Abstract
Soil bacteria produce a diverse array of antibiotics, yet our understanding of the specific roles of antibiotics in the ecological and evolutionary dynamics of microbial interactions in natural habitats remains limited. Here, we show a significant role for antibiotics in mediating antagonistic interactions and nutrient competition among locally coexisting Streptomycete populations from soil. We found that antibiotic inhibition is significantly more intense among sympatric than allopatric Streptomycete populations, indicating local selection for inhibitory phenotypes. For sympatric but not allopatric populations, antibiotic inhibition is significantly positively correlated with niche overlap, indicating that inhibition is targeted toward bacteria that pose the greatest competitive threat. Our results support the hypothesis that antibiotics serve as weapons in mediating local microbial interactions in soil and suggest that coevolutionary niche displacement may reduce the likelihood of an antibiotic arms race. Further insight into the diverse roles of antibiotics in microbial ecology and evolution has significant implications for understanding the persistence of antibiotic inhibitory and resistance phenotypes in environmental microbes, optimizing antibiotic drug discovery and developing strategies for managing microbial coevolutionary dynamics to enhance inhibitory phenotypes.
Keywords: antibiotic, coevolution, competition, resistance
Introduction
Antibiotic-producing microbes are a common component of soil communities. Antibiotics have been traditionally perceived to be important to antagonistic species interactions and to confer a fitness benefit to producers in competitive habitats (Williams et al., 1989; Riley and Gordon, 1999; Martinez et al., 2009a). More recently, antibiotics have been suggested to function predominantly as signaling molecules that may mediate competitive, mutualistic, pathogenic or commensal species interactions (Goh et al., 2002; Ng et al., 2003; Lin et al., 2005; Yim et al., 2007; Fajardo and Martinez, 2008; Martinez, 2008; Aminov, 2009; Romero et al., 2011). At subinhibitory concentrations, antibiotics modulate the transcription of a wide variety of bacterial genes, including genes that influence nutrient acquisition, virulence, motility, antibiotic production and biofilm formation (Bagge et al., 2004; Hoffman et al., 2005; Linares et al., 2006; Mitova et al., 2008; Ryan and Dow, 2008; Cummins et al., 2009; Kaplan et al., 2012). Although it has been argued that antibiotics are likely to be present predominantly at subinhibitory concentrations in natural habitats, and thus that antibiotics may act only rarely as ‘weapons', there is little empirical data on antibiotic concentrations in soil or aquatic environments.
Although broad consensus supports the concept that antibiotics have diverse roles in natural communities, we have limited understanding of the natural history and evolutionary dynamics of antibiotic phenotypes. Globally, antibiotics are central to the management of infectious diseases, and scientists are challenged to discover novel antibiotics while managing the spread of antibiotic resistance in clinical populations. Unfortunately, our lack of understanding of the ecology and evolutionary biology of antibiotic phenotypes in natural habitats significantly constrains our abilities to develop deliberate strategies for antibiotic exploration, manage indigenous soil microbial communities for enhanced inhibitory phenotypes and minimize accumulation of antibiotic resistance in environmental microbes. Multiple models have been proposed for describing the competitive and coevolutionary dynamics of antibiotic-producing microbes (Czaran et al., 2002; Czaran and Hoekstra, 2003; Wloch-Salamon et al., 2008; Laskaris et al., 2010), and antagonistic interactions have been suggested to exhibit coevolutionary arms race or polymorphism dynamics. Such dynamics can result in reciprocal accumulation of inhibitory and resistance phenotypes in interacting populations over time. Although these models assume that antibiotics confer consistent fitness advantages to the producer while imposing significant fitness costs on nonproducing organisms within local communities, we have limited experimental evidence to support or refute these assumptions. Further information on the dynamics of antibiotic phenotypes in microbial populations is needed to shed light on the roles of antibiotics in natural communities and on the evolutionary or coevolutionary trajectories contributing to the accumulation of antibiotic inhibitory and resistance phenotypes in the environment.
Our work explores the dynamics of antibiotic inhibitory and resistance phenotypes among soil-borne Streptomycetes in natural habitats. Streptomycetes are notable as producers of the majority (>70%) of naturally occurring antibiotics (Tanaka and Omura, 1990; Watve et al., 2001; Challis and Hopwood, 2003; Baltz, 2007). Streptomycetes are Gram positive, filamentous bacteria that are excellent saprophytes, prolific producers of extracellular enzymes and ubiquitous in soil and marine sediments (Gontang et al., 2007; Chater et al., 2010). Streptomycetes are the source of many clinically significant antibiotics and have been investigated in agricultural settings for their capacities to suppress plant pathogens (Bressan and Figueiredo, 2007; Hiltunen et al., 2009; Karimi et al., 2012; Kinkel et al., 2012; Meschke et al., 2012; Otto-Hanson et al., 2013). Because of their capabilities at producing bioactive compounds and in response to the tremendous therapeutic value of many of these compounds, antibiotic production in Streptomycetes has been studied extensively in vitro. In general, antibiotic production is highly variable among individual Streptomycete isolates in terms of both the amounts and the identities of antibiotics produced, and most Streptomycete isolates produce multiple antibiotics (Omura et al., 2001; Bentley et al., 2002; Challis and Hopwood, 2003). Patterns of antibiotic resistance are likewise decidedly variable among Streptomycete isolates, and as a consequence, inhibitory interactions among Streptomycete isolates tend to be highly specific (Davelos et al., 2004; Vetsigian et al., 2011).
In this work we evaluate the hypotheses that antibiotic inhibitory and resistance phenotypes confer local fitness benefits and that antibiotic inhibitory phenotypes mediate competitive and coevolutionary interactions within local soil communities. Specifically, we evaluated inhibitory intensities, resistance frequencies and relationships between nutrient use (niche) overlap and inhibition among sympatric and allopatric Streptomycete isolates from diverse locations. Differences in sympatric vs allopatric inhibition intensities and resistance frequencies provide insight into both the fitness benefits and potential roles of inhibitory interactions in mediating local population interactions. Furthermore, these data shed light on the potential for reciprocal selection of inhibitory and resistance phenotypes among locally coexisting populations, consistent with coevolutionary arms race or polymorphisms dynamics. Finally, relationships between niche overlap and antibiotic inhibition among sympatric vs allopatric populations provide crucial information on the significance of inhibitory interactions to mediating nutrient competition among coexisting Streptomycetes as well as possible alternatives to coevolutionary arms race dynamics.
Materials and methods
Soil sample collection and processing
We evaluated inhibitory intensities, resistance frequencies and relationships between nutrient use (niche) overlap and inhibition among sympatric and allopatric Streptomycete isolates from seven locations representing temperate and tropical habitats. Sympatric isolates were defined as those from the same soil core, and allopatric isolates were those from different cores; a single core was evaluated at each location. The sampling sites encompassed diverse soil characteristics, microbial communities and environmental conditions. Soil samples were collected at Cedar Creek Ecological Science Reserve (CCESR) in east-central Minnesota (MN1, MN3 and MN5) and the Konza Prairie (KS), two National Science Foundation Long-Term Ecological Research (LTER) sites. In addition, soils were collected at Fort Sherman, Santa Clara and Volcan Baru) in Panama. Soil corers (10 cm × 1 cm or 2.5 × 10 cm, with isolates collected from both samples) were used to collect soil at each location. Samples were transported back to the laboratory and maintained at 12 °C until processing. Soil from each sample was placed under two layers of sterile cheesecloth to dry overnight and 5 g subsamples from each location were subsequently placed into 50 ml centrifuge tubes containing 10 ml of buffered phosphate solution (0.5 M K2HPO4, 0.4 M KH2PO4, pH 7.0). Tubes were shaken for 1 h on a reciprocal shaker (4 °C, 250 r.p.m.). Resulting soil suspensions were dilution plated onto oatmeal agar. Plates were incubated at 28 °C for 7 days. Colonies exhibiting characteristic Streptomycete colony morphology were picked, purified and spore suspensions of each isolate were maintained in 20% glycerol at −80 °C. In total, from 9 to 10 Streptomycete isolates were selected randomly from each sample. Specifically, colonies exhibiting characteristic Streptomycete colony morphology were subcultured, purified and spore suspensions of each isolate were maintained in 20% glycerol at −80 °C. A total of 69 isolates from seven locations (soil cores) were considered in this work. For every isolate, 16S sequence analyses were performed to determine phylogeny (Schlatter et al., submitted).
Assaying inhibitory and resistance phenotypes
Inhibitory interactions were evaluated for all possible pairwise isolate combinations for a total of 4692 interactions. To test for inhibition among isolate pairs, 10 ul of spore stock suspension (∼108 colony forming unit per ml) of each isolate was dotted (a single droplet placed on the plate surface) onto starch-casein agar and grown for 3 days at 28 °C. Dotted isolates were killed by inverting the uncovered petri plates over 4 ml of chloroform in a watch glass for 1 h. Watch glasses were removed and plates were aerated in a flow hood for 30 min to permit the evaporation of chloroform. Plates were subsequently overlaid with 15 ml of 1% water agar and inoculated with 100 ul of a test isolate (∼108 colony-forming units per ml) spread uniformly over the surface of the agar; this results in a dense bacterial lawn over the plate surface in the absence of any inhibitory isolates. Plates were incubated at 28 °C for 3 days. The size of any zone of growth inhibition of the overlaid isolate surrounding any dotted isolate was measured in millimeters from the edge of the dotted colony to the edge of the cleared zone; each inhibition zone was measured in two locations, at right angles to one another. Each isolate pair was replicated three times among Minnesota isolates and two times among isolates from all other locations. Among global isolate pairs, inconsistent inhibition phenotypes (difference in zone size between measurements was greater than 2 mm) were repeated a third time. Inhibitory interactions were defined as interactions in which inhibition of one isolate by another was evident (zone size >1.0 mm). Interactions were categorized as resistant in the absence of any inhibition zone. The intensity of inhibition was defined as the mean zone size of inhibition of one isolate by another, averaged over all replicates for that isolate combination.
Characterizing nutrient utilization
Nutrient use profiles (niche) were evaluated for each isolate for 95 single carbon sources using Biolog SF-P2 plates as described previously (Schlatter et al., 2009). Briefly, spore suspensions (OD590=0.22) were used to inoculate Biolog plates following the manufacturer's instructions. Plates were incubated for 3 days at 28 °C and the growth on each carbon source was evaluated by measuring the absorbance of each well at 590 nm using a Multiskan EX microplate reader. The absorbance of the water control well was subtracted from the reading for each well.
Data analyses
Pairwise inhibitory interactions among isolates were used to determine the specific inhibition and resistance profiles for each isolate and the proportion of inhibitory interactions among isolates from each location. Sympatric isolate pairs were defined as Streptomyces from the same soil core and allopatric isolates were from different soil cores. Niche overlap for every isolate pair was calculated using the formula:
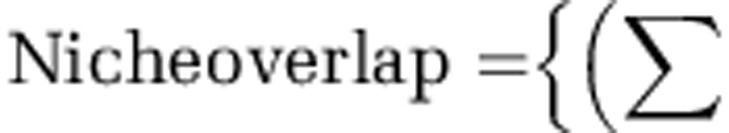 Min absorbance a, b/total absorbance a+
Min absorbance a, b/total absorbance a+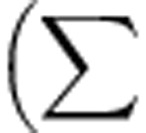 Min absorbance a, b
Min absorbance a, b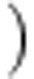 /total absorbance b/2
/total absorbance b/2
where (Min absorbance a, b) is the minimum absorbance value for a pair of Streptomycete isolates (a and b) on a given nutrient; these values are summarized overall n=95 nutrients. The total absorbance for an isolate a or b is the sum of absorbance values for that isolate over all n=95 nutrients. Thus, niche overlap for any isolate pair (a and b) is the mean proportion of total nutrient use (absorbance values) which overlaps between the two isolates. All statistical analyses were conducted in SAS (SAS Institute Inc., Cary, NC, USA) or R.
Results and discussion
Inhibitory interactions among sympatric and allopatric isolates
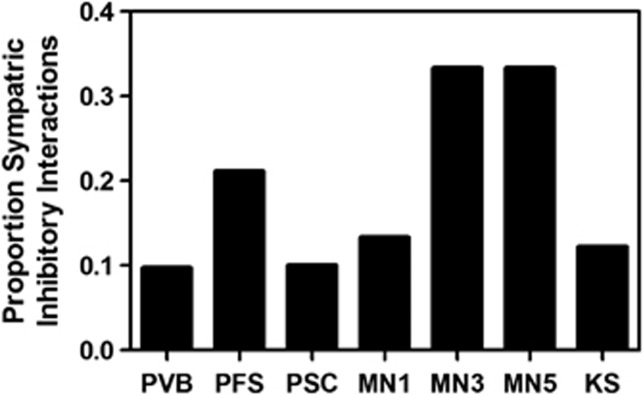
Frequencies of inhibition among sympatric isolates varied significantly among locations, ranging from 10% (10% of possible pairwise isolate interactions were inhibitory among isolates from Panama Volcan Baru and Panama Santa Clara) to 33% (MN3 and MN5), suggesting that the significance of inhibition to microbial interactions varies across the landscape (Figure 1). Among individual isolates, there was an extraordinary diversity of distinct inhibitory phenotypes. Most isolates (86%) were unique with respect to the specific profile of isolates they inhibited. Consistent with previous work, individual Streptomycete isolates resisted inhibition consistently better than they inhibited others (Davelos et al., 2004; D'Costa et al., 2006), and on average were resistant to 83% of all other isolates. The greater accumulation of resistance vs inhibitory phenotypes suggests that there are consistently greater fitness benefits and/or smaller fitness costs for resistance than inhibitory phenotypes. For example, there is evidence from clinical settings that compensatory mutations may significantly reduce the costs to maintaining antibacterial resistance phenotypes, suggesting the possibility that, once acquired, there may be little loss of antibiotic resistance beyond genetic drift (Salyers and Amabile-Cuevas, 1997; Davies and Davies, 2010). Such ‘fossil resistance' may contribute significantly to the total resistance profile of soil microbes. The greater frequency of resistance may also reflect a significant role for general resistance mechanisms that confer protection against multiple distinct antibiotics (for example, efflux pumps) (Martinez et al., 2009b). Finally, resistance phenotypes may plausibly enhance microbial fitness in ways unrelated to protection against antibiotics (Martinez et al., 2009b; Allen et al., 2010; Davies and Davies, 2010), potentially contributing to the maintenance of resistance phenotypes in the absence of antibiotic selection. Regardless of the specific mechanisms leading to the accumulation of resistance capacities within individual isolates, the data are consistent with a substantial role for antibiotic resistance phenotypes in the fitness of Streptomycetes across their geographic range.
Figure 1.
Frequency of inhibitory interactions among sympatric isolate pairs for each of the seven sampling locations. Differences in the frequency of sympatric inhibitory interactions varied significantly among locations (χ2=28.3; P<0.001). MN1, MN3 and MN5 represent Minnesota 1, 3 and 5, respectively; KS, Konza Prairie; PVB, Panama Volcan Baru; PFS, Panama Fort Sherman and PSC, Panama Santa Clara. There were n=9 to 10 isolates from each location, resulting in a total of n=72–90 sympatric interactions per location (n=612 sympatric interactions total).
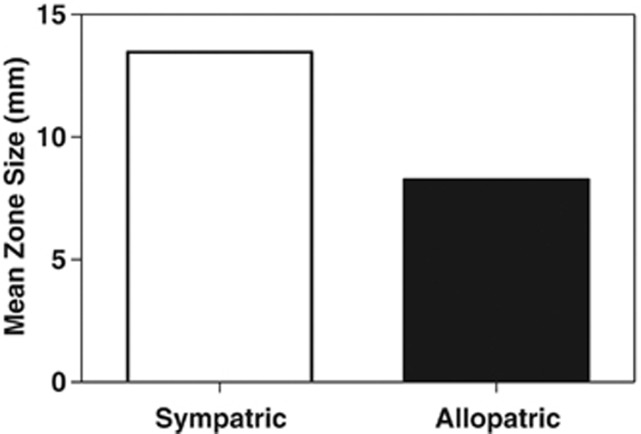
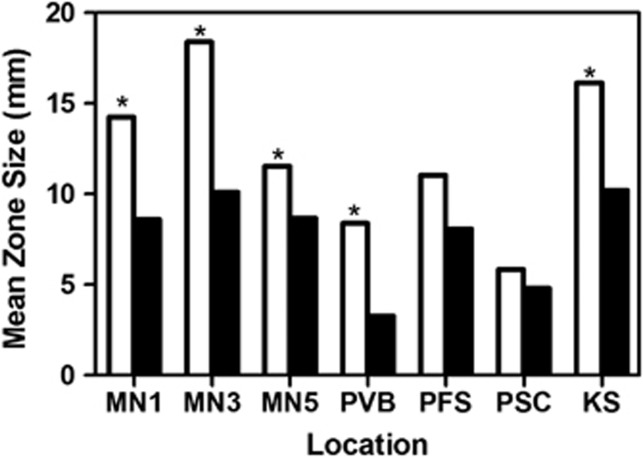
The broad variation in antibiotic inhibitory and resistance phenotypes among isolates within and among locations suggests significant potential for selection. Sympatric and allopatric inhibition were evaluated to determine whether there is evidence for local selection of inhibitory phenotypes. If the production of antibiotics that are highly effective against sympatric competitors confers a specific fitness benefit to the producer, then the intensity of antibiotic inhibition should be greater among sympatric than allopatric Streptomycetes. Among all inhibitory interactions, the intensity of antibiotic inhibition was significantly greater among sympatric than among allopatric isolates (Figure 2). Within individual locations, inhibition was also always greater against sympatric than allopatric isolates (seven of seven locations), although differences were statistically significant in only five of the seven locations (Figure 3). These data suggest that antibiotics that are strongly inhibitory toward coexisting isolates provide a local fitness benefit, and that antibiotic phenotypes are under local selection. Variation in the intensity of antibiotic inhibition among locations suggests shifts in the significance of competitive interactions and, presumably, antibiotics to Streptomycete fitness across the landscape. Populations having highly inhibitory phenotypes (for example, location MN3) may experience the greatest fitness costs due to competitive interactions and, consequently, strongest selection for inhibitory phenotypes. In contrast, communities having less inhibitory local interactions (for example, Panama Santa Clara), may be locations in which competitive interactions are less important to fitness, perhaps reflecting a significant influence of the physical environment on fitness. For example, Panama Santa Clara was located in a dunegrass stand on the edge of a hot, dry, sandy beach site along the Pacific coast in Panama. Alternatively, communities with less inhibitory interactions may be sites where antibiotics serve other functions (for example, may act as signals rather than as weapons in mediating interactions), and thus there is little production of antibiotics at inhibitory concentrations, or habitats where temperature and moisture stress, rather than species interactions, limit fitness, thus minimizing the benefits of inhibitory phenotypes. Although variation in the dynamics of antibiotic-mediated interactions and the significance of antibiotic inhibition to Streptomycete fitness may lead to diverse local coevolutionary dynamics across the landscape (Thrall and Burdon, 2003; Thompson, 2005; Nuismer and Thompson, 2006), overall the data provide compelling evidence for the significance of antibiotic inhibitory phenotypes to local interactions and Streptomyces fitness in soil.
Figure 2.
Intensity of antibiotic inhibition (mean zone size) among all inhibitory isolate pairs from the same (sympatric, n=118 interactions) and different (allopatric, n=667 interactions) locations. Mean zone size differed significantly among sympatric vs allopatric isolate pairs (P<0.0001, Welch two-sample t-test).
Figure 3.
Intensity of antibiotic inhibition (mean zone size) among all inhibitory isolate pairs from the same (sympatric, open bars) and different (allopatric, solid bars) locations. There were n=9 to 10 isolates from each location, and a total of n=72–90 sympatric interactions and 540–590 allopatric interactions for each location. Pairs of bars from the same location marked with an asterisk differ significantly (Welch two-sample t-test t=−6.5; P<0.05). MN1, MN3 and MN5 represent Minnesota 1, 3 and 5, respectively; KS, Konza Prairie; PVB, Panama Volcan Baru; PFS, Panama Fort Sherman and PSC, Panama Santa Clara.
Resistance interactions among sympatric and allopatric isolate pairs
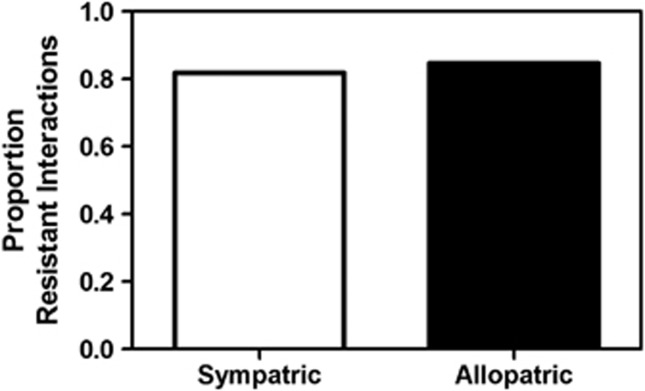
In contrast to the inhibitory phenotypes, there was no consistent evidence among locations for the accumulation of resistance to locally coexisting isolates. Specifically, comparing the frequency of resistance to antibiotics produced by sympatric vs allopatric Streptomycetes, there was greater resistance to sympatric than allopatric isolates in four of the seven locations, and greater resistance to allopatric than sympatric isolates in three of the seven locations, although differences were statistically significant in only two locations (data not shown). When data from all locations were combined, there was no significant difference in the frequencies of resistance to allopatric vs sympatric isolates (Figure 4). The apparent lack of locally adapted resistance suggests a number of possibilities about the evolutionary or coevolutionary dynamics of antibiotic inhibitory and resistance phenotypes within soil Streptomycete communities. Although one possible interpretation is that resistance does not confer a fitness benefit in local species interactions, this seems unreasonable given the accumulation of resistance among Streptomycete isolates from all locations. Instead, the lack of locally adapted resistance may reflect different cost:benefit ratios of antibiotic production (high cost, high benefit) vs resistance (low cost, high benefit). Low fitness costs or a high frequency of compensatory mutations that reduce costs of resistance may lead to the retention of resistance in local habitats long after an antibiotic has been ‘lost' from the population (Bottger et al., 1998; Lenski and Riley, 2002; Luciani et al., 2009; Brandis et al., 2012; Dillon and Parti, 2012; Sousa et al., 2012). Specifically, although resistance phenotypes may be under strong local selection in the presence of a novel antibiotic, there may be a correspondingly rapid loss of antibiotic production capacity in the presence of high frequencies of resistance to that antibiotic. Novel antibiotic inhibitory phenotypes may be highly transient within populations (Cordero et al., 2012), reflecting the metabolic dexterity of Streptomyces (Tanaka and Omura, 1990; Challis and Hopwood, 2003), the high costs of antibiotic production (Williams and Vickers, 1986; although note Garbeva et al., 2011) and the frequencies of local resistance. This may generate an unbalanced coevolutionary dynamic in which local adaptation of inhibitory phenotypes is stronger than local adaptation of resistance, as resistance against antibiotics no longer produced by local competitors may be retained within the population. Consequently, evidence for local selection for resistance may be difficult to detect. General resistance mechanisms may also minimize the likelihood of locally adapted resistance. Finally, experimental constraints may further limit the detection of locally adapted resistance within Streptomycete populations. In particular, inhibition phenotypes were measured at a single time point and in simple pairwise combinations. For some Streptomycete isolates (but not others), there may be antibiotics that are not produced until later in the growth cycle or only in response to a specific signal. In this case, we may detect resistance to antibiotics that are in fact significant to local species interactions but undetected by our assays. Selection for resistance may also be imposed by other non-Streptomycete bacteria and fungi. More extensive, time-series analyses of antibiotic inhibitory and resistance phenotypes within coexisting populations under diverse cultural conditions are needed to determine the extent to which novel resistance phenotypes proliferate in response to new inhibitory phenotypes as predicted by coevolutionary models.
Figure 4.
Frequency of non-inhibitory (resistant) interactions among all possible isolate combinations from the same (sympatric, n=612 interactions) and different (allopatric, n=4080 interactions) locations. The frequency of resistant interactions was not significantly different between sympatric vs allopatric isolate pairs (χ2=0.501; P=0.48).
Niche overlap and inhibition among sympatric and allopatric isolates
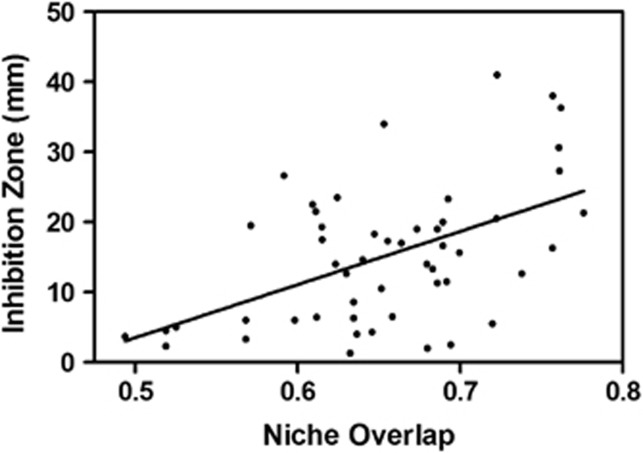
Finally, to explore the role of antibiotics in mediating competitive interactions among Streptomycetes, we considered the relationships between niche (nutrient use) overlap and antibiotic inhibition among Streptomycetes. Specifically, do inhibitory interactions target locally coexisting nutrient competitors? Among inhibitory isolate pairs, there was a significant positive correlation between niche overlap and the intensity of antibiotic inhibition among sympatric (R=0.29; P=0.0013) but not allopatric isolate pairs (R=0.009; P=0.478). Thus, isolates from the same location were better at inhibiting isolates with which they had large niche overlap than isolates with which they exhibited little niche overlap. In contrast, there was no relationship between niche overlap and the intensity of inhibition among Streptomycetes from different locations in soil, indicating that the niche-inhibitory phenotype relationship reflects local selection and not physiological tradeoffs. Local selection for inhibitory phenotypes is significantly related to niche overlap and presumably the intensity of competition between isolates from the same location in soil. The strength of this relationship varied and was not always significant within locations: among distinct locations, niche overlap explained from 7% to 60% of the total variation in inhibition zone sizes for sympatric isolate pairs within a location (for example, Figure 5). These data are consistent with the hypothesis that antibiotics mediate nutrient competition among Streptomycete populations within some local communities. However, the data also suggest that the significance of antibiotics to competitive interactions vary across the landscape, with communities ranging from those in which competition is ‘managed' predominantly by niche differentiation to communities that are strongly antagonistic. This may reflect habitat variation (for example locations with little nutrient diversity and thus limited potential for niche differentiation may be most likely to support antagonistic interactions), or the evolutionary potential of local populations. Perhaps most importantly, these data offer a distinct alternative to a coevolutionary arms race among interacting Streptomycetes. Among isolates with little niche overlap, there is relatively less accumulation of inhibitory capacity. Thus, niche differentiation may significantly reduce the strength of selection for strong inhibitory phenotypes, and coevolutionary character displacement (Thompson, 2005) may minimize the potential for a coevolutionary arms race among sympatric Streptomycete populations in soil.
Figure 5.
Relationships between mean niche overlap and mean inhibition zone size among sympatric inhibitory isolate combinations from three locations (PSC, MN1 and MN3; sympatric interactions only; R=0.51, P<0.0001; n=51 independent isolate combinations).
We conclude that antibiotic inhibitory phenotypes are under significant local selection and mediate nutrient competition in local soil communities. However, the relatively greater accumulation of resistance than inhibitory phenotypes among Streptomyces isolates in natural habitats suggests that an escalating coevolutionary arms race may represent a cost-prohibitive trajectory for antibiotic-producers or that coevolutionary polymorphisms may be unbalanced at best. The significant positive association of inhibition with nutrient overlap among locally coexisting Streptomycetes suggests that antibiotics are likely to act as ‘weapons' against sympatric competitors, but that coevolutionary displacement may provide a significant means for minimizing fitness costs of interspecies resource conflicts and the likelihood of a coevolutionary arms race.
This work sheds light on the roles of antibiotics in microbial interactions and coevolutionary dynamics within complex soil communities. However, because this study emphasizes inhibition and resistance among isolate pairs, these results are likely to underestimate both the complexity of Streptomycete coevolutionary dynamics and the potential outcomes of multispecies antibiotic inhibitory and resistance interactions in natural communities (Chait et al., 2012). Deeper understanding of the diverse roles of antibiotics in species interactions in soil, the dynamics of antibiotic inhibition in natural habitats and especially the factors that may determine the potential for a coevolutionary arms race vs coevolutionary differentiation are crucial for understanding the long-term trajectories of antibiotic-producing microbes in soil. Recent work suggests the potential for soil edaphic characteristics, nutrient availability or nutrient diversity in soil, physical environmental stress and phylogeny to predict microbial inhibitory activities and coevolutionary interactions in soil populations (Schlatter et al., 2009; Bakker et al., 2010; Kinkel et al., 2011; Bailey and Kassen, 2012; Otto-Hanson et al., 2013). More detailed understanding of the precise roles of these factors in mediating microbial species interactions and coevolution in soil will contribute significantly to the search for novel antibiotic biochemistries, to enhanced insight into the maintenance of antibiotic resistance genes in environmental microbes and to management of disease suppressive activity of indigenous soil microbes (Martinez et al., 2011; Kinkel et al., 2011; Kinkel et al., 2012).
Acknowledgments
This work was supported by the United States. National Science Foundation Microbial Observatories Program Grant 9977907, the United States Department of Agriculture Microbial Observatories Grant 2006-35319-17445 and the United State National Science Foundation Long-Term Ecological Research Grant 0620652. This project was also supported by Agriculture and Food Research Initiative Competitive Grant 2011-67019-30200 from the USDA National Institute of Food and Agriculture. Jennifer Flor, Dale Jensen, Lindsey Hanson and Lizzie Brodeen contributed technical support.
References
- Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- Aminov RI. The role of antibiotics and antibiotic resistance in nature. Environ Microbiol. 2009;11:2970–2988. doi: 10.1111/j.1462-2920.2009.01972.x. [DOI] [PubMed] [Google Scholar]
- Bagge N, Schuster M, Hentzer M, Ciofu O, Givskov M, Greenberg EP, et al. Pseudomonas eruginosa biofilms exposed to imipenem exhibit changes in global gene expression and β-lactamase and alginate production. Antimicrob Agents Chemother. 2004;48:1175–1187. doi: 10.1128/AAC.48.4.1175-1187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SF, Kassen R. Spatial structure of ecological opportunity drives adaptation in bacterium. Amer Nat. 2012;180:270–283. doi: 10.1086/666609. [DOI] [PubMed] [Google Scholar]
- Bakker MG, Glover JD, Mai JG, Kinkel LL. Plant community effects on the diversity an pathogen suppressive activity of soil streptomycetes. Appl Soil Ecol. 2010;46:35–42. [Google Scholar]
- Baltz RH. Antimicrobials from actinomycetes: back to the future. Am Soc Microbiol. 2007;2:125–131. [Google Scholar]
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Bottger EC, Springer B, Pletschette M, Sander P. Fitness of antibiotic resistant microorganisms and compensatory mutations. Nature Med. 1998;4:1343–1344. doi: 10.1038/3906. [DOI] [PubMed] [Google Scholar]
- Brandis G, Wrande M, Lilijas L, Hughes D. Fitness-compensatory mutations in rifampicin-resistant RNA polymerase. Mol Microbiol. 2012;85:142–151. doi: 10.1111/j.1365-2958.2012.08099.x. [DOI] [PubMed] [Google Scholar]
- Bressan W, Figueiredo JE. Efficacy and dose-response relationship in biocontrol of Fusarium disease in maize by Streptomyces spp. Eur J Plant Pathol. 2007;120:311–316. [Google Scholar]
- Chait R, Vetsigian K, Kishony R. What counters antibiotic resistance in nature. Nat Chem Biol. 2012;8:2–5. doi: 10.1038/nchembio.745. [DOI] [PubMed] [Google Scholar]
- Challis GL, Hopwood DA. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci USA. 2003;100:14555–14561. doi: 10.1073/pnas.1934677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater KF, Biro S, Lee KJ, Palmer T, Schrempf H. The complex extracellular biology of Streptomyces. FEMS Microbiol Rev. 2010;34:171–198. doi: 10.1111/j.1574-6976.2009.00206.x. [DOI] [PubMed] [Google Scholar]
- Cordero OX, Wildschutte H, Kirkup B, Proehl S, Ngo L, Hussain F, et al. Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science. 2012;337:1228–1231. doi: 10.1126/science.1219385. [DOI] [PubMed] [Google Scholar]
- Cummins JF, Reen JF, Bayesse C, Mooij MJ, O'Gara F. Subinhibitory concentrations of the cationic antimicrobial peptide colistin induce the Pseudomonas quinolone signal in Pseudomonas aeruginosa. Microbiol. 2009;155:2826–2837. doi: 10.1099/mic.0.025643-0. [DOI] [PubMed] [Google Scholar]
- Czaran TL, Hoekstra RF, Pagle L. Chemical warfare between microbes promotes biodiversity. Proc Natl Acad Sci USA. 2002;99:786–790. doi: 10.1073/pnas.012399899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaran TL, Hoekstra RF. Killer-sensitive coexistence in metapopulations of micro-organisms. Proc Roy Soc B: Biol Sci. 2003;270:1373–1378. doi: 10.1098/rspb.2003.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- Davelos AL, Kinkel LL, Samac DA. Spatial variation in the frequency and intensity of antibiotic interactions among streptomycetes from prairie soil. Appl Environ Microbiol. 2004;70:1051–1058. doi: 10.1128/AEM.70.2.1051-1058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Molec Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon JR, Parti RP. Fluoroquinolone resistance in Neisseria gonorrhoeae: fitness cost or benefit. J Infect Dis. 2012;205:1775–1777. doi: 10.1093/infdis/jis281. [DOI] [PubMed] [Google Scholar]
- Fajardo A, Martinez JL. Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol. 2008;11:161–167. doi: 10.1016/j.mib.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Garbeva P, Tyc O, Remus-Emsermann MNP, van der Wal A, Vos M, Silby M, et al. No apparent costs for facultative antibiotic production by the soil bacterium Pseudomonas fluorescens Pf0-1. PLoS One. 2011;6:E27266. doi: 10.1371/journal.pone.0027266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh E-B, Yim G, Tsui W, McClure J, Surette MG, Davies J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci USA. 2002;99:17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontang EA, Fenical W, Jensen PR. Phylogenetic diversity of gram-positive bacteria cultured from marine sediments. Appl Environ Microbiol. 2007;73:3272–3282. doi: 10.1128/AEM.02811-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen LH, Ojanpera T, Kortemaa H, Richter E, Lehtonen MJ, Valkonen JP. Interactions and biocontrol of pathogenic Streptomyces strains co-occurring in potato scab lesions. J Appl Microbiol. 2009;106:199–212. doi: 10.1111/j.1365-2672.2008.03992.x. [DOI] [PubMed] [Google Scholar]
- Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SL. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- Kaplan JB, Izano EA, Gopal P, Karwacki MT, Kim S, Bose JL, et al. Low levels of β-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. MBio. 2012;3:e00198–12. doi: 10.1128/mBio.00198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi E, Sadeghi A, Abbaszade Dehaji P, Dalvand Y, Omidvari Biocontrol activity of salt tolerant Streptomyces isolates against phytopathogens causing root rot of sugar beet. Bio Sci Tech. 2012;22:333–349. [Google Scholar]
- Kinkel LL, Bakker MG, Schlatter DC. A coevolutionary framework for managing disease-suppressive soils. Annu Rev Phytopathol. 2011;49:47–67. doi: 10.1146/annurev-phyto-072910-095232. [DOI] [PubMed] [Google Scholar]
- Kinkel LL, Schlatter DS, Bakker MB, Arenz B. Streptomyces competition and coevolution in relation to disease suppression. Res Microbiol. 2012;163:490–499. doi: 10.1016/j.resmic.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Laskaris P, Tolba S, Calvo-Bado L, Wellington L. Coevolution of antibiotic production and counter-resistance in soil bacteria. Environ Microbiol. 2010;12:783–796. doi: 10.1111/j.1462-2920.2009.02125.x. [DOI] [PubMed] [Google Scholar]
- Lin JT, Connelly MB, Amolo C, Otan S, Yaver DS. Global transcriptional response of Bacillus subtilis to treatment with subinhibitory concentrations of antibiotics that inhibit protein synthesis. Antimicrob Agents Chemother. 2005;49:1915–1926. doi: 10.1128/AAC.49.5.1915-1926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares JF, Gustafsson I, Baquero F, Martinez JF. Antibiotics as intermicrobial signaling agents instead of weapons. Proc Natl Acad Sci USA. 2006;103:19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski R, Riley MA. Chemical warfare from an ecological perspective. Proc Natl Acad Sci USA. 2002;99:556–558. doi: 10.1073/pnas.022641999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani F, Sisson SA, Jiang H, Francis AR, Tanaka MM. The epidemiological fitness cost of drug resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2009;106:14711–14715. doi: 10.1073/pnas.0902437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JL. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008;321:365–367. doi: 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- Martinez JL, Baquero F, Andersson DI. Beyond serial passages: new methods for predicting the emergence of resistance to novel antibiotics. Curr Opin Pharm. 2011;11:439–445. doi: 10.1016/j.coph.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Martinez JL, Fajardo A, Garmendia L, Hernandez A, Linares JF, Martinez-Solano L, et al. A global view of antibiotic resistance. FEMS Microbiol Rev. 2009a;33:44–65. doi: 10.1111/j.1574-6976.2008.00142.x. [DOI] [PubMed] [Google Scholar]
- Martinez JL, Sanchez MB, Martinez-Solano L, Hernandez A, Garmendia L, Farjardo A, et al. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol Rev. 2009b;33:430–449. doi: 10.1111/j.1574-6976.2008.00157.x. [DOI] [PubMed] [Google Scholar]
- Meschke H, Walter S, Schrempf H. Characterization and localization of prodiginines from Streptomyces lividans suppressing Verticillium dahliae in the absence or presence of Arabidopisis thaliana. Environ Microbiol. 2012;14:940–952. doi: 10.1111/j.1462-2920.2011.02665.x. [DOI] [PubMed] [Google Scholar]
- Mitova MI, Lang G, Wiese J, Imhoff JF. Subinhibitory concentrations of antibiotics induce phenazine production in a marine Streptomyces sp. J Nat Prod. 2008;71:824–827. doi: 10.1021/np800032a. [DOI] [PubMed] [Google Scholar]
- Ng W-L, Kazmierczak KM, Robertson GT, Gilmour R, Winkler ME. Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J Bacteriol. 2003;185:359–370. doi: 10.1128/JB.185.1.359-370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuismer SL, Thompson JN. Coevolutionary alternation in antagonistic interactions. Evol. 2006;60:2207–2217. [PubMed] [Google Scholar]
- Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto-Hanson LK, Grabau Z, Rosen C, Salomon CE, Kinkel LL. Pathogen variation and urea influence selection and success of Streptomyces mixtures in biocontrol. Phytopathol. 2013;103:34–42. doi: 10.1094/PHYTO-06-12-0129-R. [DOI] [PubMed] [Google Scholar]
- Riley MA, Gordon DM. The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 1999;7:129–133. doi: 10.1016/s0966-842x(99)01459-6. [DOI] [PubMed] [Google Scholar]
- Romero D, Traxler MF, Lopez D, Kolter R. Antibiotics as signal molecules. Chem Rev. 2011;111:5492–5505. doi: 10.1021/cr2000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RP, Dow JM. Diffusible signals and interspecies communication in bacteria. Microbiol. 2008;154:1845–1858. doi: 10.1099/mic.0.2008/017871-0. [DOI] [PubMed] [Google Scholar]
- Salyers AA, Amabile-Cuevas CF. Why are antibiotic resistance genes so resistant to elimination. Antimicrob Agents Chemother. 1997;41:2321–2325. doi: 10.1128/aac.41.11.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlatter D, Fubuh A, Xiao K, Hernandez D, Hobbie S, Kinkel L. Resource amendments influence density and competitive phenotypes of Streptomyces in soil. Microb Ecol. 2009;57:413–420. doi: 10.1007/s00248-008-9433-4. [DOI] [PubMed] [Google Scholar]
- Sousa A, Magalhaes S, Gordo I. Cost of antibiotic resistance and the geometry of adaptation. Mol Biol Evol. 2012;29:1417–1428. doi: 10.1093/molbev/msr302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Omura S. Metabolism and products of actinomycetes: an introduction. Actinomycetologica. 1990;4:13–14. [Google Scholar]
- Thompson JN. The Geographic Mosaic of Coevolution. Univ Chicago Press: Chicago, IL, USA; 2005. [Google Scholar]
- Thrall PH, Burdon JJ. Evolution of virulence in a plant host-pathogen metapopulation. Science. 2003;299:1735–1737. doi: 10.1126/science.1080070. [DOI] [PubMed] [Google Scholar]
- Vetsigian K, Jajoo R, Kishony R. Structure and evolution of Streptomyces interaction networks in soil and in silico. PLoS One. 2011;9:e1001184. doi: 10.1371/journal.pbio.1001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watve MG, Tickoo R, Jog MM, Bhole BD. How many antibiotics are produced by the genus Streptomyces. Arch Microbiol. 2001;176:386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- Williams DH, Stone MJ, Hauck PR, Rahman SK. Why are secondary metabolites (natural products) biosynthesized. J Nat Prod. 1989;42:1189–1208. doi: 10.1021/np50066a001. [DOI] [PubMed] [Google Scholar]
- Williams ST, Vickers JC. The ecology of antibiotic production. Microb Ecol. 1986;12:43–52. doi: 10.1007/BF02153221. [DOI] [PubMed] [Google Scholar]
- Wloch-Salamon DM, Gerla D, Hoekstra RF, de Visser JAG. Effect of dispersal and nutrient availability on the competitive ability of toxin-producing yeast. Proc Roy Soc B: Biol Sci. 2008;275:535–541. doi: 10.1098/rspb.2007.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim G, Wang HMH, Davies J. Antibiotics as signaling molecules. Phil Trans Roy Soc B-Biol Sci. 2007;362:1195–1200. doi: 10.1098/rstb.2007.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]