Abstract
An important feature of the common DNA oxidation product 8;oxo;7,8;dihydroguanine (OG) is its susceptibility to further oxidation to produce guanidinohydantion (Gh) and spiroiminodihydantoin (Sp) lesions. In the presence of amines, G or OG oxidation produces hydantoin amine adducts. Such adducts may form in cells via interception of oxidized intermediates by protein;derived nucleophiles or naturally occurring amines that are tightly associated with DNA. Gh and Sp are known to be substrates for base excision repair (BER) glycosylases; however, large Sp;amine adducts would be expected to be more readily repaired by nucleotide excision repair (NER). A series of Sp adducts differing in size of the attached amine were synthesized to evaluate the relative processing by NER and BER. The UvrABC complex excised Gh, Sp and the Sp;amine adducts from duplex DNA, with the greatest efficiency for the largest Sp;amine adducts. The affinity of UvrA with all of the lesion duplexes was found to be similar, whereas the efficiency of UvrB loading tracked with the efficiency of UvrABC excision. In contrast, the human BER glycosylase NEIL1 exhibited robust activity for all Sp;amine adducts irrespective of size. These studies suggest that both NER and BER pathways mediate repair of a diverse set of hydantoin lesions in cells.
Introduction
Cellular respiration and the inflammatory response generate reactive oxygen and nitrogen species (RONS) resulting in DNA modifications that contribute to premature aging, mutagenesis, and carcinogenesis.1;3 Of the various oxidized bases that may be formed, 8;oxo;7,8; dihydroguanine (OG) has been the most extensively studied1,2,4 and shown to be highly mutagenic in the absence of repair.1 A distinct feature of OG is its susceptibility to further oxidation that leads to formation of the hydantoin lesions, guanidinohydantoin (Gh) and the two diastereomers of spiroiminodihydantoin (Sp1 and Sp2) (Figure1).4;8 Gh and Sp have been established as the major products of G and OG oxidation by peroxynitrite, peroxyl radicals, and hypochlorous acid, reactive species that are present in cells during an inflammatory response.9;20 Recently, Gh and Sp lesions were detected in the liver and colon tissue of Rag2−/− mice at levels 100;fold less than OG.3 Oxidative conditions are also known to mediate formation of DNA; protein cross links (DPC), DNA;DNA cross links, and DNA;polyamine adducts.21;25In vitro, covalent cross;links between DNA and proteins have been shown to form under oxidative conditions when the DNA contains the OG lesion.21,24 Indeed, oxidation with a number of biologically relevant oxidants in the presence of DNA binding proteins has been shown to produce hydantoin;protein cross;links.26 These previous studies suggest that in a cellular environment a wide variety of hydantoin lesion structures may be present.
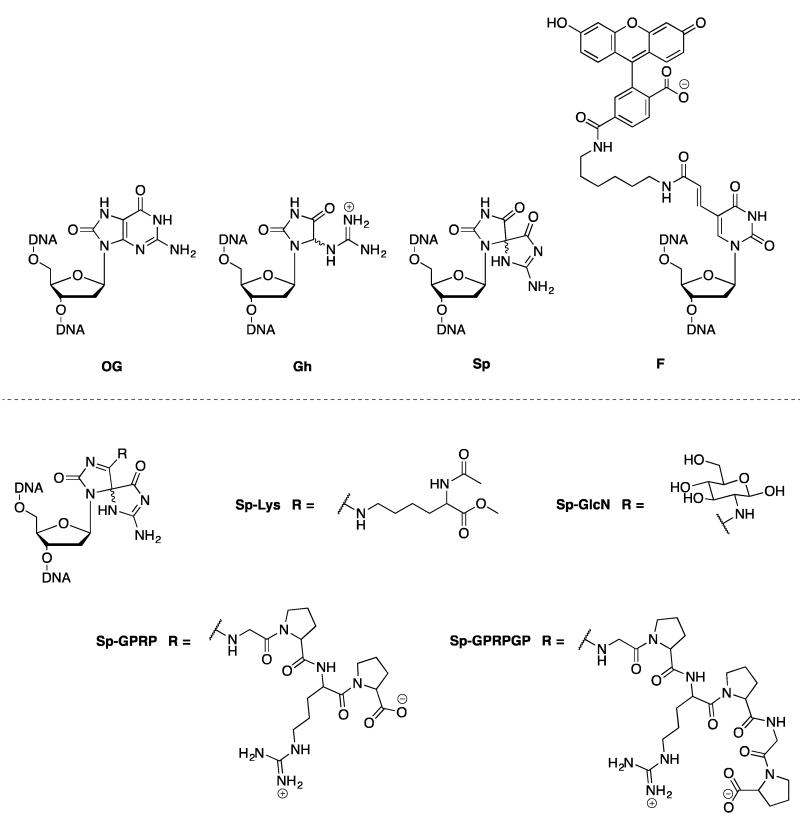
Figure 1. Structures of lesions used as substrates for NER and BER in this study.
Polymerase primer extension and cellular mutation assays using synthetic DNA oligonucleotides containing Gh or Sp5,9 have shown that these lesions are highly mutagenic resulting in G → C and G → T transversion mutations.27;31 The mutagenic consequences of oxidized DNA bases are mitigated in part by base excision repair (BER). The key players in BER are the DNA glycosylases, which are responsible for searching the genome for aberrant bases and initiating repair by extruding of the damaged nucleobase out of the helix and catalyzing N;glycosidic bond cleavage to release the modified base. The hydantoins Gh and Sp have been shown to be in vitro substrates for several BER glycosylases including bacterial Fpg and Nei, Mimivirus (Mv) Nei1 and mammalian “Nei;like” (NEIL) enzymes.32;40 The hydantoins Gh and Sp were previously shown to be excellent in vitro substrates for the human glycosylase NEIL1.1,36,41,42 In addition, the Sp lesion was detected in Nei deficient E. coli cells treated with the potent oxidant chromate.43 Together this evidence suggests that the hydantoin lesions may be important substrates for BER in vivo. Surprisingly, analysis of the DNA mutational spectrum of lung tumors from Neil1;/; Nthl1;/; mice was distinct from that expected for Gh and Sp, which suggests that there are alternative repair mechanisms to counteract the mutagenicity of these lesions.44
An alternative pathway to consider for repair of hydantoins is nucleotide excision repair (NER). NER is known to repair bulky and helix;destabilizing DNA adducts such as cis-syn thymine;thymine cyclobutane dimers, DPC and bulky alkylated bases.45,46 The prokaryotic NER pathway is initiated by the UvrABC proteins, which respond in a coordinated fashion to locate the lesion and excise out a lesion;containing oligonucleotide.45,47,48 To delineate the contributions of NER and BER for hydantoin lesion repair, we prepared a series of Sp;amine adducts of varying size (Figure 1). The Sp;amine adducts used herein were designed to increase the size of the attached adduct and are derived from lysine, glucosamine, and short peptides. Our expectation was that the larger more bulky Sp;amine adducts would be poor substrates for BER glycosylases, but better substrates for the UvrABC complex. Single;turnover (STO) experiments with UvrABC revealed robust excision activity for the Sp;amine adduct;containing duplex DNA that was similar to that observed with the standard UvrABC substrate, a fluorescein;modified T (F).49,50 Notably, the Sp and Gh lesions were also substrates for NER. The BER glycosylases Nei, Fpg, and NEIL1 were also shown to mediate the removal of Sp;amine adduct bases from DNA. Surprisingly, NEIL1 was able to cleave all Sp;amine adducts tested with rates comparable to that of the parent lesion Sp. Taken together, these results suggest that both repair pathways can mitigate the potent mutagenic properties of a diverse set of hydantoin lesions.
Experimental Section
General methods and procedures (reagents, instrumentation, enzyme purification, hydroxyl radical footprinting, Maxam;Gilbert sequencing) are listed in the Supporting information.
Substrate DNA Preparation for NER Assays
The following 50 base pair duplex was used in the incision assays: 5′;d(GAC TAC GTA CTG TTA CGG CTC CAT CXC TAC CGC AAT CAG GCC AGA TCTGC) * 3′;d(CTG ATG CAT GAC AAT GCC GAG GTA GYG ATG GCG TTA GTC CGG TCT AGA CG), where X= OG, Gh, Sp or F and Y= A, C, T or G. Oligonucleotides (50 nt) containing the lesions were generated by ligation of shorter oligonucleotides.51 The oligonucleotides used for the ligation were 5`;d(GAC TAC GTA CTG TTA CG) and 5′;d(GCT CCA TCX CTA CCG CAA TCA GGC CAG ATC TGC) on the template strand, 5′;d(TGG CCT GAT TGC GGT AGA GAT GGA GCC GTA ACA GTA.
The 51 base pair duplex used in the incision assays had the following sequence: 5′; d(GAC TAC GTA CTG TTA CGG CTC CAT CXG CTA CCG CAA TCA GGC CAG ATC TGC) * 3′;d(CTG ATG CAT GAC AAT GCC GAG GTA GYC GAT GGC GTT AGT CCG GTC TAG ACG), where X = Sp;Lys, Sp;GlcN, Sp;GPRP or Sp;GPRPGP, and Y = A, C, T or G. The synthesis of the adducts was as follows: the 51 nt OG lesion strand (10 MM, 1 nmole) was mixed in the presence of the chosen primary amine (2 mM, 200 nmole) in a 20 mM sodium phosphate (pH 7.4) solution maintained at 45 °C. After a 30 min pre;incubation, the oxidation reaction was initiated by the addition of Na2IrCl6 (200 MM, 20 nmoles) and incubated for an additional 30 min. The reaction was terminated by addition of Na2EDTA (2 mM, 200 nmoles). The mixtures were purified as described previously and the identities for each adduct were determined by ESI;MS.25 UvrABC incision assays were conducted with both duplexes that contained 32P; labeled lesion (X);containing strand. The X;containing strand (2.5 pmol) was radiolabeled either on the 5′;end using [γ;32P];ATP with T4 kinase or on the 3′;end using [α;32P] cordycepin;5′;triphosphate with terminal transferase at 37 °C. The labeled strand was then annealed with 10% excess of the complement by heating to 90°C for 10 min in annealing buffer (20 mM Tris;HCl, pH 7.6, 10 mM EDTA, and 150 mM NaCl) and then allowed to cool slowly overnight.
Substrate DNA Preparation for BER Assays
Gh;, Sp1;, Sp2;, and Sp;amine adduct; containing oligonucleotides were synthesized as described previously.25,52 The following duplex sequence was used in the base excision assays: 5′;d(TGT TCA TCA TGC GTC XTC GGT ATA TCC CAT) * 3′;d(ACA AGT AGT ACG CAG YAG CCA TAT AGG GTA), where X= Gh, Sp1, Sp2, or Sp;amine adducts (Sp;Lys, Sp;GlcN, Sp;GPRP or Sp;GPRGP) and Y= A, C G, or T. For glycosylase assays, the X;containing strand was 5′;end labeled using [γ;32P];ATP with T4 kinase and then mixed with unlabeled X;containing oligonucleotide to yield a solution that contained 5% labeled X;oligonucleotide. The mixture was then annealed with 20% excess of the Y;containing complement strand by heating to 90 °C for 10 min in annealing buffer (20 mM Tris;HCl, pH 7.6, 10 mM EDTA, and 150 mM NaCl) and then allowed to cool slowly overnight.
UvrABC Incision Assay
Single;turnover experiments, where [Enzyme] > [DNA], were performed to evaluate the incision activity of the UvrABC complex. 51,53 In each case the final reaction volume was 100 μL with a final labeled DNA duplex concentration of 2 nM. The duplex was incubated with 20 nM UvrA, 100 nM UvrB, and 50 nM UvrC, in assay buffer (50 mM Tris;HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 5 mM DTT, and 1 mM ATP) at 55°C. Before addition to the reaction, the individual Uvr enzyme solutions were incubated for 10 minutes at 65°C to activate the enzymes. Reaction time;courses were initiated by addition of UvrC. Aliquots (8 ML) were taken at various time points ranging from 1 minute to 4 hours and quenched by the addition of 2 μL of 100 mM EDTA (pH 8) and incubation at 90 °C for 5 minutes. Reactions were chilled on ice and 10 μL of formamide denaturing dye (99.9% formamide, 0.025% xylene cyanol, 0.025% bromophenol) was added prior to electrophoresis. The samples were electrophoresed on a 15% denaturing polyacrylamide gel in TBE running buffer (89 mM Tris;HCl, 89 mM boric acid, 2 mM EDTA, pH 7.6) at 800 V for 3 hours to separate the DNA fragments arising from the cleaved product and the 50 or 51;nucleotide substrate. The gel was visualized using storage phosphor autoradiography and quantitated using ImageQuant.
Base Excision Glycosylase Assays
Single;turnover (STO) experiments, where [Enzyme] > [DNA], were performed using the 30 base pair duplex sequence to evaluate the glycosylase activity of BER glycosylases. 19,35,36,41,42,54 In all cases, the final reaction volume was 100 μL with a final DNA duplex concentration of 20 nM. In qualitative experiments to evaluate the glycosylases capable of excising Sp;amine adducts, the lesion;containing duplex was incubated with either 200 nM NEIL1, 200 nM Fpg, 200 nM E3Q Fpg, 200 nM Nei, 300 nM E3Q NEIL1, or 100 nM hOGG1 in assay buffer [20 mM Tris;HCl, pH 7.6, 10 mM EDTA, 0.1 mg/mL BSA, and NaCl (150 mM for NEIL1, 30 mM for Fpg, E3Q Fpg, and Nei, or 70 mM NaCl for hOGG1)] at 37 °C for 20 minutes. Enzyme concentrations are listed as active concentrations for NEIL1, Fpg and hOGG1, and as total protein concentration for Nei and E3Q NEIL1. For NEIL1 reactions under STO conditions, a Rapid Quench Flow instrument (RQF;3) from Kintek was utilized to determine kinetic parameters. The NEIL1 enzyme was mixed with 20 nM final DNA duplex for time points ranging from 0.2 seconds to 2 minutes and quenched with 0.5 M NaOH. Denaturing PAGE provided separation of the 15;nucleotide DNA fragment arising from the cleaved product and the 30;nucleotide fragment originating from the substrate. Gels were imaged using storage phosphor autoradiography and band intensities were quantified with ImageQuant to provide binding plots using GraFit (v. 5.0).
Gel Mobility Shift Assay
Electrophoretic mobility shift assays (EMSA) were performed to determine the Kd values of UvrA for lesion;containing DNA. 53 For binding studies, the labeled lesion;containing strand was annealed to a complement DNA strand that positioned A opposite the lesion strand. Reactions (20 ML) contained 200 pM DNA duplex that was 32P ;5′ end labeled on the lesion;containing X;strand in buffer (50 mM Tris;HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 5 mM DTT, and 1 mM ATP), and UvrA concentrations ranging from 0.5 nM to 200 nM. Samples of the protein/DNA mixture were incubated at 55 °C for 20 minutes followed by the addition of 2 μL of 80% glycerol. Bound versus unbound DNA was visualized using electrophoresis on a 4% nondenaturing poylacrylamide gel (80:1 acrylamide:bisacrylamide ratio) at room temperature in TBE running buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA) containing 10 mM MgCl2 and 1 mM ATP at 100 V for 2;3 hours. Gels were dried and exposed to a storage phosphor screen overnight. Kd values were determined by fitting the data (percent bound substrate vs. log [UvrA]) using a one;site binding isotherm (GraFit 5.0). The Kd values were determined from data generated from at least three separate experiments (typically five) using separate diluted UvrA in each experiment.
The loading of UvrB by UvrA was examined by incubating 5′;labeled DNA duplex (200 pM) with UvrA (0.5 nM) in the presence of 100 nM UvrB in buffer (50 mM Tris;HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 5 mM DTT, and 1 mM ATP) at 55 °C for 20 minutes. After addition of 2 μL of 80% glycerol, samples were electrophoresed on a 4% gel as described above.53 The percent UvrB;DNA complex was determined from quantitation of the storage phosphor autoradiogram. At least three separate experiments were performed and the values averaged to provide the %UvrB loaded onto the lesion;DNA.
RESULTS
Substrate design, synthesis, purification and characterization
Insight into the biological consequences of Gh and Sp lesions has emerged in part due to the ability to synthesize DNA oligonucleotides containing these lesions at a defined location by oxidation of OG with the one;electron oxidant sodium hexachloroiridate(IV).5,9 Appropriate oxidation conditions were used to produce oligonucleotides containing primarily Gh or Sp; subsequent separation of the desired lesion oligonucleotide from other products was performed using high performance liquid chromatography (HPLC).52 A 33;nt fragment was used for the Ir(IV) oxidation reaction to make the Gh or Sp;containing oligonucleotide which was then ligated to a 17;nt strand using a 36;nt complement sequence as a template.51 In this 33;nt sequence, the two diastereomers of Sp could not be separated via HPLC and therefore, the duplex substrates containing Sp are a mixture of diastereomers. The overall yield of the ligation and purification of the 50;nt strands containing OG, Gh, and Sp was 50%, 20%, and 10%, respectively. After gel purification, the ligated 50 nt product was annealed to the appropriate 50 nt complement strand to position the lesion opposite C, G, A or T. The Sp;amine adduct; containing DNA duplexes were generated by Ir(IV) oxidation of a 51;nt strand containing a central OG lesion in the presence of the chosen primary amine. The amines were selected to be biologically relevant water;soluble amines, such as glucosamine, lysine and short peptides. The peptide sequences featured a N;terminal amino group to serve as nucleophile and rigid, bulky components (proline) as well as arginine for enhanced water solubility and DNA affinity. Spermine and spermidine adducts were previously synthesized; however, these adducts undergo intramolecular rearrangement and decomposition reactions, making them unsuitable choices for the studies reported herein.23 HPLC purification was used to separate the amine;adduct from other products as a mixture of diastereomers. The identity for each adduct was determined by ESI;MS: 51;mer OG (calcd mass = 15,612.2 Da; exp mass = 15,612.8 Da), 51;mer Sp;Lys (calcd mass = 15,813.9 Da, exp mass = 15,812.8 Da), 51;mer Sp;GlcN (calcd mass = 15,789.3 Da, exp mass = 15,788.8 Da), 51;mer Sp;GPRP (calcd mass = 16,034.7 Da, exp mass = 16,035.2 Da), and 51;mer Sp;GPRPGP (calcd mass = 16,188.8 Da, exp mass = 16,189.6 Da).
Incision Activity of UvrABC with DNA substrates containing OG, Gh, Sp, F, and Sp-amine adducts
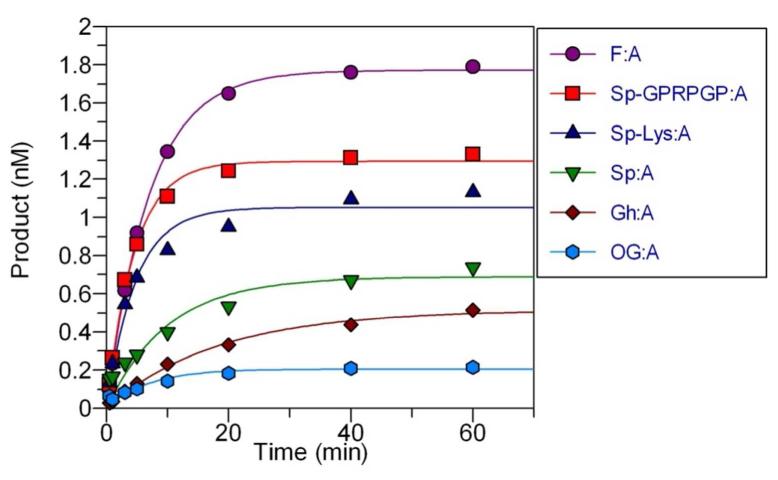
The incision activity of the UvrABC complex was evaluated using recombinant Bacillus caldotenax UvrA and UvrB, and Thermatoga maritima UvrC enzymes with duplex substrates containing the lesion X (X = OG, Gh, Sp, F, Sp;Lys, Sp;GlcN, Sp;GPRP, or Sp;GPRPGP) paired with all of the four natural bases (A, G, C, T). The general method involved 5′;end; labeling the X;containing strand using [γ;32P];ATP in order to monitor the extent of excision 5′ to the X;nucleotide. Alternatively, 3′;end;labeling of the X;containing strand was achieved with [α;32P] cordycepin;5′triphosphate to monitor the cleavage 3′ to the X;nucleotide. Reactions were performed under single turnover (STO) conditions, where [Enzyme]>[DNA], and the data was fit to a single exponential equation to determine the observed rate of product formation as a function of time (kobs). The excision activity of UvrABC with Gh, Sp, the Sp;amine adducts, and OG was compared to the standard NER substrate, fluorescein;dT (F);containing DNA and the conditions were optimized to give maximal levels of excision similar to that reported previously. 51,53,55 Representative reaction progress curves for the lesions base;paired with A are shown in Figure 2. The observed rates of product formation as a function of time for the reaction of UvrABC with all lesion:A;containing duplexes and the control F:A containing duplex were found to be similar (0.2 ± 0.1 min−1) (Table 1). However, distinct differences were observed with respect to the maximal amount of excision mediated by UvrABC with the series of lesion:A; containing duplexes. Indeed, the reaction proceeded to 88 ± 2% product formation with the fluorescein;containing duplex, whereas, experiments using Gh and Sp reached 23 ± 4% and 32 ± 3% completion after 4 hours, respectively. In contrast, the reactions with OG reached only 10 ± 1% completion. Excision reactions with the Sp;amine adducts Sp;Lys, Sp;GlcN, Sp;GPRP and Sp;GPRPGP proceeded to 51 ± 1%, 62 ± 4%, 62 ± 3% and 62 ± 4% completion, respectively. The extent of product formation and the observed rates of the Sp;amine adducts, F, Gh, Sp, and OG from all possible natural base pair contexts were within error for each lesion (data not shown), indicating a lack of preference for the opposite base during UvrABC substrate processing. Although the rates of incision are similar for OG, F and all hydantoin lesions, a comparison of the maximal extent of substrate cleavage reveals that the Sp;amine adducts are good substrates for UvrABC. Indeed, the extents of Sp;amine adduct removal are similar to that reported for excision of DNA;peptide cross;links by UvrABC. 55
Figure 2. Representative reaction profiles of UvrABC with hydantoin lesion-containing DNA.
Reactions conditions consisted of 20 nM UvrA, 100 nM UvrB, and 50 nM UvrC, 2 nM DNA duplex in assay buffer (50 mM Tris;HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 5 mM DTT, and 1 mM ATP) at 55 °C. Reactions and plots with UvrABC and DNA containing Sp;GPRP:A and Sp;GlcN:A were essentially identical to the reaction with Sp;GPRPGP:A.
Table 1.
Observed rates (kobs) and extent of product formed in single-turnover (STO) reactions (%) with UvrABC, equilibrium dissociation constants (Kd) determined for UvrA, and fraction of UvrB DNA complex (%) with lesion-containing duplexes.
| Base Pair | kobs(min−1)a | Product formed under STO (%) |
Kd (nM) b | DNA bound in stable complex with UvrB (%)c |
|---|---|---|---|---|
|
|
|
|||
| F:A | 0.2 ± 0.1 | 88 ± 2 | 7 ± 5 | 81 ± 7 |
| Sp:A | 0.1 ± 0.1 | 32 ± 3 | 13 ± 7 | 9 ± 4 |
| Cih: A | 0.1 ± 0.1 | 23 ± 4 | 20 ± 10 | 7 ± 3 |
| OG:A | 0.1 ± 0.1 | 10 ± 1 | 23 ± 7 | 5 ± 2 |
| Sp-Lys:A | 0.2 ± 0.1 | 51 ± 1 | 2 ± 1 | 38 ± 2 |
| Sp-GlcN:A | 0.2 ± 0.1 | 62 ± 4 | 3 ± 1 | 22 ± 1 |
| Sp-GPRP:A | 0.2 ± 0.1 | 62 ± 3 | 2 ± 1 | 54 ± 1 |
| Sp-GPRPGP:A | 0.2 ± 0.1 | 62 ± 4 | 2 ± 1 | 61 ± 5 |
Errors reported arc the standard deviation of the average of at least three reactions,
kobsdetermined using a single exponential rate equation at 55°C .
Kdvalues and % DNA bound by UvrB determined using EMSA at 55°C (see methods).
DNA bound by UvrB (%) was determined from gel quantitation with [UvrA] = 0.5 nM
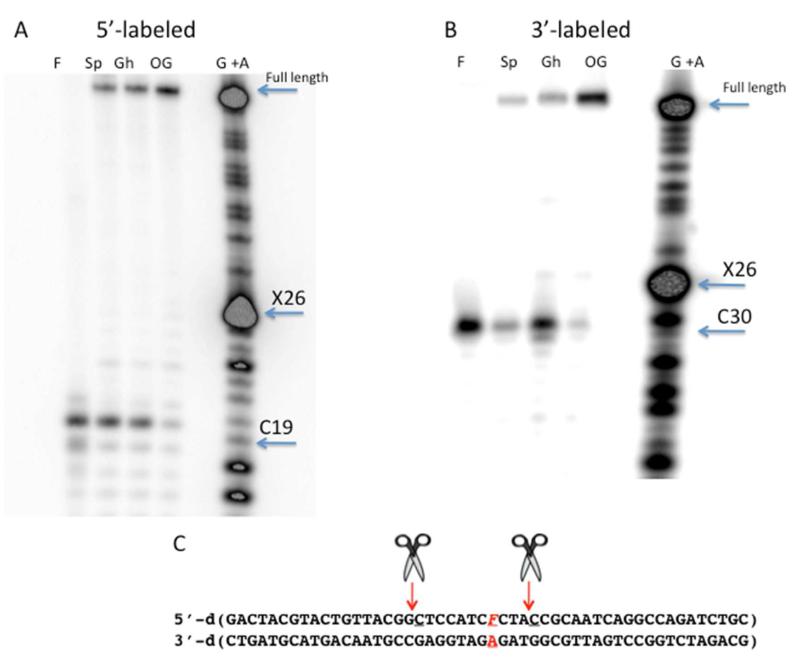
In order to determine the sites of UvrABC backbone cleavage relative to the lesion, the bands for UvrABC incision were compared to Maxam;Gilbert G +A sequencing reactions (Figure 3A, B and C).54,56 These experiments were performed using both 5′; and 3′;end;labeled duplex. In the case of the 3′;labeling, the UvrABC incision site maps the most closely to C30, which is located four nucleotides from the lesion. In the case of the 5′;incision site, the UvrABC product band appears between C19 and T20 in the Maxam;Gilbert sequencing lane likely due to presence of a phosphate end in the later. Due to slower migration of a DNA fragment with a hydroxyl;end versus a phosphate end, the cleavage site for UvrABC was assigned to the phosphodiester 5′ of C19. Importantly, the site of phosphodiester backbone cleavage by UvrABC both 5′ and 3′ to the lesion was the same with F, OG, Gh, Sp and the Sp;amine adducts (SI Figure 1).
Figure 3. Location of UvrABC incision sites near the lesion site.
(A) Storage phosphor autoradiogram of UvrABC 5′;side incision site on 5′;32P;F, OG, Gh, Sp;containing strand of 50 bp duplex substrate. The Maxam;Gilbert G + A sequencing reaction (lane G + A) was used to determine the location of the lesion site (X26) and the nucleotide cleaved by UvrABC (C19). Reactions containing 2 nM DNA duplex with 20 nM UvrA, 100 nM UvrB, and 50 nM UvrC were incubated for four hours at 55°C. (B) Storage phosphor autoradiograph of the same experiment using 3′;end;labling to visualize 3′;cleavage site. Highlighted nucleotides are the lesion site (X 26) and the UvrABC 3′incision site (C30). (C) The sequence of the 50 bp duplex containing F:A with nucleotides that are hydrolyzed by UvrABC indicated with arrows and scissors. Sites of UvrABC incision were the same for all lesion;containing duplexes.
Equilibrium Dissociation constants (Kd) of UvrA with lesion containing DNA
Prokaryotic NER is initiated by UvrA, which forms a heterotetramer with UvrB (UvrA2 YUvrB2) 45,57;60 to locate and bind damaged DNA. To determine if the reduced amount of product formation for the hydantoin lesions and OG relative to the fluorescein standard substrate was due to inefficient recognition of the lesion base by UvrA, electrophoretic mobility shift assays (EMSA) were employed to determine the equilibrium dissociation constants (Kd) (SI Figure 2). EMSA analysis was performed under conditions where [DNA;duplex] < Kd. The [DNA;UvrA complex] was determined as a function of [UvrA], and the data was fit using a one; site binding isotherm. UvrA binding experiments conducted with lesion;containing oligonucleotides revealed that UvrA recognizes and binds F, Gh, Sp, OG, and the Sp;amine adducts when base paired to adenine (Table 1). The Kd of UvrA for fluorescein containing DNA was determined to be 7 ± 5 nM. The Kd values of UvrA for Gh and Sp;containing duplexes were 20 ± 10 nM and 13 ± 7 nM, respectively, while the Kd value of UvrA for OG;containing DNA was found to be 23 ± 7 nM. UvrA was shown to possess the highest binding affinity for the Sp; amine adducts in which the values determined were as follows: Sp;Lys, 2 ± 1 nM; Sp;GlcN, 3 ± 1 nM; Sp;GPRP, 2 ± 1 nM; and Sp;GPRPGP, 2 ± 1 nM. However, the measured Kd values for the Sp;amine adducts were all within error of the fluorescein;dT adduct.
Formation of UvrB·DNA pre-incision complex measured via electrophoretic mobility shift assay
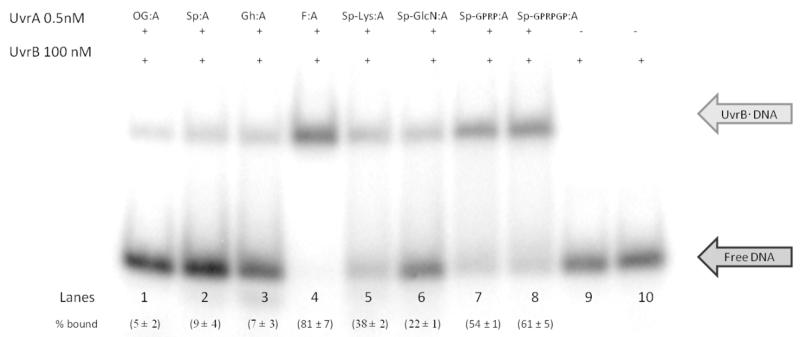
The loading of UvrB onto the site of damage by UvrA and the formation of the stable UvrBYDNA pre;incision complex is an important step in the damage repair mechanism of NER.45 The UvrB;DNA pre;incision complex recruits UvrC to carry out the 5′ and 3′ incision to remove the damaged DNA fragment. The extent of formation of a stable UvrB hydantoin;containing DNA complex was evaluated using EMSA. Failure to form a stable UvrBYDNA pre;incision complex despite successful UvrA recognition may be the cause of the reduced UvrABC; mediated excision observed with Gh, Sp and OG substrates relative to F. EMSA were performed in the presence of 100 nM UvrB, using a low concentration of DNA;duplex (below Kd for UvrA) to examine the formation of DNA;UvrB complex as a function of [UvrA]. Efficient formation of UvrBYDNA pre;incision complex was detected with F:A containing DNA [(81 ± 7)% DNA bound] even at the lowest concentrations of UvrA (0.5 nM) (Figure 4). Using this concentration of UvrA, reduced formation of UvrBYDNA pre;incision complex was observed with Gh [(7 ± 3)%] and Sp[(9 ± 4)%]. Similarly, a small fraction of OG;containing DNA [(5 ± 2)%] was bound to UvrB under these same conditions. Significantly higher amounts of the UvrBYDNA pre;incision complex were detected with Sp;Lys [(38 ± 2)%], Sp;GlcN [(22 ± 2)%], Sp;GPRP [(54 ± 2)%], and Sp;GPRPGP [(61 ± 5)%] relative to OG, Gh, and Sp. The extent of UvrB; lesion DNA complex observed via EMSA indicates efficiency of loading of UvrB onto DNA and formation of a stable complex. Notably, the amount of UvrB;DNA complex observed with OG, Gh, and Sp;containing DNA relative to that observed for Sp;GlcN, Sp;Lys, Sp;GPRP, Sp; GPRPGP and F correlates well with the extent of product formation from the UvrABC incision assay experiments.
Figure 4. Representative EMSA storage phosphor autoradiogram illustrating the formation of the UvrB*DNA pre-incision complex with F, OG, Gh, Sp, and Sp-amine adducts containing DNA.
Reaction mixtures contained 200 pM DNA duplex, 100 nM UvrB, 0.5 nM UvrA, 50 mM Tris;HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 5 mM DTT, and 1 mM ATP, incubated at 55 °C for 20 min.
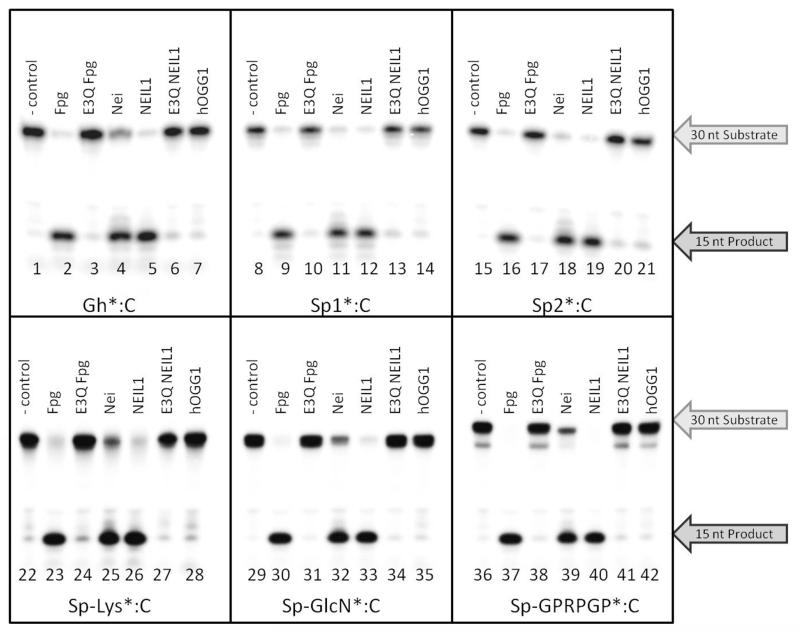
Activity of BER glycosylases with DNA substrates containing Gh, Sp1, Sp2 and bulky Sp-amine adducts
The glycosylase activity of Nei, Fpg, E3Q Fpg, hOGG1, NEIL1 and E3Q NEIL1 was evaluated using conditions of single turnover (STO, [E] > [DNA substrate]) with a 30;bp duplex containing Gh, Sp1, Sp2, Sp;Lys, Sp;GlcN, and Sp;GPRPGP lesions base paired to C at a single time point of 20 minutes (Figure 5). The reactions were performed in a manner similar to that previously published. 35,36,42,54,61 Briefly, the protocol involved 5′;end;labeling the X;containing strand with [γ;32P];ATP, and resolution of reaction products with denaturing PAGE to quantify the extent of strand cleavage at the X;nucleotide after quenching with 0.5 M NaOH. The base treatment ensured that cleavage of the phosphodiester backbone occurred at all abasic sites produced by the glycosylase.36 These experiments revealed that Nei, Fpg and NEIL1 were able to remove the Sp;amine adducts while the hOGG1 glycosylase exhibits minimal activity for Gh, Sp1, Sp2 or the Sp;amine adducts above background levels (Figure 5). The catalytically inactive variants E3Q Fpg and E3Q NEIL1 were unable to mediate base release confirming the requirement for enzyme catalysis of N;glycosidic bond cleavage.
Figure 5. Sp-amine adduct removal by human and bacterial BER glycosylases.
Qualitative glycosylase assays were performed with Fpg, E3Q Fpg, Nei, NEIL1, E3Q NEIL1 and hOGG1 using the 30 base pair duplex containing Gh, Sp1, Sp2, Sp;Lys, Sp;GlcN, and Sp;GPRPGP lesions base paired to C. 35,36,54,61 The asterisk indicates the lesion;containing strand containing the 5′;32Phosphate. The enzyme reactions were performed at 37 °C and the reactions were quenched with 0.5 M NaOH after 20 minutes. The Sp;GPRPGP oligonucleotide degraded slightly to approximately 3% Sp, as evident in the slightly faster mobility in the substrate band (Lanes 36;42).
Measurements of the rate of base removal mediated by Fpg under STO conditions showed a decrease in rate of 4 to 5;fold for the Sp;amine adducts compared to Sp alone (SI Figure 3). In contrast, manual glycosylase assays for removal of Sp;amine adducts and Sp by NEIL1 indicated that the reactions were complete in the first time;point (20 sec). Initial experiments used edited NEIL1 (Arg at position 242), which is the form of NEIL1 most commonly studied. 41 However, due to previous work that has shown differences in activity between edited versus unedited NEIL1 (Lys at position 242), 41 a more detailed analysis was carried out with both enzyme forms using STO conditions with a 30;bp duplex containing the lesion (Gh, Sp1,Sp2, Sp;Lys, Sp;GlcN, and Sp;GPRP) in all four possible natural base;pairing contexts (Table 2, SI Tables 1 and 2). Strand cleavage was monitored using a rapid;quench flow instrument and the resulting progress curves were fit to either a single; or double; exponential equation to determine rates for the glycosylase reactions. The measured rates for removal of Gh, Sp1, and Sp2 follow the previously reported trend for a different 30;bp sequence, in which all three hydantoin lesions are excellent substrates for both NEIL1 isoforms.36 Of the three lesions, Sp1 was the most rapidly removed diastereomer. Remarkably, all bulky Sp;amine adduct lesions in all base pair contexts were removed efficiently by both isoforms of NEIL1 (edited and non;edited). In most base pair contexts the reactions were processed at such a high rate (>500 min;1) that it was only possible to estimate a lower limit. A complete table of kinetic values for the glycosylase activity with the lesions used in this work in all base pair contexts is available in Supporting Information (SI Tables 1 and 2). Base excision by edited NEIL1 from duplex substrates containing the hydantoin lesion paired with A were within experimentally accessible rate limits, and therefore measurable rate constants for removal of Sp;Lys (370 ± 70 min;1), Sp;GlcN (210 ± 90 min;1), and Sp;GPRP (330 ± 80 min;1) were determined that are on the same order of magnitude as the rate of removal of Sp1 in the corresponding duplex substrate (320 ± 30 min;1). Representative reaction profiles for edited NEIL1 with DNA duplexes containing Sp1:A and Sp;GPRP:A are shown in SI Figure 4, and the rate constants for Sp1:A, Sp2:A, Gh:A, Sp;Lys:A, Sp;GlcN:A, and Sp;GPRP:A are listed in Table 2.
Tabic 2.
Rate constants for NEIL1 with hydantoins determined under single-turnover conditions
| Base Pair | kg’ (min−1) | kg” (min−1) |
|---|---|---|
| Spl:A | 320 ± 30 | NA |
| Sp2:A | 47 ± 5 | NA |
| Gh:A | 130 ± 10 | NA |
| Sp-Lys:A | 370 ± 70 | 7 ± 1 |
| Sp-GlcN:A | 210 ± 90 | 3 ± 1 |
| Sp-GPRP:A | 330 ± 80 | 9 ± 2 |
Rate Constants determined at 37°C under STO conditions with hydantoin lesions base paired to A in a 30 base pair duplex. Reactions proceeded to >75% completion. Data for Sp amine adducts was fit with a double-exponential model generating kg’ and kg”. Errors reported are the standard deviations of the average of at least three independent trials. NA represents not applicable. The capacity represents the % of each rate in the double rate fits and the capacity for kg’ with Sp-Lys:A is 50 ± 2%, 41 ± 2% for Sp-(JlcN:A. and 36 ± 2% for Sp-GPRP.A and kg” with Sp-Lys:A is 50 ± 2%. 59 ± 2% for Sp-GlcN:A. and 64 ± 2% for Sp-GPRP:A.
Hydroxyl Radical Footprinting with E3Q NEIL1
Hydroxyl radical footprinting was used to define the contact region of the catalytically inactive E3Q NEIL1 protein on both lesion and complementary strand of the DNA duplex. Hydroxyl radicals were generated using Fe(II)*EDTA to initiate 2′;deoxyribose cleavage in a manner similar to previously reported work performed by our laboratory. 35 The 30;bp duplexes containing Gh, Sp1, Sp2, Sp;Lys, Sp;GlcN, and Sp;GPRPGP lesions paired with G or A were incubated with increasing amounts of E3Q NEIL1 followed by addition of Fe(II)*EDTA. In each case, either the lesion;containing strand or the complementary strand contained a 5′;[32P]; phosphate label. The end;labeled oligonucleotides were also subjected to G+A Maxam;Gilbert sequencing in order to determine the nucleotides that were protected by E3Q NEIL1 DNA interactions. Representative storage phosphor autoradiograms for the footprinting experiments with E3Q NEIL1 with Sp;GlcN:G containing DNA and a histogram showing the extent of protection at specific nucleotides are shown in SI Figures 5 and 6. In each case, a protected region was observed in the presence of the enzyme that spans approximately 17 ;20 nucleotides on both strands of the DNA duplex, regardless of the lesion identity or the base pair context. The protection from hydroxyl radical cleavage provided by E3Q NEIL1 on the G;containing strand is much more modest than that on the lesion;containing strand, suggesting more localization of the enzyme on the lesion;strand. Notably, there was an increase in the hydroxyl radical cleavage of the deoxyribose of the lesion nucleotide (2.4;fold) as well as a G nucleotide 5 bps 5′ of the lesion in the presence of E3Q NEIL1. This enzyme;induced hyper;reactivity at the lesion site and nearby suggests increased access to hydroxyl radical upon formation of the E3Q NEIL1;DNA complex.
DISCUSSION
NER typically targets large bulky lesions; whereas, BER is generally entrusted to repair more subtle base modifications. However the work presented herein with the UvrABC proteins shows that NER is able to recognize the bulky Sp;amine adducts quite efficiently, but also effectively acts on the small hydantoin lesions Gh and Sp. Remarkably, this work also demonstrates that the BER glycosylase NEIL1 is capable additionally of removing large and bulky Sp;amine base modifications as efficiently as the small parent lesion Sp. Indeed, despite the distinct mechanisms of DNA repair of NER and BER, this work underscores the similarity in mechanisms used to recognize a diverse set of damaged DNA substrates. Moreover, the overlapping mechanisms targeting hydantoin and hydantoin;amine adducts is likely important to ensure removal of these potently mutagenic and toxic lesions.
Our studies revealed that the UvrABC system excises the small base lesions, Gh, Sp and OG, as well as the large Sp;amine adducts. Similar rates of excision were measured for all substrates tested; however, the overall level of excised product differed among the lesion; containing substrates. Fluorescein;adducted dT, that served as a control for an excellent substrate, was the most efficiently processed substrate with reactions proceeding nearly to completion [(88 ± 2)%]. The Sp;amine adducts were also efficiently processed with reactions reaching ~ 60% completion, suggesting that the more bulky Sp;amine adducts are the most preferred hydantoin lesions for repair via NER. The difference in amount of excised product did not correlate with UvrA binding to the lesion;duplex based on the similar equilibrium dissociation constants (Kd) measured for UvrA with all of the damaged substrates. Notably, however, the extent of formation of a stable UvrBYDNA pre;incision complex correlated with the level of lesion excision, with a lesser fraction of UvrB bound to more poorly processed substrates OG, Gh, and Sp than the more efficiently processed substrates F, Sp;GlcN, Sp;Lys, Sp;GPRP, and Sp;GPRPGP. These results are consistent with the known sequence of events in NER that requires initial lesion duplex binding by UvrA followed by recruitment of UvrB to form the pre; incision complex.57,58,60 The pre;incision complex with UvrB recruits UvrC to catalyze the phosphodiester incision 5′ and 3′ to the lesion. We suggest that the formation of the stable pre; incision complex is the limiting factor in determining the fraction of substrate converted to product and this process is sensitive to the type of lesion.45 However, once the UvrB;DNA complex is formed, recruited UvrC mediates phosphodiester cleavage at a similar rate regardless of lesion identity.
The ability to effectively recognize these structurally related lesions that differ considerably in size highlights the flexible damage recognition mechanism of NER.45 Most studies suggest that the thermodynamic destabilization of B;form DNA induced by lesions is an important factor in both prokaryotic and eukaryotic NER processing.62 Previous work has shown that insertion of a β;hairpin motif of UvrB into the DNA duplex facilitates the ATPase activity of UvrB, and its associated helicase activity, which allows UvrB to take possession of the lesion from UvrA and to locate the lesion on the correct strand. 63,64 It is believed that the lesion containing strand is captured underneath the β;hairpin, in which the base 3′ to the damaged nucleotide is flipped into a highly conserved nucleotide;binding pocket in the process of forming the UvrB preincision complex.65,66 The stability of the UvrB;DNA pre;incision complex is likely connected to the ease of insertion of the β;hairpin into the duplex at the site of damage, aided by base stacking interactions with key tyrosine residues, which in turn is modulated by the structural and thermodynamic features of the lesion;containing duplex.62 Our results are consistent with this proposal: reduced activity and formation of UvrB;DNA pre; incision complex was measured for OG, a planar, non;helix;distorting lesion compared to the non;planar, helix;destabilizing spirocyclic Sp and its amine adducts.29,67,68 Accommodating the Sp amine adducts within the DNA duplex would be expected to require widening of the duplex and perturbations in both DNA base stacking and base;pairing that result in lower duplex stability.67,69 The compromised local base pair stability would facilitate insertion of the β;hairpin by UvrB into the DNA duplex and subsequent capture of the damaged DNA strand into the active site.
This work also establishes that the Sp;amine adducts are substrates for bacterial BER glycosylases Nei and Fpg and the human BER glycosylase NEIL1. In the case of Fpg, the efficiency of removal of the Sp;amine adducts was reduced relative to that observed for the parent lesion Sp. In contrast, the Sp;amine adducts are removed from duplex DNA at rates similar to Gh, Sp1, and Sp2, for both edited and non;edited NEIL1. This underscores that the Sp;amine adducts, despite their steric bulk, are excellent substrates for NEIL1. These new hydantoin substrates join an already broad list of substrates that have been identified for NEIL1.70 Of the oxidized bases removed by NEIL1, 2,6;diamino;4;hydroxy;5; formamidopyrimidine (FapyG) and 4,6;diamino;5;formamidopyrimidine (FapyA) are well; established substrates for NEIL1.44,71;74 Oxidized pyrimidines such as thymine glycol (Tg) and 5;hydroxy;uracil (5;OH;U) are also removed by NEIL1; however, notably, NEIL1 shows little activity towards OG.34,36,41,73,75;77 In addition to small base lesions, NEIL1 has been shown to remove bulky psoralen;base adducts, as well as psoralen;induced interstrand crosslinks in three; stranded DNA structures. 78,79 Importantly, the work herein shows quantitatively that the extent of removal of large Sp;amine adducts is similar to the parent Sp lesion. These results suggest that the NEIL1 active site is flexible and open in order to accommodate large adducts to the base and efficiently remove them.
In the analysis of the glycosylase assays of NEIL1 with the amine adducts (eg. Sp;Lys), the data fit better to a double;exponential equation with two distinct rates. These two different rates are likely due to differential rates of processing of the two Sp;amine adduct diastereomers. We have previously shown that NEIL1 preferentially removes one diastereomer (Sp1 over Sp2) relative to the other.36 The large attached amine may be more readily accommodated in the orientation presented by one diastereomer than the other. The preferred anti/syn glycosidic bond orientation and influence of the opposite base;pair would also be expected to be distinct for the two diastereomers. 29,67,69,80 Indeed, a slower rate of processing of all of the hydantoin lesions was observed in base;pairing contexts with A relative to C. The influence of the opposite base with NEIL1 may be indirect by influencing base;pair stability or lesion conformation.
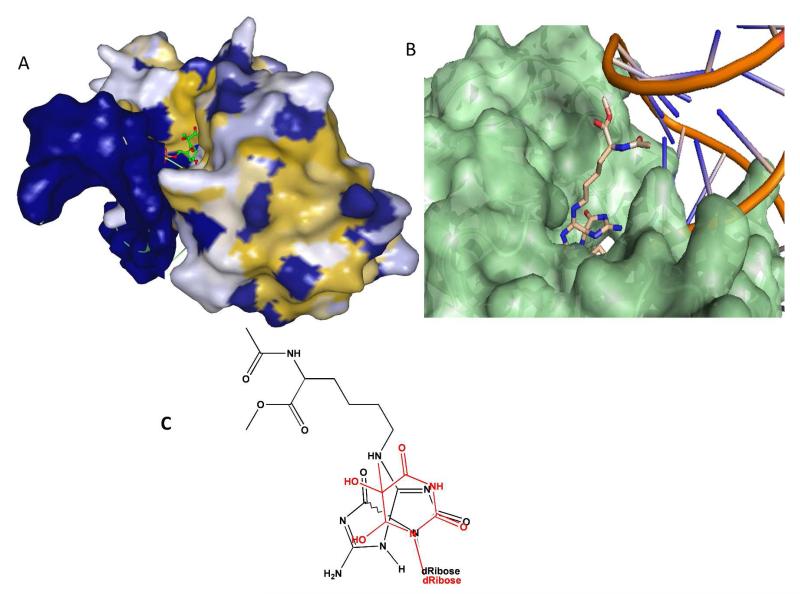
Recently, the first co;crystal structures of Mimivirus Nei1 (MvNei1), a viral ortholog of NEIL1, bound to damage;containing DNA were elucidated. 81 Examination of the X;ray structure of the complex of an inactive variant of MvNei1(E3Q) bound to a thymine glycol (Tg); containing duplex shows that the C5 and C6 positions of Tg are solvent exposed, with only water mediated amino acid interactions (Figure 6A). To provide insight into how a glycosylase (e.g. NEIL1) may be able to recognize bulky Sp;amine adducts, a structure was generated in which S; Sp;Lys was modeled into the active site of this lesion;containing MvNei1 structure (Figure 6B). The modeled Sp;Lys was positioned so the “pseudo;thymine” A ring of Sp;Lys would overlap with the oxidized pyrimidine ring of Tg (Figure 6C). This model reveals that the A ring of Sp; Lys can orient in a similar position as the Tg, allowing the B ring of Sp;Lys and the lysine chain of the adduct to be accommodated in the open space of the complex. This model suggests that despite their significant bulk, Sp;amine adducts may be easily accommodated in the mvNei1 (and presumably NEIL1) active site. Consistent with this model, Fe(II)*EDTA footprinting experiments using the catalytically inactive E3Q NEIL1 demonstrated increased reactivity of the hydantoin lesion nucleotide with hydroxyl radicals, suggesting increased accessibility of the nucleotide sugar to hydroxyl radical in the presence of the enzyme. 35
Figure 6. Structural rationale for potent activity of NEIL1 on Sp-amine adducts.
(A) Structure of MvNei1E3Q bound to Tg;containing DNA. The enzyme;DNA structure surface is mapped according to hydrophobicity (ranging from yellow (high) to blue (Low)) with Tg structure shown as ball and stick model. The enzyme has very few amino acid interactions with the Tg lesion, which appears to be solvent exposed. (B) Modeled S;Sp;Lys in active site: A ring of Sp;Lys in similar orientation as Tg, while the B ring and Lys adduct are accommodated in open space at back of binding pocket. (C) Structures of Tg (red) overlaid on structure of Sp;Lys adduct. Figures in (A) and (B) were generated using the MvNei;Tg structure in the protein database (PDB 3VK8) reported by Doublie and co;workers. 81
Interestingly, the crystal structures of E3Q MvNei1 bound to duplex DNA containing Tg or 5;hydroxyuracil (5;OHU) exhibit minimal protein amino acid interactions with the pyrimidine lesions in the lesion binding site.81 In fact, 5;OHU was found to adopt both anti or syn glycosidic bond orientations, suggesting minimal stabilization of the lesion in the active site. Additionally, the authors confirmed that mutation of two amino acid residues that make contacts to the lesion base (E6A and Y253F) did not significantly reduce glycosylase activity, revealing that these amino acids are not strictly required for removal of aberrant bases. The authors replaced the Tg in the E3Q MvNeil;Tg structure with an undamaged T to illustrate that lack of obvious amino acid interactions in the active site to provide for discrimination for the damaged base. Since MvNei1 does not cleave thymine, this suggests that MvNei1 uses an alternative method to recognize substrates prior to base flipping into the active site. These observations taken together suggested that this crystal structure captures a glimpse of a step that is subsequent to the one allowing for discrimination between damaged and undamaged DNA bases. The results suggest that key features of damage recognition by this glycosylase involve sensing local conformational distortions in the phosphodiester backbone and base pair stability. Notably, these damage recognition features are reminiscent of those used in the NER pathway.
The ability of NEIL1 to process large Sp;peptide adducts suggests that NEIL1 may be capable of excising DPC formed via oxidative reactions. Hydantoin*protein cross;links can be generated from oxidation with numerous biologically relevant oxidants in the presence of DNA binding proteins.21,22,25,26 The mechanism of repair of DPC may involve proteolytic digestion to aid in removal of the majority of the protein adduct followed by repair initiated by NER, BER or cooperation between the two pathways. 46,55,82;85 In addition, NEIL1 has been suggested to influence NER;mediated repair of (5′R); and (5′S);8,5′;cyclo;2′;deoxyadenosine lesions based on accumulation of these NER substrates in Neil1;/; mice.86 Since NEIL1 is not capable of cleaving these cyclopurine adducts, NEIL1 was postulated to play a role in the initial lesion recognition and recruitment of NER.86 Of note, cells from patients with Cockayne’s syndrome (CS) have been shown to have decreased ability to repair OG, 8;oxo;7,8;dihydro;adenine (OA), and Tg base lesions generated during oxidative stress. 87;92 This finding was surprising since the mutated genes associated with CS (CSA & CSB) code for two transcription;coupled repair NER; specific factors.93,94 It has also been shown in vitro that CSB stimulates NEIL1 glycosylase activity, and that CSB and NEIL1 co;immunoprecipitate and co;localize in HeLa cells. These results suggest that defective interactions between the mutant Cockayne syndrome NER proteins and the BER glycosylase NEIL1 may be involved in the pathogenesis of CS. 87 These observations, taken together with the overlapping substrate spectrum of the two repair pathways, suggest that coordination events between NER and BER may be critical for preservation of the genome.
CONCLUSION
We have demonstrated that the helix;distorting hydantoin lesions derived from guanine oxidation, as well as their amine;adducted structures, are substrates for both the BER and NER pathways. Even though hydantoin lesions are present in low concentrations compared to OG, their high mutagenic potential, ability to stall replication forks and induce strand slippage makes their presence in cells more detrimental than OG. Moreover, Sp;amine;containing DNA;protein crosslinks would be expected to be extremely toxic to cells.2 Undoubtedly, maintenance of genomic integrity and cell survival requires efficient repair of hydantoin lesions. These studies also reveal that both NEIL1 and the UvrABC nuclease system use similar strategies that rely heavily on the local duplex stability to recognize a wide variety of structurally diverse lesions within DNA. During periods of high oxidative stress and oxidative DNA damage, the action of both BER and NER pathways would accelerate repair. Overlapping lesion specificity among DNA repair pathways would also be an evolutionary advantage, particularly for the highly mutagenic lesions Gh and Sp. 31
Supplementary Material
ACKNOWLEDGEMENTS
We thank the National Institutes of Health (R01CA090689 to S.S.D and C.J.B, 1R01ES019566 to B.V.H.) for support of this work. We also thank Professors Susan Wallace and Sylvie Doublie (University of Vermont) for the NEIL1 and Nei expression plasmids, and Ms. Amelia Manlove for help in preparing figures and editing the manuscript. We also thank Dr. Sucharita Kundu for performing initial experiments that showed that Sp;amine adducts are substrates for NEIL1.
Footnotes
ASSOCIATED CONTENT
Supporting information: Kinetic data tables for edited and non;edited NEIL1 (SI Tables 1;2), EMSA experiments with UvrA (SI Figure 2), representative PAGE autoradiograms for UvrABC excision sites (SI Figure 1), representative kinetic plots for Sp;amine adduct removal by Fpg and NEIL1 (SI Figures 3 & 4), representative PAGE autoradiograms of footprinting with E3Q NEIL1 (SI Figure 5) and corresponding histogram (SI Figure 6) and Supplementary Methods. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).David SS, O’Shea VL, Kundu S. Nature. 2007;447:941. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Neeley WL, Essigmann JM. Chem. Res. Toxicol. 2006;19:491. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- (3).Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. Proc. Natl. Acad. Sci. U .S. A. 2012;109:E1820. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Burrows CJ, Muller JG, Kornyushyna O, Luo W, Duarte V, Leipold MD, David SS. Environ. Health Perspect. 2002;110(Suppl 5):713. doi: 10.1289/ehp.02110s5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Luo W, Muller JG, Rachlin EM, Burrows CJ. Org. Lett. 2000;2:613. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- (6).Munk BH, Burrows CJ, Schlegel HB. J. Am. Chem. Soc. 2008;130:5245. doi: 10.1021/ja7104448. [DOI] [PubMed] [Google Scholar]
- (7).Fleming AM, Burrows CJ. Chem. Res. Toxicol. 2013;26:593. doi: 10.1021/tx400028y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Fleming AM, Muller JG, Dlouhy AC, Burrows CJ. J. Am. Chem. Soc. 2012;134:15091. doi: 10.1021/ja306077b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Luo W, Muller JG, Rachlin EM, Burrows CJ. Chem. Res. Toxicol. 2001;14:927. doi: 10.1021/tx010072j. [DOI] [PubMed] [Google Scholar]
- (10).Suzuki T, Masuda M, Friesen MD, Ohshima H. Chem. Res. Toxicol. 2001;14:1163. doi: 10.1021/tx010024z. [DOI] [PubMed] [Google Scholar]
- (11).Suzuki T, Ohshima H. FEBS Lett. 2002;516:67. doi: 10.1016/s0014-5793(02)02503-6. [DOI] [PubMed] [Google Scholar]
- (12).Suzuki T, Friesen MD, Ohshima H. Chem. Res. Toxicol. 2003;16:382. doi: 10.1021/tx025638y. [DOI] [PubMed] [Google Scholar]
- (13).Adam W, Arnold MA, Grune M, Nau WM, Pischel U, Saha-Moller CR. Org Lett. 2002;4:537. doi: 10.1021/ol017138m. [DOI] [PubMed] [Google Scholar]
- (14).Crean C, Geacintov NE, Shafirovich V. Angew. Chem. Int .Ed. Engl. 2005;44:5057. doi: 10.1002/anie.200500991. [DOI] [PubMed] [Google Scholar]
- (15).Crean C, Lee YA, Yun BH, Geacintov NE, Shafirovich V. ChemBiochem. 2008;9:1985. doi: 10.1002/cbic.200800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Joffe A, Geacintov NE, Shafirovich V. Chem. Res. Toxicol. 2003;16:1528. doi: 10.1021/tx034142t. [DOI] [PubMed] [Google Scholar]
- (17).Niles JC, Wishnok JS, Tannenbaum SR. Org. Lett. 2001;3:963. [PubMed] [Google Scholar]
- (18).Suzuki T, Masuda M, Friesen MD, Fenet B, Ohshima H. Nucleic Acids Res. 2002;30:2555. doi: 10.1093/nar/30.11.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Yu H, Venkatarangan L, Wishnok JS, Tannenbaum SR. Chem. Res. Toxicol. 2005;18:1849. doi: 10.1021/tx050146h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Niles JC, Wishnok JS, Tannebaum SR. Chem. Res. Toxicol. 2004;17:1510. doi: 10.1021/tx0400048. [DOI] [PubMed] [Google Scholar]
- (21).Johansen ME, Muller JG, Xu X, Burrows CJ. Biochemistry. 2005;44:5660. doi: 10.1021/bi047580n. [DOI] [PubMed] [Google Scholar]
- (22).Xu X, Muller JG, Ye Y, Burrows CJ. J. Am. Chem. Soc. 2008;130:703. doi: 10.1021/ja077102a. [DOI] [PubMed] [Google Scholar]
- (23).Hosford ME, Muller JG, Burrows CJ. J. Am. Chem. Soc. 2004;126:9540. doi: 10.1021/ja047981q. [DOI] [PubMed] [Google Scholar]
- (24).Hickerson RP, Chepanoske CL, Williams SD, David SS, Burrows CJ. J. Am. Chem. Soc. 1999;121:9901. [Google Scholar]
- (25).Schibel AE, An N, Jin Q, Fleming AM, Burrows CJ, White HS. J. Am. Chem. Soc. 2010;132:17992. doi: 10.1021/ja109501x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Solivio MJ, Nemera DB, Sallans L, Merino EJ. Chem. Res. Toxicol. 2012;25:326. doi: 10.1021/tx200376e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kornyushyna O, Burrows CJ. Biochemistry. 2003;42:13008. doi: 10.1021/bi0350755. [DOI] [PubMed] [Google Scholar]
- (28).Aller P, Ye Y, Wallace SS, Burrows CJ, Doublie S. Biochemistry. 2010;49:2502. doi: 10.1021/bi902195p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kornyushyna O, Berges AM, Muller JG, Burrows CJ. Biochemistry. 2002;41:15304. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- (30).Duarte V, Muller JG, Burrows CJ. Nucleic Acids Res. 1999;27:496. doi: 10.1093/nar/27.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. Biochemistry. 2003;42:9257. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- (32).Leipold MD, Muller JG, Burrows CJ, David SS. Biochemistry. 2000;39:14984. doi: 10.1021/bi0017982. [DOI] [PubMed] [Google Scholar]
- (33).Leipold MD, Workman H, Muller JG, Burrows CJ, David SS. Biochemistry. 2003;42:11373. doi: 10.1021/bi034951b. [DOI] [PubMed] [Google Scholar]
- (34).Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. DNA Repair (Amst) 2005;4:41. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- (35).Krishnamurthy N, Muller JG, Burrows CJ, David SS. Biochemistry. 2007;46:9355. doi: 10.1021/bi602459v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Krishnamurthy N, Zhao X, Burrows CJ, David SS. Biochemistry. 2008;47:7137. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Hazra TK, Muller JG, Manuel RC, Burrows CJ, Lloyd RS, Mitra S. Nucleic Acids Res. 2001;29:1967. doi: 10.1093/nar/29.9.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Tretyakova NY, Wishnok JS, Tannenbaum SR. Chem. Res. Toxicol. 2000;13:658. doi: 10.1021/tx000083x. [DOI] [PubMed] [Google Scholar]
- (39).Liu M, Bandaru V, Holmes A, Averill AM, Cannan W, Wallace SS. Protein Exp. Pur. 2012;84:130. doi: 10.1016/j.pep.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Bandaru V, Zhao X, Newton MR, Burrows CJ, Wallace SS. DNA Repair. 2007;6:1629. doi: 10.1016/j.dnarep.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Yeo J, Goodman RA, Schirle NT, David SS, Beal PA. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20715. doi: 10.1073/pnas.1009231107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Zhao X, Krishnamurthy N, Burrows CJ, David SS. Biochemistry. 2010;49:1658. doi: 10.1021/bi901852q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Hailer MK, Slade PG, Martin BD, Sugden KD. Chem. Res. Toxicol. 2005;18:1378. doi: 10.1021/tx0501379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Chan MK, Ocampo-Hafalla MT, Vartanian V, Jaruga P, Kirkali G, Koenig KL, Brown S, Lloyd RS, Dizdaroglu M, Teebor GW. DNA Repair (Amst) 2009;8:786. doi: 10.1016/j.dnarep.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Truglio JJ, Croteau DL, Van Houten B, Kisker C. Chem. Rev. 2006;106:233. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- (46).Minko IG, Zou Y, Lloyd RS. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1905. doi: 10.1073/pnas.042700399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).VanHouten B. Microbiol. Rev. 1990;54:18. [Google Scholar]
- (48).Van Houten B, McCullough A. Ann. N. Y. Acad. Sci. 1994;726:236. doi: 10.1111/j.1749-6632.1994.tb52822.x. [DOI] [PubMed] [Google Scholar]
- (49).Wang H, DellaVecchia MJ, Skorvaga M, Croteau DL, Erie DA, Van Houten B. J. Biol. Chem. 2006;281:15227. doi: 10.1074/jbc.M601476200. [DOI] [PubMed] [Google Scholar]
- (50).Croteau DL, DellaVecchia MJ, Wang H, Bienstock RJ, Melton MA, Van Houten B. J Biol Chem. 2006;281:26370. doi: 10.1074/jbc.M603093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Imoto S, Bransfield LA, Croteau DL, Van Houten B, Greenberg MM. Biochemistry. 2008;47:4306. doi: 10.1021/bi7021427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Zhao X, Muller JG, Halasyam M, David SS, Burrows CJ. Biochemistry. 2007;46:3734. doi: 10.1021/bi062214k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Skorvaga M, Theis K, Mandavilli BS, Kisker C, Van Houten B. J. Biol. Chem. 2002;277:1553. doi: 10.1074/jbc.M108847200. [DOI] [PubMed] [Google Scholar]
- (54).McKibbin PL, Kobori A, Taniguchi Y, Kool ET, David SS. J. Am. Chem. Soc. 2012;134:1653. doi: 10.1021/ja208510m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Minko IG, Kurtz AJ, Croteau DL, Van Houten B, Harris TM, Lloyd RS. Biochemistry. 2005;44:3000. doi: 10.1021/bi0478805. [DOI] [PubMed] [Google Scholar]
- (56).Sambrook J, Russell DW. Molecular Cloning, A Laboratory Manual. 3 ed. Vol. 1. Cold Spring Harbor Laboratory Press; Plainview, NY: 2001. [Google Scholar]
- (57).Zou Y, Van Houten B. EMBO. J. 1999;18:4889. doi: 10.1093/emboj/18.17.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Zou Y, Luo C, Geacintov NE. Biochemistry. 2001;40:2923. doi: 10.1021/bi001504c. [DOI] [PubMed] [Google Scholar]
- (59).Webster MPJ, Jukes R, Zamfir VS, Kay CWM, Bagnéris C, Barrett T. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Pakotiprapha D, Samuels M, Shen K, Hu JH, Jeruzalmi D. Nat. Struct. Mol. Biol. 2012;19:291. doi: 10.1038/nsmb.2240. [DOI] [PubMed] [Google Scholar]
- (61).Chu AM, Fettinger JC, David SS. Bioorg. Med. Chem. Lett. 2011;21:4969. doi: 10.1016/j.bmcl.2011.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Liu Y, Reeves D, Kropachev K, Cai Y, Ding S, Kolbanovskiy M, Kolbanovskiy A, Bolton JL, Broyde S, Van Houten B, Geacintov NE. DNA Repair (Amst) 2011;10:684. doi: 10.1016/j.dnarep.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Truglio JJ, Karakas E, Rhau B, Wang H, DellaVecchia MJ, Van Houten B, Kisker C. Nat. Struct. Mol. Biol. 2006;13:360. doi: 10.1038/nsmb1072. [DOI] [PubMed] [Google Scholar]
- (64).Hughes C, Wang H, Ghodke H, Simons M, Towheed A, Peng Y, Van Houten B, Kad N. Nucleic Acids Res. 2013;41:4901. doi: 10.1093/nar/gkt177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Malta E, Verhagen CP, Mollenaar GF, Fillipov DV, van der Marel GA, Goosen N. DNA Repair. 2008;7:1647. doi: 10.1016/j.dnarep.2008.06.011. [DOI] [PubMed] [Google Scholar]
- (66).Malta E, Moolenar GF, Goosen N. J. Biol. Chem. 2006;281:2184. doi: 10.1074/jbc.M508901200. [DOI] [PubMed] [Google Scholar]
- (67).Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. Biochemistry. 2005;44:13342. doi: 10.1021/bi050790v. [DOI] [PubMed] [Google Scholar]
- (68).Yennie CJ, Delaney S. Chem. Res. Toxicol. 2012;25:1732. doi: 10.1021/tx300190a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. Biochemistry. 2005;44:6043. doi: 10.1021/bi0473657. [DOI] [PubMed] [Google Scholar]
- (70).Prakash A, Doublie S, Wallace SS. Prog. Mol. Biol. Trans. Sci. 2012;110:71. doi: 10.1016/B978-0-12-387665-2.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Proc. Natl. Acad. Sci. U .S. A. 2002;99:3523. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Hu J, de Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, Dizdaroglu M, Bohr VA. J Biol Chem. 2005;280:40544. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- (73).Jaruga P, Birincioglu M, Rosequist TA, Dizdaroglu M. Biochemistry. 2004;43:15909. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- (74).Roy LM, Jaruga P, Wood TG, McCullough AK, Dizdaroglu M, Lloyd RS. J. Biol. Chem. 2007;282:15790. doi: 10.1074/jbc.M610626200. [DOI] [PubMed] [Google Scholar]
- (75).Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapanti SK, Mitra S, Izumi T. J. Biol. Chem. 2002;277:30417. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- (76).Bandaru V, Sunkara S, Wallace SS, Bond JP. DNA Repair. 2002;1:517. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- (77).Takao M, Kanno S, Kobayashi K, Zhang Q-M, Yonei S, van der Horst GTJ, Yasui A. Proc. Natl. Acad. Sci. U.S.A. 2002;277:42205. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- (78).Couve S, Mace-Aime G, Rosselli F, Saparbaev M. J. Biol. Chem. 2009;284:11963. doi: 10.1074/jbc.M900746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Couve-Privat S, Mace-Aime G, Rosselli F, Saparbaev M. Nucleic Acid Res. 2007;35:5672. doi: 10.1093/nar/gkm592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Chinyengetere F, Jamieson ER. Biochemistry. 2008;47:2584. doi: 10.1021/bi701502t. [DOI] [PubMed] [Google Scholar]
- (81).Imamura K, Averill A, Wallace SS, Doublie S. J. Biol. Chem. 2012;287:4288. doi: 10.1074/jbc.M111.315309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Desai SD, Zhang H, Rodriguez-Bauman A, Yang JM, Wu X, Gounder MK, Rubin EH, Liu LF. Mol. Cell Biol. 2003;23:2341. doi: 10.1128/MCB.23.7.2341-2350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Mao Y, Desai SD, Ting CY, Hwang J, Liu LF. J. Biol. Chem. 2001;276:40652. doi: 10.1074/jbc.M104009200. [DOI] [PubMed] [Google Scholar]
- (84).Quievryn G, Zhitkovich A. Carcinogenesis. 2000;21:1573. [PubMed] [Google Scholar]
- (85).Baker DJ, Wuenschell G, Xia L, Termini J, Bates SE, Riggs AD, O’Connor TR. J. Biol. Chem. 2007;282:22592. doi: 10.1074/jbc.M702856200. [DOI] [PubMed] [Google Scholar]
- (86).Jaruga P, Xiao Y, Vartanian V, Lloyd RS, Dizdaroglu M. Biochemistry. 2010;49:1053. doi: 10.1021/bi902161f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Muftuoglu M, de Souza-Pinto NC, Dogan A, Aamann M, Stevnsner T, Rybanska I, Kirkali G, Dizdaroglu M, Bohr VA. J. Biol. Chem. 2009;284:9270. doi: 10.1074/jbc.M807006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Tuo J, Jaruga P, Rodriguez H, Dizdaroglu M, Bohr VA. J. Biol. Chem. 2002;277:30832. doi: 10.1074/jbc.M204814200. [DOI] [PubMed] [Google Scholar]
- (89).Sunesen M, Stevnsner T, Brosh RM, Jr., Dianov GL, Bohr VA. Oncogene. 2002;21:3571. doi: 10.1038/sj.onc.1205443. [DOI] [PubMed] [Google Scholar]
- (90).Stevnsner T, Nyaga S, de Souza-Pinto NC, van der Horst GT, Gorgels TG, Hogue BA, Thorslund T, Bohr VA. Oncogene. 2002;21:8675. doi: 10.1038/sj.onc.1205994. [DOI] [PubMed] [Google Scholar]
- (91).Kyng KJ, May A, Brosh RM, Jr., Cheng WH, Chen C, Becker KG, Bohr VA. Oncogene. 2003;22:1135. doi: 10.1038/sj.onc.1206187. [DOI] [PubMed] [Google Scholar]
- (92).Hanawalt PC. Science. 1994;266:1957. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- (93).Hanawalt PC. Nature. 2000;405:415. doi: 10.1038/35013197. [DOI] [PubMed] [Google Scholar]
- (94).Friedberg EC, Henning KA. Mutat. Res. 1993;289:47. doi: 10.1016/0027-5107(93)90129-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.