Abstract
The nonessential process of peptidoglycan synthesis during Bacillus subtilis sporulation is one model to study bacterial cell wall biogenesis. SpoVD is a class B high-molecular-weight penicillin-binding protein that is specific for sporulation. Strains lacking this protein produce spores without the peptidoglycan cortex layer and are heat sensitive. The detailed functions of the four different protein domains of SpoVD are unknown, and the observed phenotype of strains lacking the entire protein could be an indirect defect. We therefore inactivated the transpeptidase domain by substitution of the active-site serine residue. Our results demonstrate that endospore cortex synthesis depends on the transpeptidase activity of SpoVD specifically.
Keywords: peptidoglycan, penicillin-binding protein, endospore
Introduction
The cell wall in bacteria determines shape and provides structural support and protection to the cell. Peptidoglycan is the major structural component of the bacterial cell wall. It consists of glycan strands that are cross-linked via short peptide chains. Biosynthesis of peptidoglycan can be divided into three stages: (1) synthesis of precursor molecules in the cytoplasm, (2) transport of lipid II across the membrane and (3) incorporation of disaccharide-peptide units into nascent peptidoglycan at the outer side of the cytoplasmic membrane (Foster & Popham, 2002; Typas et al., 2012). The incorporation of disaccharide-peptide units is achieved through the action of penicillin-binding proteins (PBPs), which catalyze transglycosylation and transpeptidation reactions (Scheffers, 2007; Typas et al., 2012). Bacterial cell wall synthesis is an effective target for many antibiotics in clinical use, such as penicillins and cephalosporins (Bugg et al., 2011). However, peptidoglycan synthesis and cell wall morphogenesis are still far from understood at the molecular level. The complex macromolecular structure of peptidoglycan and the essential nature of many proteins involved in its synthesis make experimental studies difficult.
Peptidoglycan synthesis occurs during sporulation in Bacillus subtilis in the form of spore cortex synthesis that takes place in the intermembrane space of the forespore (Eichenberger, 2012). Sporulation takes several hours to complete and involves a series of morphological changes. Upon initiation of sporulation, the cell divides asymmetrically to form the smaller forespore and the larger mother cell. Subsequently, the forespore becomes engulfed by the mother cell in a phagocytosis-like process, which results in the formation of a double-membrane-enclosed forespore in the mother cell cytoplasm. Upon completion of engulfment, the cortex layer is assembled in the forespore intermembrane compartment and the multilayered protein coat is formed on the surface of the forespore. Finally, the mature spore is released via lysis of the mother cell (Piggot & Hilbert, 2004; Eichenberger, 2012).
Cortex synthesis, unlike vegetative cell wall synthesis, is not essential for cell viability and growth. Therefore, peptidoglycan synthesis during sporulation enables analysis of mutants defective in enzymes that otherwise are essential for growth and offers an experimental system to elucidate cell wall assembly. Heat resistance of spores depends on the presence of the cortex layer (Todd et al., 1986), and loss of heat resistance provides a convenient assay in screening for mutants defective in cortex synthesis.
SpoVD is a class B high-molecular-weight PBP that is essential for spore cortex synthesis (Daniel et al., 1994). SpoVD has four domains: an N-terminal single transmembrane segment (c. 46 residues) followed by a domain of an unknown function (c. 180 residues), a transpeptidase domain (c. 339 residues) and C-terminal PASTA domain (c. 60 residues). The detailed functions of the domains are unknown, but presumably the transpeptidase domain catalyzes the formation of peptide cross-links between glycan strands in nascent cortex. The activity of the transpeptidase domain of SpoVD seems regulated by a dithiol-based redox switch not previously reported for any PBP (Liu et al., 2010).
Previous studies on the role of SpoVD in cortex synthesis were carried out using various mutants in which the spoVD gene was insertionally inactivated (Daniel et al., 1994; Fay et al., 2010; Liu et al., 2010), leading to absence of the SpoVD protein. SpoVD is believed to form a peptidoglycan synthesis multiprotein complex together with SpoVE (Fay et al., 2010) and other not yet identified proteins. SpoVE is a putative lipid II flippase that recently was shown to depend on SpoVD for stability against degradation in sporulating cells (Fay et al., 2010). SpoVE- and SpoVD-deficient B. subtilis mutants show identical phenotypes, that is, form heat-sensitive spores completely lacking the cortex layer. The object of the work reported here was to elucidate whether the synthesis of cortex depends on the transpeptidase activity of SpoVD or on some other function of this membrane protein.
Materials and methods
Bacterial strains and growth media
Used bacterial strains are listed in Table 1. Escherichia coli TOP10 was used to propagate plasmid DNA. Escherichia coli strains were grown at 37 °C in LB medium or on LB agar plates (Sambrook & Russell, 2001). Bacillus subtilis strains were grown at 30 or 37 °C in LB medium, nutrient sporulation medium with phosphate (NSMP) (Fortnagel & Freese, 1968), growth medium and resuspension medium for induction of sporulation (Nicholson & Setlow, 1990), Spizizen's minimal medium (SMM) (Harwood & Archibald, 1990) or on tryptose blood agar base (TBAB) plates (Difco). Antibiotics were used when appropriate at the following concentrations: ampicillin 100 μg mL−1 for E. coli, and spectinomycin 100 μg mL−1, erythromycin 1 μg mL−1, chloramphenicol 3–5 μg mL−1, tetracycline 15 μg mL−1 for B. subtilis. TBAB medium supplemented with 1% (w/v) soluble starch was used to test amylase activity of B. subtilis colonies.
Table 1.
Strains and plasmids used in this work
| Strain or plasmid | Genotype/Description | Origin/Reference |
|---|---|---|
| E. coli | ||
| TOP10 | F- mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ−; StrR | Invitrogen |
| B. subtilis | ||
| 1A1 | trpC2 | BGSC* |
| LMD100 | trpC2 spoVDΩpLEB2; CmR | This work |
| LMD101 | trpC2 ΔspoVD | This work |
| LMD104 | trpC2 ΔspoVD amyE::PspoVD-spoVD-mCherry; SpR | This work |
| LMD115 | trpC2 ΔspoVD amyE:: PspoVD -spoVD(Ser294Ala)-mCherry; SpR | This work |
| Plasmids | ||
| pJM103-I-SceI | Suicide integration vector pJM103 (Perego, 1993) with I-SceI restriction site; ApR, CmR | Perego (1993) |
| pBKJ223 | I-SceI expression vector; ApR, TcR | Janes & Stibitz (2006) |
| pDG1730 | amyE integration vector; ApR, SpR, EryR | Guerout-Fleury et al. (1996) |
| pKS-mCherry-E-T3 | E. coli vector for generating gene fusions with mCherry; ApR | N. Ausmees |
| pLEB1 | 289 bp region upstream of spoVD cloned into pJM103-I-SceI; ApR, CmR | This work |
| pLEB2 | 336 bp region downstream of spoVD cloned into pLEB1; ApR, CmR | This work |
| pLEB5 | pKS-mCherry-E-T3 with a 2.0-kb fragment containing PspoVD –spoVD | This work |
| pLEB6 | pDG1730 with a 2.8-kb fragment containing PspoVD -spoVD-mCherry gene fusion; ApR, SpR, EmR | This work |
| pLEB19 | pDG1730 with a 2.8-kb fragment containing PspoVD -spoVD(Ser294Ala)-mCherry; AmpR, SpR, EmR | This work |
Bacillus Genetic Stock Center, Columbus, OH.
Ap, ampicillin; Cm, chloramphenicol; Em, erythromycin; Sp, spectomycin, Str, streptomycin; Tc, tetracycline.
DNA techniques
DNA manipulation was performed by standard methods (Sambrook & Russell, 2001). Plasmid DNA from E. coli was isolated using the Quantum Miniprep (BioRad) or QIAfilter Midi (QIAGEN) plasmid purification kit. Chromosomal DNA from B. subtilis was isolated according to the procedure described by Marmur (Marmur, 1963). PCR was carried out using Phusion high-fidelity DNA polymerase (Finnzymes). Supporting Information, Table S1, shows the sequences of oligonucleotides used to amplify DNA using either B. subtilis chromosomal DNA or plasmid DNA as template. DNA ligation was performed using T4 DNA ligase (New England Biolabs) at 14 °C, over night. Ligates were precipitated prior to transformation into E. coli by electroporation (Hanahan et al., 1991). Bacillus subtilis was grown to natural competence, as described by Hoch (Hoch, 1991), and c. 0.5 μg of DNA was added to 0.5 mL competent cells. All DNA fragments cloned in plasmids were verified by sequence analysis.
Construction of plasmids
Used plasmids are listed in Table 1.
Construction of pLEB2
This plasmid was constructed in two steps. First, a fragment containing the c. 280 bp upstream region of spoVD and the three-first nucleotides of the spoVD open reading frame was amplified by PCR using primers Ewa1 and Ewa2 and B. subtilis 1A1 chromosomal DNA as template. Primers Ewa1 and Ewa2 generated restriction sites for XmaI and BamHI, respectively. Following restriction enzyme digestion, the PCR product was ligated into pJM103-I-SceI cut with the same enzymes, resulting in plasmid pLEB1. Next, a fragment containing the three last nucleotides of spoVD and the c. 330 bp downstream region of spoVD was amplified using primers Ewa3 and Ewa4. The PCR product was digested with BamHI and SphI and inserted into pLEB1 cut with the same enzymes, resulting in plasmid pLEB2.
Construction of pLEB6
Primers Ewa9 and Ewa10 were used to amplify a 2068-bp fragment of the B. subtilis 1A1 chromosome comprising the promoter region and the coding sequence of spoVD (without the stop codon). These primers introduced restriction sites for KpnI and XhoI. The PCR fragment was digested and inserted into KpnI/XhoI-digested pKS-mCherry-E-T3, resulting in plasmid pLEB5. The resulting plasmid encodes a SpoVD-mCherry fusion protein with a linker (LEVDGIDKLDDP). The PspoVD-spoVD-mCherry in-frame gene fusion was amplified from pLEB5 with primers Ewa5 and Ewa13, generating a 2800-bp fragment flanked by EcoRI and BamHI sites. After digestion with EcoRI and BamHI, the PCR product was cloned into EcoRI/BamHI-digested pDG1730, giving plasmid pLEB6.
Construction of pLEB19
The codon for the active-site serine (Ser294) residue of the transpeptidase domain of SpoVD in pLEB6 was changed to encode alanine by site-directed mutagenesis (Phusion® Site-Directed Mutagenesis, Finnzymes) using primers Ewa30 and Ewa31, resulting in plasmid pLEB19.
Construction of B. subtilis strains
All B. subtilis strains described in this work are derivatives of 1A1 (Table 1). Bacillus subtilis LMD101 deleted for spoVD was constructed based on a method described for Bacillus anthracis by Janes and Stibitz (Janes & Stibitz, 2006) with slight modifications. Briefly, pLEB2 (carrying a I-SceI restriction site) was transformed into B. subtilis 1A1, resulting in the integration of the entire plasmid at the spoVD locus by a single crossover event (Campbell-type recombination). The obtained B. subtilis strain, LMD100, was then transformed with pBKJ223 which encodes the I-SceI endonuclease. SceI cleavage generates a double-stranded break in the chromosomal DNA, which can be repaired by homologous recombination resulting in our case in either a markerless deletion of spoVD or retained spoVD. Tetracycline-resistant transformants were scored for loss of the allelic exchange plasmid by patching single colonies onto TBAB plates containing chloramphenicol. Chloramphenicol-sensitive clones were then passed two times on TBAB plates without tetracycline in order to lose pBJ223. Chromosomal DNA was isolated from transformants that were chloramphenicol- and tetracycline-sensitive. The spoVD in-frame deletion was confirmed by PCR (using primers Ewa1 and Ewa4) and DNA sequence analysis. The constructed strain was named LMD101. Bacillus subtilis LMD101 was transformed with pLEB6 and pLEB19 resulting in strains LMD104 and LMD115, respectively.
Light microscopy
A 100 μl sample was taken from the culture of sporulating cells. The cells were collected by centrifugation and suspended in such a volume of phosphate-buffered saline that the density of cells was appropriate for microscopy. Five microlitre of the cell suspension was added on an agarose pad on a microscopy glass slide and covered with a cover slip. Phase contrast and fluorescence images were acquired using a Zeiss Axio Imager.Z1 microscope equipped with X-Cite 120 Illumination (EXFO Photonic Solutions Inc.) and a 9100-02 EM-CCD camera (Hamamatsu Photonics). Eight hundred millisecond exposure time was typically used to obtain fluorescence images of cells. Finally, images were processed in Volocity 5.5 and saved in TIFF format.
Immunoblot analysis
Proteins were fractionated by SDS-PAGE and transferred to a PVDF blotting membrane (Immobilon P, Millipore) using a wet blot. Transfer buffer was 20 mM Tris, 150 mM glycine buffer containing 20% (v/v) methanol. Anti-dsRed antiserum from rabbit (Clontech) was used at a dilution of 1 : 1000. Immunodetection was carried out by chemiluminescence using anti-rabbit secondary antibodies conjugated to horseradish peroxidase (GE Healthcare) in 1 : 3000 dilution and Super Signal® West Pico Chemiluminescent substrate (Pierce Chem. Co.).
Spore assay
Spores were prepared either by growth in NSMP medium as described by Erlendsson et al. (2004) or by the resuspension method (Nicholson & Setlow, 1990). Sporulation efficiency was analyzed by heating 5 mL of the culture at 80 °C for 10 min. Serial dilutions of heated and unheated samples were spread on TBAB plates. The number of colonies was counted after incubation of the plates at 37 °C over night, and the spore yield was calculated.
Bocillin FL-binding assay
The Bocillin FL-binding assay to detect PBPs was performed as described before (Zhao et al., 1999) with the following modifications. Membrane samples containing 3 mg mL−1 of proteins were incubated with 25 μM Bocillin FL (Invitrogen) for 30 min at 35 °C. The reaction was stopped by adding equal volume of 2× SDS sample buffer followed by incubation of samples for 30 min at 40 °C. The proteins (30 μg in each well) were separated by SDS-PAGE on a 8% (w/v) polyacrylamide gel. Bocillin FL-labeled PBPs were detected using a ChemiDoc™ MP Imaging System (Bio-Rad) (excitation light 455–485 nm, emission light filter 532AE28). Subsequently, the gel was stained with Coomassie Brilliant Blue in order to confirm equal amount of proteins loaded in each well.
Miscellaneous methods
Electron microscopy of endospores was performed as described before (Erlendsson et al., 2004).
Membranes from cells collected at different stages of sporulation were isolated as described previously (Hederstedt, 1986). Protein concentrations were determined using the BCA Protein Assay Kit (Thermo Scientific).
Results and discussion
Construction and complementation of an in-frame spoVD deletion in the chromosome of B. subtilis
For convenient analysis of spoVD mutant alleles in B. subtilis, we first constructed a strain, LMD101, which lacks the entire ORF of the spoVD gene. The spoVD gene lies in a cluster of genes involved in cell wall synthesis (Daniel & Errington, 1993; Daniel et al., 1994). Strain LMD101 contains a markerless in-frame spoVD deletion to avoid possible polar effects on downstream genes. Strain LMD101, as expected, forms heat-sensitive spores (Table 2) without cortex layer (Fig. 1b).
Table 2.
Sporulation efficiency of B. subtilis strains
| Strain | Relevant property | CFU mL−1 preheat | CFU mL−1 postheat | % sporulation* |
|---|---|---|---|---|
| 1A1 | Wild type | 3.5 × 108 | 2.7 × 108 | 76 |
| LMD101 | SpoVD− | 1.7 × 107 | < 10 | < 6 × 10−7 |
| LMD104 | SpoVD−, SpoVD-mCherry | 1.1 × 109 | 6.7 × 108 | 55 |
| LMD115 | SpoVD−, SpoVD(Ser296Ala)-mCherry | 1.7 × 108 | < 10 | < 6 × 10−8 |
Cells were grown in NSMP medium for 2 days at 37 °C. Heat resistance of cells was assayed by incubation at 80 °C for 10 min. Sporulation efficiency was calculated as colony-forming units (CFU) after heating the culture divided by CFU of not heated culture. Sporulation assays were performed at least three times per strain. Representative results are shown.
Figure 1.
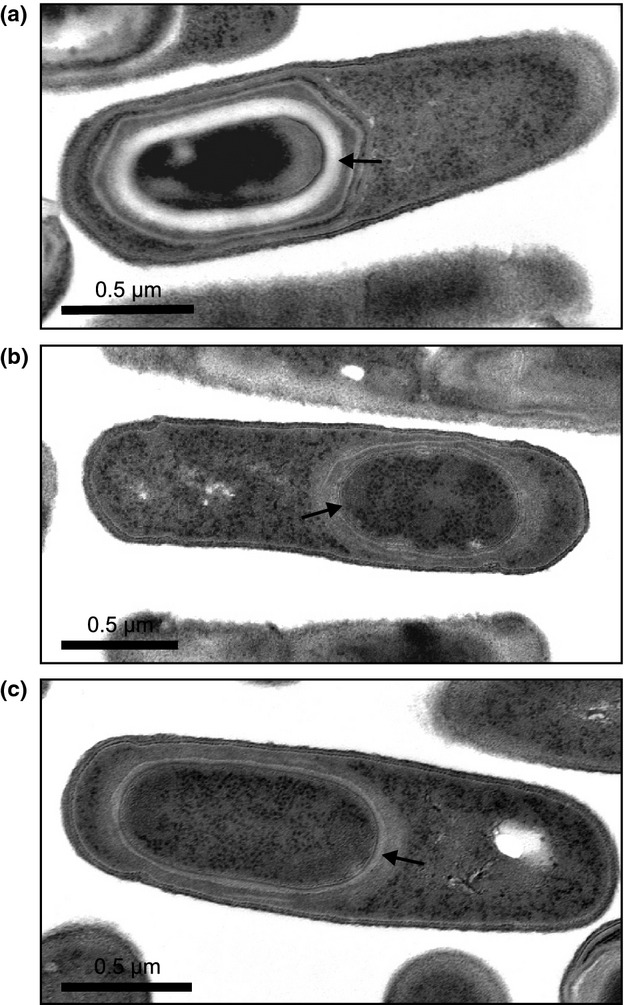
Electron micrographs showing the morphology of the Bacillus subtilis endospore in the mother cell of strains: (a) LMD104 (wild-type SpoVD-mCherry), (b) LMD101 (lacks SpoVD) and (c) LMD115 (contains Ser294Ala substitution in SpoVD-mCherry). The cortex layer in the spore of LMD104 is indicated by an arrow. In the case of the mutant strains, an arrow indicates lack of cortex. Scale bar is 0.5 μm.
To complement the deletion mutant and to study the subcellular localization of SpoVD, a C-terminal translational fusion of mCherry to SpoVD was constructed. The gene fusion was placed under the native spoVD promoter and inserted into the amyE locus of strain LMD101. The obtained strain, LMD104, formed heat-resistant spores (Table 2) with normal cortex layer (Fig. 1a) showing that the fusion protein is functional. Presence of the full-length fusion protein (c. 100 kDa) in membranes was confirmed by immunoblot analysis with antibodies directed against mCherry (Fig. 2, strain LMD104). No degradation of fusion protein, for example, cleavage between SpoVD and mCherry, was detected in cell extracts using immunoblot (data not shown).
Figure 2.
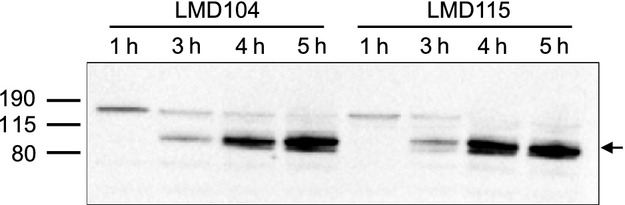
Immunoblot for SpoVD in Bacillus subtilis strains LMD104 (wild-type SpoVD-mCherry) and LMD115 (active-site SpoVD-mCherry mutant). Cells were sporulated by resuspension at 30 °C, and samples were taken at hourly intervals after the resuspension as indicated. Isolated membranes, 15 μg of proteins, were loaded in each lane. The samples were probed with anti-dsRed serum. SpoVD-mCherry (c. 100 kDa) is indicated by an arrow. The antigen band seen at > 115 kDa is background independent of mCherry. Molecular mass markers, in kDa, are indicated.
Presence of endospore cortex is dependent on the transpeptidase activity of SpoVD
To determine whether the lack of cortex in spoVD-negative mutants is an indirect effect, for example, a result of disruption of a protein complex (where SpoVD might act as a scaffold for other proteins), a result of SpoVE degradation or is due to lack of SpoVD enzyme activity, we mutated the active-site serine residue of the transpeptidase domain in SpoVD. This serine is essential for transpeptidase activity (Goffin & Ghuysen, 1998).
Plasmid pLEB6, which carries the spoVD-mCherry gene fusion, was used as a template for site-directed mutagenesis, and the codon for the active-site serine (Ser294) was changed to encode alanine. The mutant gene variant was inserted into the amyE locus in the chromosome of strain LMD101. The obtained SpoVD-mCherry active-site mutant strain, LMD115, was found to form heat-sensitive spores (Table 2). Electron microscopic examination of LMD115 sporulating cells showed lack of the cortex layer in spores (Fig. 1c), consistent with the heat sensitivity of the spores.
Normal temporal expression and presence of the full-length mutant fusion protein (c. 100 kDa) in the membrane fraction of sporulating LMD115 cells was confirmed by immunoblot analysis (Fig. 2, strain LMD115). Next, the enzymatic activity of the mutant protein was tested using Bocillin FL, a commercially available fluorescent penicillin derivative (Zhao et al., 1999). Isolated membranes of LMD104 and LMD115 were incubated with Bocillin FL, and proteins were fractionated by SDS-PAGE. Strain LMD104 showed a weakly fluorescent protein band corresponding to SpoVD-mCherry (Fig. 3). As expected, the mutant SpoVD-mCherry protein of LMD115 was unable to covalently bind Bocillin FL, confirming that transpeptidase activity was abolished by the Ser294Ala mutation.
Figure 3.
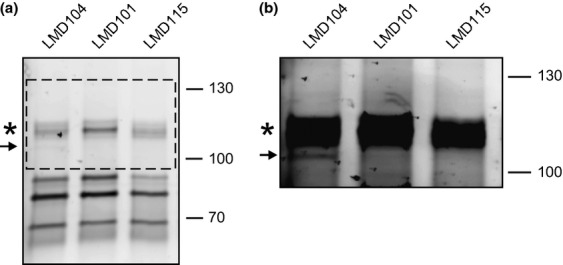
Covalent binding of the fluorescent penicillin derivative Bocillin FL to PBPs present in sporulating Bacillus subtilis cells. (a) Membranes from strains LMD104 (wild-type SpoVD-mCherry; positive control), LMD101 (SpoVD deletion mutant; negative control) and LMD115 (active-site SpoVD-mCherry mutant). Cells were sporulated by resuspension at 30 °C, and membranes were isolated from cells at 5 h of sporulation (see Fig. 2). After incubation with Bocillin FL, membrane proteins were separated by 8% SDS-PAGE. The electrophoresis was run to resolve high-molecular-weight proteins. The gel was analyzed by fluorometry. (b) shows an overexposure of the area outlined by the dashed box in panel (a) in order to reveal the relatively weak fluorescence intensity of the band corresponding to SpoVD-mCherry in LMD104. The position of Bocillin FL-labeled SpoVD-mCherry is indicated by an arrow. An asterisk indicates the position of PBP1a/b identified on the basis of published data (McPherson et al., 2001). Molecular mass markers, in kDa, are indicated on the right-hand side of the panels.
Subcellular localization of mutant SpoVD
Subcellular localization of some PBPs is reported to be dependent on their transpeptidase activity (Pinho & Errington, 2005; Costa et al., 2008). We therefore asked whether the lack of cortex in strain LMD115 is a result of mislocalization of the mutant SpoVD protein. The subcellular localization of the SpoVD(Ser294Ala)-mCherry in sporulating B. subtilis cells was compared with that of the wild-type variant using fluorescence microscopy. The fluorescence signal of both SpoVD(Ser294Ala)-mCherry and the functional SpoVD-mCherry was enriched at the forespore (Fig. 4). The results demonstrate that the localization of SpoVD to the forespore is not dependent on its transpeptidase activity, and the lack of cortex in the SpoVD(Ser294Ala) mutant is not a result of mislocalization of SpoVD.
Figure 4.
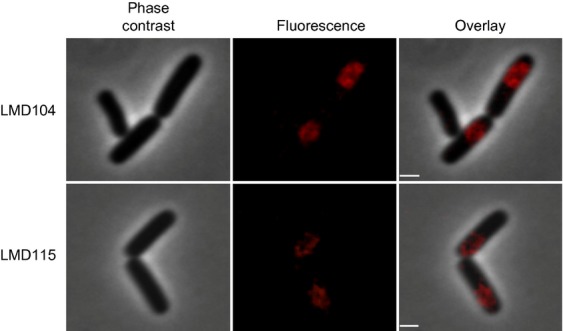
Subcellular localization of wild-type (LMD104) and Ser294Ala mutant (LMD115) SpoVD-mCherry in Bacillus subtilis sporulating cells. Cells were sporulated by resuspension at 30 °C, and phase contrast and mCherry fluorescence images of cells were taken at 5 h after the start of sporulation. Scale bar is 1 μm.
Conclusion
Our major finding is that the presence of cortex in B. subtilis endospores is dependent on SpoVD transpeptidase activity specifically. This suggests that other enzymes with transpeptidase activity that are expressed during sporulation, that is, Pbp2d and Pbp4b, (Pedersen et al., 2000; McPherson et al., 2001; Wei et al., 2004), cannot compensate for lack of SpoVD transpeptidase activity. One should note that the endospore germ cell wall layer, situated underneath the cortex layer in spores (not seen in our electron micrographs), is formed also in SpoVD-defective mutants because the heat-sensitive spores germinate normally. This layer seemingly forms during engulfment (Tocheva et al., 2013). The detailed role of SpoVD transpeptidase activity in cortex synthesis remains unknown. SpoVD could, for example, bind lipid II delivered across the outer forespore membrane by the activity of SpoVE and then in cooperation with other PBPs with transglycosylase activity incorporate the delivered disaccharide-peptide unit into a nascent peptidoglycan strand as previously pointed out by Fay and coworkers (Fay et al., 2010).
Acknowledgments
We thank Claes von Wachenfeldt and Nora Ausmees for generously providing plasmids pJM103-I-SceI, pBKJ223 and pKS-mCherry-E-T3. Special thanks to Stuart Cantlay and Sebastian Wasserstrom for the help with initial fluorescence microscopy experiments, Fernando Sorroche for comments on the manuscript and Rita Wallén for the expert help with electron microscopy. This work was supported by grants from the Swedish Research Council, 621-2010-5672, and the Research School in Pharmaceutical Sciences (FLÄK).
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Oligonucleotides used in this work.
References
- Bugg TDH, Braddick D, Dowson CG, Roper DI. Bacterial cell wall assembly: still an attractive antibacterial target. Trends Biotechnol. 2011;29:167–173. doi: 10.1016/j.tibtech.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Costa T, Priyadarshini R, Jacobs-Wagner C. Localization of PBP3 in Caulobacter crescentus is highly dynamic and largely relies on its functional transpeptidase domain. Mol Microbiol. 2008;70:634–651. doi: 10.1111/j.1365-2958.2008.06432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel RA, Errington J. DNA sequence of the murE-murD region of Bacillus subtilis 168. J Gen Microbiol. 1993;139:361–370. doi: 10.1099/00221287-139-2-361. [DOI] [PubMed] [Google Scholar]
- Daniel RA, Drake S, Buchanan CE, Scholle R, Errington J. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J Mol Biol. 1994;235:209–220. doi: 10.1016/s0022-2836(05)80027-0. [DOI] [PubMed] [Google Scholar]
- Eichenberger P. Genomics and cellular biology of endospore formation. In: Graumann P, editor. Bacillus: Cellular and Molecular Biology. Norfolk, UK: Caister Academic Press; 2012. pp. 319–350. [Google Scholar]
- Erlendsson LS, Moller M, Hederstedt L. Bacillus subtilis StoA is a thiol-disulfide oxidoreductase important for spore cortex synthesis. J Bacteriol. 2004;186:6230–6238. doi: 10.1128/JB.186.18.6230-6238.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay A, Meyer P, Dworkin J. Interactions between late-acting proteins required for peptidoglycan synthesis during sporulation. J Mol Biol. 2010;399:547–561. doi: 10.1016/j.jmb.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortnagel P, Freese E. Analysis of sporulation mutants. II. Mutants blocked in the citric acid cycle. J Bacteriol. 1968;95:1431–1438. doi: 10.1128/jb.95.4.1431-1438.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster S, Popham D. Structure and synthesis of cell wall, spore cortex, teichoic acids, S-layers, and capsules. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and its Closest Relatives: from Genes to Cells. Washington, DC: American Society for Microbiology; 2002. pp. 21–41. [Google Scholar]
- Goffin C, Ghuysen JM. Multimodular penicillin binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Jessee J, Bloom FR. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
- Harwood CR, Archibald AR. Growth maintenance and general techniques. In: Harwood CR, Cutting SM, editors. Molecular Biological Methods for Bacillus. Chichester, UK: John Wiley & Sons Ltd; 1990. pp. 1–23. [Google Scholar]
- Hederstedt L. Molecular properties, genetics, and biosynthesis of Bacillus subtilis succinate dehydrogenase complex. Methods Enzymol. 1986;126:399–414. doi: 10.1016/s0076-6879(86)26040-1. [DOI] [PubMed] [Google Scholar]
- Hoch JA. Genetic analysis in Bacillus subtilis. Methods Enzymol. 1991;204:305–320. doi: 10.1016/0076-6879(91)04015-g. [DOI] [PubMed] [Google Scholar]
- Janes BK, Stibitz S. Routine markerless gene replacement in Bacillus anthracis. Infect Immun. 2006;74:1949–1953. doi: 10.1128/IAI.74.3.1949-1953.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YM, Moller MC, Petersen L, Soderberg CAG, Hederstedt L. Penicillin-binding protein SpoVD disulphide is a target for StoA in Bacillus subtilis forespores. Mol Microbiol. 2010;75:46–60. doi: 10.1111/j.1365-2958.2009.06964.x. [DOI] [PubMed] [Google Scholar]
- Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. Methods Enzymol. 1963;6:726–738. [Google Scholar]
- McPherson DC, Driks A, Popham DL. Two class A high-molecular-weight penicillin-binding proteins of Bacillus subtilis play redundant roles in sporulation. J Bacteriol. 2001;183:6046–6053. doi: 10.1128/JB.183.20.6046-6053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson WL, Setlow P. Sporulation germination and outgrowth. In: Harwood CR, Cutting SM, editors. Molecular Biological Methods for Bacillus. Chichester, UK: John Wiley & Sons Ltd; 1990. pp. 391–450. [Google Scholar]
- Pedersen LB, Ragkousi K, Cammett TJ, Melly E, Sekowska A, Schopick E, Murray T, Setlow P. Characterization of ywhE, which encodes a putative high-molecular-weight class A penicillin-binding protein in Bacillus subtilis. Gene. 2000;246:187–196. doi: 10.1016/s0378-1119(00)00084-6. [DOI] [PubMed] [Google Scholar]
- Perego M. Integrational vectors for genetic manipulation in Bacillus subtilis. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and Other Gram-positive Bacteria: Biochemistry, Physiology, and Molecular Genetics. Washington, DC: American Society for Microbiology; 1993. pp. 615–624. [Google Scholar]
- Piggot PJ, Hilbert DW. Sporulation of Bacillus subtilis. Curr Opin Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Pinho MG, Errington J. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol Microbiol. 2005;55:799–807. doi: 10.1111/j.1365-2958.2004.04420.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Scheffers D-J. The cell wall of Bacillus subtilis. In: Graumann P, editor. Bacillus: Cellular and Molecular Biology. Norfolk, UK: Caister Academic Press; 2007. pp. 331–373. [Google Scholar]
- Tocheva EI, López-Garrido J, Hughes HV, Fredlund J, Kuru E, VanNieuwenhze MS, Brun YV, Pogliano K, Jensen GJ. Peptidoglycan transformations during Bacillus subtilis sporulation. Mol Microbiol. 2013;88:673–686. doi: 10.1111/mmi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JA, Roberts AN, Johnstone K, Piggot PJ, Winter G, Ellar DJ. Reduced heat resistance of mutant spores after cloning and mutagenesis of the Bacillus subtilis gene encoding penicillin-binding protein 5. J Bacteriol. 1986;167:257–264. doi: 10.1128/jb.167.1.257-264.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei YP, McPherson DC, Popham DL. A mother cell-specific class B penicillin-binding protein, PBP4b, in Bacillus subtilis. J Bacteriol. 2004;186:258–261. doi: 10.1128/JB.186.1.258-261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao GS, Meier TI, Kahl SD, Gee KR, Blaszczak LC. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob Agents Chemother. 1999;43:1124–1128. doi: 10.1128/aac.43.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotides used in this work.


