Abstract
The number of proteasomal substrates that are degraded without prior ubiquitylation continues to grow. However, it remains poorly understood how the proteasome recognizes substrates lacking a ubiquitin (Ub) signal. Here we demonstrated that the Ub-independent degradation of Rpn4 requires the 19S regulatory particle (RP). The Ub-independent degron of Rpn4 was mapped to an N-terminal region including the first 80 residues. Inspection of its amino acid sequence revealed that the Ub-independent degron of Rpn4 consists of an intrinsically disordered domain followed by a folded segment. Using a photo-crosslinking-label transfer method, we captured three 19S RP subunits (Rpt1, Rpn2 and Rpn5) that bind the Ub-independent degron of Rpn4. This is the first time that specific 19S RP subunits have been identified interacting with a Ub-independent degron. This study provides insight into the mechanism by which Ub-independent substrates are recruited to the 26S proteasome.
Keywords: Proteasome, Protein degradation, Ubiquitin-independent, Degron, Rpn4
1. Introduction
Recent studies have shown that some substrates are degraded by the proteasome without prior ubiquitylation. These Ub-independent substrates include the polyamine biosynthetic enzyme ornithine decarboxylase (ODC) [1,2], several proto-oncoproteins and tumor suppressors [3], the human thymidylate synthase (hTS) [4], the gap junction protein connexin43 [5], the BIMEL pro-apoptotic BH3-only protein [6], the transcriptional coactivator SRC-3 [7], the yeast transcription factor Rpn4 [8], the hepatitis C virus F and core proteins [9,10], and others [11,12]. The identification of Ub-independent substrates from various cellular pathways in different organisms suggests a significant contribution of the Ub-independent degradation pathway to the regulation of protein homeostasis in the cell.
Although the number of Ub-independent substrates continues to grow, current knowledge of the mechanism underlying Ub-independent protein degradation is limited. Some Ub-independent substrates, mainly native unstructured, misfolded, or damaged proteins, are degraded by the 20S proteasome by a “default” mechanism [13]. Other substrates, however, need the help of proteasome activators for their degradation. For example, the 19S RP is required for the Ub-independent degradation of ODC [2]. The proteasome activators are presumably responsible for recruiting substrates [14–16], but it remains an open question how the proteasome activators recognize substrates lacking a Ub tag. This is even more mysterious for the 19S RP, which consists of at least 19 different subunits. Which one of its subunits recruits Ub-independent substrates?
Rpn4 is the transcription factor that activates the yeast Saccharomyces cerevisiae proteasome genes [17,18]. Our early work has shown that Rpn4 is degraded by two pathways: one is Ub-dependent, whereas the other is Ub-independent (8). Here we demonstrated that the Ub-independent degradation of Rpn4 is mediated by the 26S but not the 20S proteasome. We mapped the Ub-independent degron of Rpn4 to its N-terminal region including the first 80 amino acids. Using a photo-crosslinking-label transfer method, we identified three 19S RP subunits (Rpt1, Rpn2 and Rpn5) that bind the Ub-independent degron of Rpn4. This study provides a foundation for investigation of the molecular mechanism by which Ub-independent substrates are delivered to the 26S proteasome.
2. Materials and methods
2.1. Yeast strains and plasmids
The yeast strains used in this study were listed in Supplemental Materials and Methods. Details of plasmid constructs are available upon request. Briefly, for protein purification from Escherichia coli, C-terminally FLAG-tagged Rpn4 and Rpn41–80-DHFRha, and C-terminally His-tagged Rpn41–80 and Rpn41–120 were expressed from the pET11d vector (Novagen). For in vivo degradation analysis, C-terminally 3ha-tagged Rpn4Δ211–229 was expressed from its own promoter in the pRS313 vector. C-terminally ha-tagged lysine-less Rpn4 N-terminal fragment (K0Rpn41–229) was expressed from the CUP1 promoter in pRS313. The DHFR fusions with various N-terminal fragments of Rpn4 were expressed from the CUP1 promoter in the pRS314 vector.
2.2. Protein purification
The procedure to purify 20S and 26S proteasomes was described in Supplemental Materials and Methods. Expression of Rpn4 and its derivatives in E. coli (BL21) was induced by 0.3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 25 °C for 3 h. Cells were suspended in buffer A (50 mM Tris.CL, pH 7.5, 150 mM NaCl, 10% glycerol, 0.2% Triton X-100) containing 0.1 mg/ml of lysozyme and 2 mM PMSF, and disrupted by sonification. Cell extracts were collected by centrifugation at 13,200 rpm for 10 min and further clarified by ultracentrifugation at 100 K rpm for 1 h. To purify His-tagged Rpn41–80 and Rpn41–120, supernatants were loaded onto 1 ml Ni–NTA agarose column equilibrated with buffer A containing 20 mM imidazole, followed by washing with 20 ml of equilibration buffer. His-tagged proteins were then eluted by step-gradient of imidazole from 40 to 200 mM in buffer B (25 mM HEPES, pH 7.8, 5 mM MgCl2, 25 mM KCl, 10% glycerol). Excess imidazole was removed by ultrafiltration (3 kDa MWCO, Millipore). FLAG-tagged Rpn4 and Rpn41–80-DHFRha were purified using a similar approach as purification of FLAG-tagged proteasomes.
2.3. In vitro degradation assay
A degradation assay was set up in a 15 μl reaction containing 4.6 μg of purified proteasomes and 50–70 ng of substrates in buffer B supplemented with 1 mM ATP. The reactions were initiated by addition of 20S or 26S proteasome, and then incubated at 30 °C for different periods. To inhibit substrate degradation, proteasomes were pre-treated with 50 μM MG-132 at RT for 10 min before applied to the degradation assays. The reaction mixture was separated by SDS–PAGE, followed by immunoblotting with an anti-FLAG antibody for Rpn4-FLAG, an anti-ha antibody for Rpn41–80-DHFRha, or an anti-His antibody for Rpn41–80-His and Rpn41–120-His. The immunoblotting signals were detected by the Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, Nebraska).
2.4. Pulse chase and cycloheximide (CHX) chase assays
Pulse chase analysis was described previously [19] and detailed in Supplemental Materials and Methods. For CHX-chase analysis, CHX was added to yeast cultures grown to OD600 of 0.8–1.2 at a final concentration of 0.2 mg/ml. An aliquot was withdrawn at each time point. Cells were lysed in equal volume of 2× SDS buffer. Cell extracts were separated by SDS–PAGE, followed by immunoblotting with an anti-ha antibody for the detection of Rpn4Δ211–229-3 ha and K0Rpn41–229-ha, or an anti-yeast α tubulin antibody (Chemicon, Temecula, CA) to verify comparable loading.
2.5. Photo-crosslinking-label transfer assay
Purified Rpn41–80-His protein (0.1 mg/ml) was incubated with 1 mM of photo-crosslinking reagent Sulfo-SBED (Pierce, Product # 33033) for 30 min in the dark. Non-reacted free crosslinker was inactivated by addition of 50 mM Tris–Cl, pH 7.5, and removed by ultrafiltration. To transfer the biotin label from Rpn41–80-His to 26S proteasome, 0.9 μg of Rpn41–80-Hisbiotin was incubated with 69 lg of purified 26S proteasome in 150 μl of buffer B containing 1 mM ATP at RT in the dark for 15 min. The mixture was then exposed to UV light (UV Stratalinker 2400, Stratagene) for 3 min. DTT (200 mM) and SDS (2%) were added to the reaction, followed by heating at 100 °C for 10 min. An aliquot (15 μl) was withdrawn and resolved by SDS–PAGE (9% gel), followed by western analysis with HRP-conjugated streptavidin and detection with the Super-Signal West Pico Chemiluminescent Substrate (Pierce Biotechnology). The remaining mixture was diluted 25 times in PBS containing 0.5% NP-40 and 0.1% SDS, and incubated with 15 μl of streptavidin agarose beads at 4 °C for 4 h. The beads were washed 5 times with pulldown buffer, and the biotin-labeled proteins were eluted by SDS sample buffer and separated by SDS–PAGE (9% gel). The proteins pulled down by streptavidin beads were visualized by SYPRO-Ruby staining. The bands aligned with the positions of proteasome subunits were cut out from the gel and subjected to LC–MS/MS analysis.
3. Results
3.1. Ub-independent degradation of Rpn4 requires the 19S RP
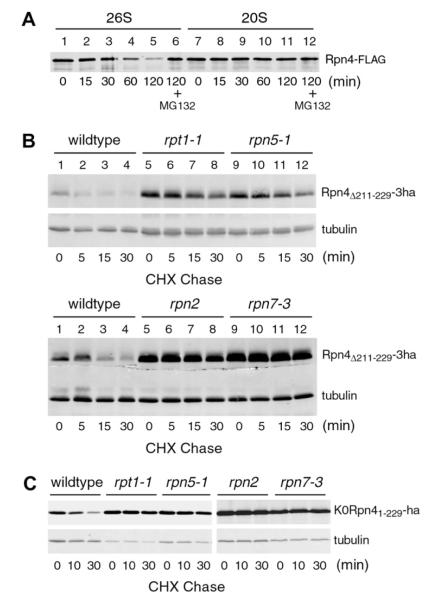
To verify our previous in vivo study revealing Ub-independent degradation of Rpn4, we decided to examine if nonubiquitylated Rpn4 could be degraded by the proteasome in vitro. In addition, we wanted to test if the 19S RP is required. To address these two questions, we purified 20S and 26S proteasomes from a yeast strain in which a FLAG tag was linked to the C-terminus of Pre1, a subunit of the 20S core. The integrity and assembly of purified 20S and 26S proteasomes were verified by their characteristic polypeptide composition on SDS–PAGE and in-gel hydrolysis of the fluorogenic peptide Suc-LLVY-AMC (Supplemental Fig. 1). The in-gel degradation assay also confirmed that both 20S and 26S proteasomes retained their proteolytic activity over the course of purification. C-terminally FLAG-tagged Rpn4 was expressed in and purified from E. coli (Supplemental Fig. 2). The Rpn4 substrate was mixed with purified 20S or 26S proteasome and incubated for different intervals from 15 to 120 min. The degradation of Rpn4 was assessed by immunoblotting analysis with an anti-FLAG antibody (Fig. 1A). Rpn4 was degraded by the 26S proteasome (lanes 1–5), which was inhibited by the proteasome inhibitor MG-132 (lane 6). In contrast, the 20S proteasome did not noticeably degrade Rpn4 (lanes 7–12). Thus, Rpn4 is degraded by the 26S proteasome without prior ubiquitylation, and the 19S RP is required.
Fig. 1.
Ub-independent degradation of Rpn4 by the 26S proteasome. (A) Rpn4 is degraded by the 26S but not the 20S proteasome. In vitro degradation assays with purified 20S or 26S proteasome were performed to measure Ub-independent degradation of Rpn4. (B and C) Both base and lid subcomplexes of the 19S RP are required for the Ub-independent degradation of Rpn4. CHX-chase assays were conducted to assess the degradation of Rpn4Δ211–229-3ha (B) and K0Rpn41–229-ha (C) in various 19S RP mutants and a wildtype strain.
We then examined if the 19S RP is essential for Ub-independent degradation of Rpn4 in vivo. Our early work has located the Ub-dependent degron of Rpn4 to residues 211–229 and shown that Rpn4Δ211–229 is degraded by a Ub-independent mechanism [19]. We transformed a low-copy vector expressing Rpn4Δ211–229 from the native RPN4 promoter into four 19S RP mutants, bearing mutations in base subunits Rpt1 (cim5–1) and Rpn2 (rpn2) and lid subunits Rpn5 (rpn5–1) and Rpn7 (rpn7–3), respectively. A triple hemagglutinin (ha) epitope tag was added to the C-terminus of Rpn4Δ211–229 for immunoblotting analysis. CHX-chase analysis showed that Rpn4Δ211–229-3ha was rapidly degraded in the wildtype strain, but substantially stabilized in all the mutants (Fig. 1B). The steady-state level of Rpn4Δ211–229-3ha, reflected at the zero-time point, was also much higher in the mutants than the wildtype strain (compare lanes 5 and 9 to 1). Similar results were obtained when K0Rpn41–229, another Ub-independent substrate derived from Rpn4, was used in the CHX-chase assay (Fig. 1C). K0Rpn41–229 is a 229-amino acid N-terminal fragment of Rpn4 with all lysines mutated to arginines [8]. Together, these results indicate that the 19S RP is essential for Ub-independent degradation of Rpn4 in vivo. Both of its subcomplexes, the base and the lid, are indispensable.
3.2. The N-terminal region of Rpn4 contains a transplantable Ub-independent degron
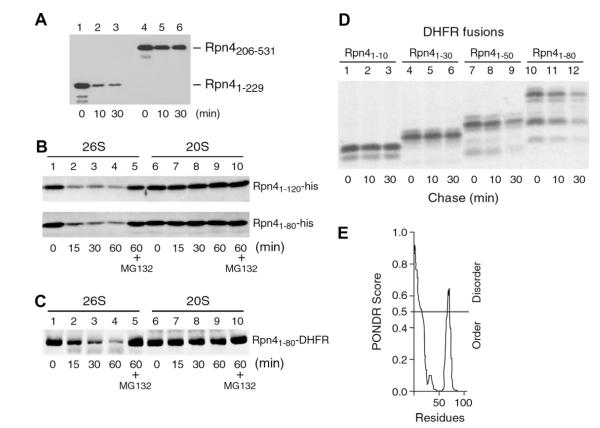
To explore the mechanism of Ub-independent degradation of Rpn4, we set out to map its Ub-independent degron. We first divided Rpn4 into two halves: the N-terminal fragment containing the first 229 amino acids and the C-terminal fragment including residues 206–531. These two fragments were expressed in and purified from E. coli and subjected to in vitro degradation assays with purified 26S proteasome (Fig. 2A). Whereas the C-terminal fragment was stable, the N-terminal fragment was degraded, indicating that the Ub-independent degron is located in the N-terminal region. We further measured the degradation of two smaller N-terminal fragments, Rpn41–80 and Rpn41–120. As shown in Fig. 2B, both fragments were degraded by the 26S proteasome (lanes 1–5). Like the full-length Rpn4, these two N-terminal fragments were not degraded by the 20S proteasome (lanes 6–10). Together, these results demonstrate that the Ub-independent degron of Rpn4 resides in the N-terminal region consisting of the first 80 amino acids.
Fig. 2.
The N-terminal 80 amino acids of Rpn4 constitute a transplantable Ub-independent 19S RP-dependent degron. (A) The N-terminal region of Rpn4 harbors a Ub-independent degron. An N-terminal fragment (Rpn41–229) and a C-terminal fragment (Rpn4206–531) of Rpn4 were applied to in vitro degradation assays with purified 26S proteasome. (B) In vitro degradation of Rpn41–80 and Rpn41–120 by the 26S but not the 20S proteasome. C-terminally His-tagged Rpn41–80 and Rpn41–120 proteins were subjected to in vitro degradation assays with purified 20S and 26S proteasomes, respectively. (C) The N-terminal 80 amino acids of Rpn4 destabilize DHFR. The Rpn41–80-DHFR fusion protein expressed in and purified from E. coli was applied to in vitro degradation assays with 20S or 26S proteasome. (D) Pulse chase analysis was carried out to examine in vivo degradation of DHFR fusion proteins carrying various N-terminal fragments of Rpn4. An ha tag was added to the C-terminus of DHFR for immunoprecipitation with an anti-ha antibody. (E) A plot generated by the PONDR algorithm shows the distribution of folded and unfolded regions for the N-terminal 80 amino acids of Rpn4.
To test if the Ub-independent degron of Rpn4 is able to destabilize heterologous proteins, we attached Rpn41–80 to the N-terminus of dihydrofolate reductase (DHFR). In vitro degradation assay showed that the Rpn41–80-DHFR fusion protein was degraded by the 26S but not 20S proteasome (Fig. 2C). The ability of Rpn41–80 to destabilize DHFR was also revealed in vivo by a pulse-chase assay (Fig. 2D, lanes 10–12). Using truncation-functional analysis, we determined that the N-terminal 10 or 30 residues of Rpn4 were not sufficient to mediate the degradation of DHFR (Fig. 2D, lanes 1–6). The N-terminal 50 residues destabilized DHFR, but to a less extent compared to Rpn41–80 (compare lanes 7–9 to 10–12). We conclude that the N-terminal 80 residues of Rpn4 constitute a transplantable Ub-independent 19S RP-dependent degron.
It has been suggested that Ub-independent substrates carry intrinsically disordered domains, which play a critical role in their degradation [13]. Since no structural data of Rpn4 is available, we inspected the secondary structure of the Ub-independent degron of Rpn4 using the PONDR VL-XT program [20]. As shown in Fig. 2E, the N-terminal 80 amino acids of Rpn4 apparently consist of a disordered N-terminal segment of 15–20 residues, followed by a folded domain.
3.3. Identification of 19S RP subunits interacting with the Ub-independent degron of Rpn4
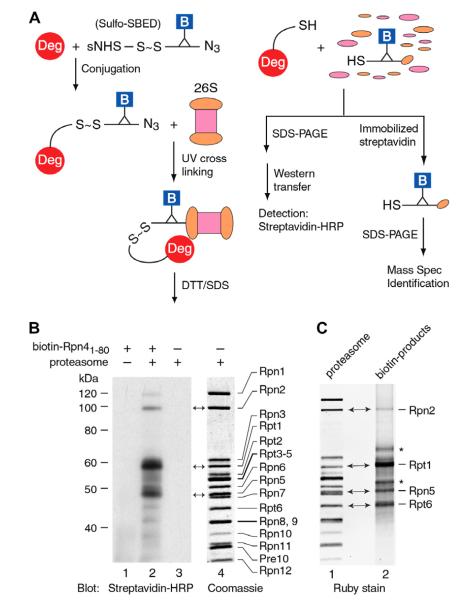
The requirement of 19S RP for Ub-independent degradation of Rpn4 suggests that one or more 19S RP subunits may serve as a receptor for the Ub-independent degron of Rpn4. To determine the relevant 19S RP subunit(s), we initially applied glutathione S-transferase (GST) pulldown and yeast two-hybrid assays by fusing the 19S RP subunits to GST or GAL4 DNA binding domain. Somewhat surprisingly, none of the GST fusion proteins was able to retain Rpn41–80 in the pulldown assay. Similarly, no interaction between the 19S RP subunits (bait) and Rpn41–80 (prey) was detected in the yeast two-hybrid assay. We suspected that the interaction between the Rpn4 Ub-independent degron and the 19S RP might be dynamic and transient. Alternatively, free 19S RP subunits might possess different conformation from their counterparts in the assembled 26S proteasome and therefore lose the ability to recognize the Ub-independent degron of Rpn4. To overcome these potential problems, we decided to use a photo-crosslinking-label transfer method to capture the 19S RP subunit(s) that bind Rpn41–80 in the context of 26S proteasome. A flowchart of this method is outlined in Fig. 3A. Sulfo-SBED is a photo-crosslinking reagent composed of three functional groups: an amine-reactive sulfo-NHS-ester on one arm, a photoactivatable arylazide on the other side, and a biotin on the handle. The sulfo-NHS-ester arm contains a cleavable disulfide bond, which allows the transfer of the biotin label from a bait protein to its prey. In our experiments, Sulfo-SBED was first conjugated to Rpn41–80 (“Deg”) through its amine group. Free crosslinker was inactivated and removed by ultrafiltration. Purified Sulfo-SBED-Rpn41–80 conjugate was then incubated with 26S proteasome. The mix was subsequently exposed to ultraviolet (UV) light, resulting in covalent linkage between the photo-reactive arylazide and the proteasome subunits in close spatial proximity to Rpn41–80. The crosslinked Rpn41–80-proteasome complex was treated with DTT and SDS to release Rpn41–80 through the cleavage of the disulfide bond and to disassemble the proteasome complex. The proteasome subunits carrying the biotin label were determined by western blot analysis with horseradish peroxidase (HRP)-conjugated streptavidin or mass spectrometry following their isolation over immobilized streptavidin beads. In a typical western blot analysis (Fig. 3B), we resolved the UV crosslinked samples by SDS–PAGE on a 9% gel, which was able to separate most of the 19S RP subunits (lane 4). The samples from three UV crosslinking reactions, containing both biotin-conjugated Rpn41–80 and 26S proteasome (lane 2), biotin-Rpn41–80 alone (lane 1), or 26S proteasome alone (lanes 3, 4), were analyzed. The latter two reactions were used as controls. After electrophoresis and transfer, the membrane was cut into two portions: one was subjected to detection with streptavidin-HRP (lanes 1–3), whereas the other was stained with Coomassie Blue to serve as a reference of the proteasome subunits (lane 4). We repeatedly detected three major biotin-labeled bands corresponding to the positions of Rpn2, Rpt1 and Rpn5 (compare lanes 2 and 4).
Fig. 3.
Identification of 19S RP subunits binding the Ub-independent degron of Rpn4. (A) A schematic of the photo-crosslinking-label transfer method. “Deg” stands for the Rpn41–80 degron. (B) Western blot analysis to identify biotinylated proteasome subunits. The assignment of the 19S RP subunits (lane 4) was referred to early publications[24,25]. (C) SYPRO Ruby staining to show the proteins isolated over streptavidin beads. The bands aligned with 19S RP subunits were cut out from the gel and subjected to LC–MS/MS. The bands marked with asterisks were not aligned with any 19S RP subunits and were not characterized further.
In parallel with the western blot analysis, we isolated biotin-labeled products through streptavidin beads. The isolates were then resolved by SDS–PAGE and visualized by SYPRO Ruby stain (Fig. 3C). Four bands were aligned with the positions of Rpn2, Rpt1, Rpn5 and Rpt6, respectively. Their identities were confirmed by LC–MS/MS (liquid chromatography coupled with tandem mass spectrometry) (Supplemental Fig. 3). It is unclear why Rpt6 was recovered from streptavidin pulldown but was not detected by streptavidin-HRP in the western blot (Fig. 3B and C). One possibility is that Rpt6 might nonspecifically bind to the streptavidin beads. We consider this possibility less likely because we did not detect the binding of free 19S RP subunits resulting from the disassembly of 26S proteasome to the streptavidin beads. It is possible that Rpt6 itself was not biotinylated, but was co-eluted with a biotin-labeled 19S RP subunit during the pulldown with streptavidin beads. Further experiments are required to address this problem. Nonetheless, based on the results of western blot analysis and LC–MS/MS, we conclude that Rpn2, Rpt1 and Rpn5 are the receptors for the Ub-independent degron of Rpn4.
4. Discussion
In this study we demonstrated that Rpn4 is degraded by the 26S proteasome in a Ub-independent manner. The Ub-independent degron of Rpn4 is located in an N-terminal region including the first 80 residues. Inspection of the amino acid sequence of the Rpn4 Ub-independent degron reveals that it likely consists of a disordered N-terminal segment of 15–20 residues, followed by a folded domain. Our previous work has shown that deletion of the N-terminal 10 amino acids of Rpn4 impairs its Ub-independent degradation [8,19]. Here we demonstrate that the first 30 residues of Rpn4 are insufficient to mediate the degradation of DHFR (Fig. 2D). These results suggest that the N-terminal unstructured segment and the following folded domain are two essential elements of the Rpn4 Ub-independent degron. Interestingly, similar to the Ub-independent degron of Rpn4, the Ub-independent degron of hTS is comprised of an intrinsically unstructured region including the first 27 amino acids and an α-helix spanning residues 31–42 [4]. Dohmen and coworkers recently reported that the N-terminal unstructured domain of yeast ODC (yODC) was able to destabilize Ura3 but not DHFR or the E. coli maltose binding protein (MBP) when it was fused to their termini [2]. Analysis of the crystal structures of these three reporter proteins reveals that the N-terminus of Ura3 leads into an α-helix, whereas the termini of DHFR and MBP lead into β strands. It appears that different Ub-independent degrons have a structural feature in common: an intrinsically unstructured region followed by an α-helical (folded) domain. This structural feature may be shared by Ub-dependent degrons as well. Lee et al. fused a Ub-dependent degron to the N-termini of various circular permutated DHFR proteins and found that the degradation of these model substrates was markedly influenced by the structural context in the vicinity of the Ub-dependent degron [21]. The substrates were rapidly degraded when the Ub-dependent degron was followed by α-helical domains. In contrast, the degradation was impaired when the Ub-dependent degron was followed β strands.
Although the number of Ub-independent substrates continues to grow and the structural features of Ub-independent degrons are gradually unveiled, little is known about the proteasome subunits that recognize Ub-independent degrons. Unlike many Ub-independent substrates, which are degraded by the 20S proteasome through a “default” mechanism [13], the Ub-independent degradation of Rpn4 is mediated by the 26S but not 20S proteasome. This observation suggests that the 19S RP is responsible for recruiting Rpn4 to the proteasome. In support of this hypothesis, we identified three 19S RP subunits including two base subunits (Rpt1 and Rpn2) and one lid subunit (Rpn5) that interact with the Ub-independent degron of Rpn4. It is of interest to note that Rpt1 but not other Rpt subunits recognizes the Rpn4 Ub-independent degron. This suggests that the six subunits of the Rpt ring, all essential for proteasomal degradation, play distinct roles in substrate engagement, i.e. substrate binding, unfolding and translocation. Our work also implies that different subunits of the lid subcomplex may be involved in different steps of Ub-independent degradation of Rpn4. While multiple lid subunits are essential for Rpn4 degradation (Fig. 1), Rpn5 appears to be the only one that binds the Rpn4 Ub-independent degron with substantial affinity.
yODC is another Ub-independent substrate whose degradation depends on the 19S RP. However, different from Rpn4 whose degradation requires both subcomplexes of the 19S RP, the degradation of yODC does not need the lid subcomplex [2]. This “discrepancy” suggests that some Ub-independent substrates may be directly recruited by the base subcomplex, while others may be delivered to the proteasome through cooperation between the base and the lid subcomplexes. In fact, the Ub-independent degron of Rpn4 is bound by both base and lid subunits. It is also possible that the lid subcomplex participates in processing certain substrates after their initial recruitment and before their translocation into the 20S core. Regardless of the exact mechanism, the finding that the lid subcomplex is required for the Ub-independent degradation of Rpn4 has an important implication. Up until now, the only biochemical function clearly assigned to the lid subcomplex is the deubiquitylation of ubiquitylated substrates by its Rpn11 subunit [22,23]. Our results argue that the lid subcomplex may play a broader role in the degradation of ubiquitylated substrates.
To our best knowledge, this is the first time that specific 19S RP subunits have been identified binding a Ub-independent degron. Further elucidation of the molecular details of the interactions between 19S RP subunits and the Rpn4 Ub-independent degron should shed more light on how the 26S proteasome recruits Ub-independent substrates. It will be also of great interest to investigate whether other Ub-independent degrons are recognized by the same or different 19S RP subunits.
Supplementary Material
Acknowledgments
We thank Yasushi Saeki and Jürgen Dohmen for yeast strains, Paul Stemmer and Joe Caruso for mass spectrometric analysis. This work was supported by a National Science Foundation grant MCB-0816974 and a fund from the Office of the Vice President for Research at Wayne State University (Y.X.).
Footnotes
Appendix A. Supplementary data Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bbrc.2012.01.152.
References
- [1].Li X, Coffino P. Regulated degradation of ornithine decarboxylase requires interaction with the polyamine-inducible protein antizyme. Mol. Cell. Biol. 1992;12:3556–3562. doi: 10.1128/mcb.12.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gödderz D, Schäfer E, Palanimurugan R, Dohmen RJ. The N-terminal unstructured domain of yeast ODC functions as a transplantable and replaceable ubiquitin-independent degron. J. Mol. Biol. 2010;407:354–367. doi: 10.1016/j.jmb.2011.01.051. [DOI] [PubMed] [Google Scholar]
- [3].Jariel-Encontre I, Bossis G, Piechaczyk M. Ubiquitin-independent degradation of proteins by the proteasome. Biochim. Biophys. Acta. 2008;1786:153–177. doi: 10.1016/j.bbcan.2008.05.004. [DOI] [PubMed] [Google Scholar]
- [4].Melo SP, Yoshida A, Berger FG. Functional dissection of the N-terminal degron of human thymidylate synthase. Biochem. J. 2010;432:217–226. doi: 10.1042/BJ20101027. [DOI] [PubMed] [Google Scholar]
- [5].Su V, Nakagawa R, Koval M, Lau AF. Ubiquitin-independent proteasomal degradation of ER-localized connexin43 mediated by CIP75. J Biol. Chem. 2010;285:40979–40990. doi: 10.1074/jbc.M110.170753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wiggins CM, Tsvetkov P, Johnson M, Joyce CL, Lamb CA, Bryant NJ. BIMEL, an intrinsically disordered protein, is degraded by 20S proteasomes in the absence of poly-ubiquitylation. J. Cell Sci. 2011;124:969–977. doi: 10.1242/jcs.058438. [DOI] [PubMed] [Google Scholar]
- [7].Li X, Lonard DM, Jung SY, Malovannaya A, Feng Q, Qin J. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by REGgamma proteasome. Cell. 2006;124:381–392. doi: 10.1016/j.cell.2005.11.037. [DOI] [PubMed] [Google Scholar]
- [8].Ju D, Xie Y. Proteasomal degradation of RPN4 via two distinct mechanisms: Ubiquitin-dependent and -independent. J. Biol. Chem. 2004;279:23851–23854. doi: 10.1074/jbc.C400111200. [DOI] [PubMed] [Google Scholar]
- [9].Moriishi K, Okabayashi T, Nakai K, Moriya K, Koike K, Murata S, et al. Proteasome activator PA28gamma-dependent nuclear retention and degradation of hepatitis C virus core protein. J. Virol. 2003;77:10237–10249. doi: 10.1128/JVI.77.19.10237-10249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yuksek K, Chen WL, Chien D, Ou JH. Ubiquitin-independent degradation of hepatitis C virus F protein. J. Virol. 2009;83:612–621. doi: 10.1128/JVI.00832-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Orlowski M, Wilk S. Ubiquitin-independent proteolytic functions of the proteasome. Arch. Biochem. Biophys. 2003;415:1–5. doi: 10.1016/s0003-9861(03)00197-8. [DOI] [PubMed] [Google Scholar]
- [12].Baugh JM, Viktorova EG, Pilipenko EV. Proteasomes can degraded a significant proportion of cellular proteins independent of ubiquitination. J. Mol. Biol. 2009;386:814–827. doi: 10.1016/j.jmb.2008.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Asher G, Reuven N, Shaul Y. 20S proteasomes and protein degradation “by default”. Bioessays. 2006;28:844–849. doi: 10.1002/bies.20447. [DOI] [PubMed] [Google Scholar]
- [14].Stadtmueller BM, Hill CP. Proteasome activators. Mol. Cell. 2011;41:8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- [17].Mannhaupt G, Schnall R, Karpov V, Vetter I, Feldmann H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999;450:27–34. doi: 10.1016/s0014-5793(99)00467-6. [DOI] [PubMed] [Google Scholar]
- [18].Xie Y, Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: A negative feedback circuit. Proc. Natl. Acad. Sci. USA. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ju D, Xie Y. Identification of the preferential ubiquitination site and ubiquitin-dependent degradation signal of Rpn4. J. Biol. Chem. 2006;281:10657–10662. doi: 10.1074/jbc.M513790200. [DOI] [PubMed] [Google Scholar]
- [20].Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting protein disorder for N–C–, and internal regions. Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- [21].Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- [22].Verma R, Aravind L, Oania R, McDonald WH, Yates JR, Koonin EV, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- [23].Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- [24].Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Saeki Y, Isono E, Toh-e A. Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods Enzymol. 2005;399:215–227. doi: 10.1016/S0076-6879(05)99014-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.