009;11:2078–2093. doi: 10.1111/j.1462-2920.2009.01929.x. [DOI] [PubMed] [Google Scholar]
- Schröder I, Kröger A, Macy JM. Isolation of the sulphur reductase and reconstitution of the sulphur respiration of Wolinella succinogenes. Arch Microbiol. 1988;149:572–579. [Google Scholar]
- Stepanauskas R. Single cell genomics: an individual look at microbes. Curr Opin Microbiol. 2012;15:613–620. doi: 10.1016/j.mib.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Stevens H, Ulloa O. Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environ Microbiol. 2008;10:1244–1259. doi: 10.1111/j.1462-2920.2007.01539.x. [DOI] [PubMed] [Google Scholar]
- Stramma L, Johnson GC, Sprintall J, Mohrholz V. Expanding oxygen-minimum zones in the Tropical Oceans. Science. 2008;320:655–658. doi: 10.1126/science.1153847. [DOI] [PubMed] [Google Scholar]
- Suzuki MT, Preston CM, Beja O, de la Torre JR, Steward GF, DeLong EF. Phylogenetic screening of ribosomal RNA gene-containing clones in Bacterial Artificial Chromosome (BAC) libraries from different depths in Monterey Bay. Microb Ecol. 2004;48:473–488. doi: 10.1007/s00248-004-0213-5. [DOI] [PubMed] [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D, et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the Dark Ocean. Science. 2011;333:1296–1300. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- Sztukowska M, Bugno M, Potempa J, Travis J, Kurtz DM. Role of rubrerythrin in the oxidative stress response of Porphyromonas gingivalis. Mol Microbiol. 2002;44:479–488. doi: 10.1046/j.1365-2958.2002.02892.x. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, et al. 2001The COG database: new developments in phylogenetic classification of proteins from complete genomes Nucleic Acids Res 2922–28.Downloaded 5 Feb 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling H, Meyerdierks A, Bauer M, Amann R, Glockner FO. Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ Microbiol. 2004a;6:938–947. doi: 10.1111/j.1462-2920.2004.00624.x. [DOI] [PubMed] [Google Scholar]
- Teeling H, Waldmann J, Lombardot T, Bauer M, Glockner FO. TETRA: a web-service and a stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences. BMC Bioinformatics. 2004b;5:163. doi: 10.1186/1471-2105-5-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoni MA, Curti B. Structure—function studies of glutamate synthases: A class of self-regulated iron-sulfur flavoenzymes essential for nitrogen assimilation. IUBMB life. 2008;60:287–300. doi: 10.1002/iub.52. [DOI] [PubMed] [Google Scholar]
- Walsh DA, Hallam SJ. John Wiley & Sons, Inc: Hoboken, NJ, USA; 2011. Bacterial community structure and dynamics in a seasonally anoxic Fjord: Saanich Inlet, British Columbia. In Molecular Microbial Ecology II: Metagenomics in Different Habitats de Bruijn FJ, (ed) [Google Scholar]
- Walsh DA, Zaikova E, Hallam SJ. Large volume (20L+) filtration of coastal seawater samples. J Vis Exp. 2009;28:1161. doi: 10.3791/1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh DA, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG, et al. Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science. 2009;326:578–582. doi: 10.1126/science.1175309. [DOI] [PubMed] [Google Scholar]
- Ward BB, Kilpatrick KA, Wopat AE, Minnich EC, Lindstrom ME. Methane oxidation in Saanich Inlet. Cont Shelf Res. 1989;9:65–75. [Google Scholar]
- Whitney FA, Freeland HJ, Robert M. Persistently declining oxygen levels in the interior waters of the eastern subarctic Pacific. Prog Oceanogr. 2007;75:179–199. [Google Scholar]
- Woyke T, Xie G, Copeland A, Gonzalez JM, Han C, Kiss H, et al. Assembling the marine metagenome, one cell at a time. PLoS One. 2009;4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JJ, Konwar KM, Hallam SJ. Microbial ecology of expanding oxygen minimum zones. Nat Rev Microbiol. 2012;10:381–394. doi: 10.1038/nrmicro2778. [DOI] [PubMed] [Google Scholar]
- Wright JJ, Lee S, Zaikova E, Walsh DA, Hallam SJ. DNA extraction from 0.22 microM Sterivex filters and cesium chloride density gradient centrifugation. J Vis Exp. 2009;31:pii 1352. doi: 10.3791/1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaikova E, Hawley A, Walsh DA, Hallam SJ. Seawater sampling and collection. J Vis Exp. 2009;28:e1159. doi: 10.3791/1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaikova E, Walsh DA, Stilwell CP, Mohn WW, Tortell PD, Hallam SJ. Microbial community dynamics in a seasonally anoxic fjord: Saanich Inlet, British Columbia. Environ Microbiol. 2010;12:172–191. doi: 10.1111/j.1462-2920.2009.02058.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.