Abstract
Enzymatic deamination of bases in DNA or RNA leads to an alteration of flow of genetic information. Adenine deaminases edit RNA (ADARs, TADs). Specialized cytidine deaminases are involved in RNA/DNA editing in lipid metabolism (APOBEC1) and in innate (APOBEC3 family) and humoral (AID) immunity. APOBEC2 is required for proper muscle development and, along with AID, was implicated in demethylation of DNA. The functions of APOBEC4, APOBEC5, and other deaminases recently discovered by bioinformatics approaches are unknown. What is the basis for the diverse biological functions of enzymes with similar enzyme structure and the same principal enzymatic reaction? AID, APOBEC1, lamprey CDA1, and APOBEC3G enzymes cause uracil DNA glycosylase-dependent induction of mutations when overproduced ectopically in bacteria or yeast. APOBEC2, on the contrary, is nonmutagenic. We studied the effects of the expression of various deaminases in yeast and bacteria. The mutagenic specificities of four deaminases, hAID, rAPOBEC1, hAPOBEC3G, and lamprey CDA1, are strikingly different. This suggests the existence of an intrinsic component of deaminase targeting. The expression of yeast CDD1 and TAD2/TAD3, human APOBEC4, Xanthomonas oryzae APOBEC5, and deaminase encoded by Micromonas sp. gene MICPUN_56782 was nonmutagenic. A lack of a mutagenic effect for Cdd1 is expected because the enzyme functions in the salvage of pyrimidine nucleotides, and it is evolutionarily distant from RNA/DNA editing enzymes. The reason for inactivity of deaminases grouped with APOBEC2 is not obvious from their structures. This can not be explained by protein insolubility and peculiarities of cellular distribution and requires further investigation.
Keywords: editing deaminases, mutagenesis, immunity, DNA repair
Enzymatic deamination of nucleic acid bases is remarkably widely and ingeniously exploited in biological systems. Cytosine to uracil deamination by cytidine deaminase (CDA) was first characterized as an important reaction in the pathways of thymidine biosynthesis and pyrimidine salvage [1]. Enzymatic deaminations of cytosine and adenosine play important roles in the regulation of nucleotide pools [2]. In humans, polymorphisms in the gene encoding for CDA lead to an altered response to cytidine analog anticancer agents [3], and a defect in adenosine deaminase leads to severe combined immunodeficiency (reviewed in [4]).
Deamination on the level of polynucleotides is even more important. Editing of DNA or RNA leads to mutations or modulates the expression of genetic information. Adenine to hypoxanthine deamination in mRNA and tRNA can articulate splicing and translation and therefore plays a critical role for the proper expression of genetic information [5]. Cytosine to uracil deamination is used for RNA editing of transcripts and for tightly regulated DNA editing in antibody diversity, protection from retroviruses, and demethylation in development [6–10].
Cytidine deaminases, CDA and CDD, are zinc-dependent evolutionarily conserved enzymes. The activation of water that attacks the C4 position of cytosine is proceeded by a zinc ion coordinated by one histidine and two or three cysteines in the enzyme active site located at the borders of α-helixes 2 and 3 (Fig. 1). The same enzymatic mechanism is utilized by RNA/DNA editing enzymes [8]. APOBEC1 is the founder of the AID/APOBEC superfamily of editing enzymes. It is a catalytic subunit of a larger editase complex that deaminates cytosine at position 6666 in the apolipoprotein B mRNA [11]. The modification creates a premature stop codon that leads to tissue-specific production of a truncated apolipoprotein. The ability to edit RNA is related to the acquisition of additional extended α-helixes at the C-terminus of APOBEC1 marked black and gray in Fig. 1. The Apobec1 knockout mice are viable, but they have abnormal lipid metabolism [12].
Fig. 1.
Similarity of the general plan of structure of Cdd1 and editing deaminases. Solved or predicted helices indicated by cylinders, strands by arrows; helix-4 distinctive for nucleic acid editing deaminases shown in black; helices conserved among TadA/Tad2p-like and AID/APOBEC clade shown in gray [29, 32]. Amino acid sequences shown for catalytic HxE and PC motifs and the conserved motif in helix-4. The secondary structure of MICPUN_56782 was predicted in the present study with the Phyre program (Protein Homology/analogY Recognition Engine; http://www.sbg.bio.ic.ac.uk/~phyre/). The prediction of new second helix shown in pale color and countered by a broken line is on the border of significance. Different structures are drawn approximately to scale and aligned around two central cysteine motif (PC(x)iC) coordinating zinc in the active site.
Structurally similar enzyme AID (Fig. 1) is involved in immunity. The immune system uses gene modification mechanisms to generate various types of high affinity antibodies by class switch recombination (CSR), immunoglobulin gene conversion (GC), and somatic hypermutation (SHM) [13, 14]. SHM leads to the accumulation of mostly point mutations in the V genes. The frequency of mutations here is up to six orders of magnitude higher than in other genomic locations [13]. Most of the mutations are base pai r substitutions, occurring with the same frequency at G-C base pairs and at A-T base pairs. Statistically preferred hotspots are DGYW motifs (D = G, T, or A; Y = pyrimidine base; W = A or T) [15] for mutations at G-C pairs and WA motifs [16] for mutations at A-T pairs (mutating bases are underlined).
CSR, SHM, and GC depend on AID [17–23]. Patients with defective AID have elevated levels of only one type of low-affinity antibodies, IgM. They suffer from recurrent bacterial infections. AID works in affinity maturation via deamination of cytosine in DNA of of variable region of immunoglobulin genes. When produced in E. coli, AID is mutagenic, and the effect is much greater when the gene for uracil DNA glycosylase (ung) is inactivated [24]. Uracil generated by AID in B-cells triggers downstream events leading to genetic instability [24–27]. Amazingly, the specificity of deaminations by AID in vitro coincides with hotspots of SHM [28]. Replication of the U-G pair generates only the transition mutations. Uracil removal by uracil DNA glycosylase leads to an abasic site (AP), which, when bypassed by a specialized DNA polymerase, will lead to transitions and transversions. An AP site may also be incised by AP endonuclease and then repaired by short patch base excision repair (BER) with the involvement of error-prone DNA polymerases with the generation of all types of base substitutions. DNA breaks, which are intermediates of these reactions, cause an induction of recombination (reviewed in [29]).
In jawless vertebrates, the role of immunoglobulins is played by variable lymphocyte receptors (VLR) [30, 31]. In mature lymphocytes, the VLR genes contain a central variable region consisting of various leucine-rich repeats (LRR). It was suggested that mature VLR genes result from a combination of various LRR modules flanking embryonic VLR genes [31]. It is likely that consecutive LRR assembly producing VLR occurs by a mechanism similar to class switch recombination in jawed vertebrates. There are two AID/APOBEC genes, PmCDA1 and PmCDA2, in the lamprey genome. Both genes are specifically expressed in lymphocytes and hematopoietic tissues, suggesting the involvement of the corresponding proteins in the immune response [32]. By analogy with CSR, cytosine deamination in DNA by these enzymes can cause DNA double-strand breaks, whose repair leads to LRR insertion in the maturing VLR gene. Experiments with unicellular organisms are consistent with this hypothesis. As in the case of AID, the PmCDA1 production in E. coli or in yeast caused uracil DNA glycosylase-suppressible C to T transitions. PmCDA1 production induced intragenic recombination in yeast for which the activity of Ung was required. These results supported the hypothesis of the recombinational mechanism of VLR formation [32].
APOBEC3-group cytidine deaminases protect cells from retroviruses, including HIV-1 [33, 34]. All retroviruses have similar architecture and are comprised of two copies of single -stranded RNA genomes packed in virions with reverse transcriptase. APOBEC3G helps to fight HIV by deamination. APOBEC3G is a strong mutator when expressed in E. coli [35]. The viruses without Vif (Virus Infectivity Factor) experience hypermutation, and all these mutations were transitions that could be explained by deamination of a minus DNA strand of the virus [36, 37]. This deamination can lead to hypermutagenesis or the destruction of the viral genome during repair of uracil [38]. The detection of multiple mutations in surviving viruses is consistent with the ability of APOBEC3G to deaminate DNA in vitro in processive fashion [39, 40].
The human APOBEC3 is located on chromosome 22 in a cluster of seven homologous genes (APOBEC3A-APOBEC3H). The products of four of these genes (APOBEC3B, G, F, C) suppress retroviruses and endogenous L1 retroelements and differ in hotspot motifs of hypermutagenesis [7, 41]. APOBEC3A suppresses retrotransposons; the functions of the 3DE and 3H genes are less clear [42]. It is possible that the different APOBEC3 family members can neutralize specific lentiviruses or other viruses [43]. From the evolutionary viewpoint, it is interesting that, in contrast to humans and chimpanzees, rodents have only one APOBEC3 gene.
Mutagenic specificity of APOBEC3G is very different from AID [39, 44, 45]. Domain swapping between AID and APOBEC3G helped to approach the molecular mechanisms for this specificity [46]; however, the precise answer awaits the solution of crystal structures of the ternary complexes of the enzymes with DNA. So far, only parts of the inactive APOBEC2 and the active domain of APOBEC3G have been solved [47–50].
Although very structurally similar, APOBEC2 is nonetheless nonmutagenic in bacteria [35] and cannot perform deamination in vitro. Knockout mice develop myopathy with anomalous muscles [51] and a lack of APOBEC2-related proteins leads to the development of a dystrophic muscle phenotype in zebrafish embryos [52]. APOBEC2 from zebrafish was also implicated in DNA demethylation during development [53].
There is one report on mutagenic effects of adenosine deaminase [54]. It has been shown that TbADA2 from trypanosoma has relaxed specificity and can deaminate “A” in RNA and “C” in ssDNA, and the expression of this adenosine deaminase was mutagenic in ung− E. coli. This observation is consistent with the similarity of cytidine and adenosine deaminases [55].
The structures of CDA from bacteria, yeast, and humans reveal a core α1β1, β2α2, β3α3, β4,} β5 arrangement [32, 56, 57] (Fig. 1). All five β-strands form a sheet that supports the parallel positioning of two α-helices that contain the His, Cys, and Glu residues that are required for zinc-coordination, proton transfer, and catalysis [57]. The crystal structures of several TadA proteins [58, 59], APOBEC2 [49], as well as the crystal and NMR structures of the APOBEC3G catalytic domain [47, 60], established secondary structures of the proteins. In all other representative families the secondary structure is predicted. A sequence–structure analysis of the representatives of the deaminase superfamily reveals unique features shared among the AID/APOBEC proteins, six α-helices and five β-strands, the characteristic deaminase “HxE-PCxxC” (where “x” is any amino acid) zinc coordination motif (shown on α-helixes 2 and 3 represented by white cylinders in Fig. 1), including a conserved tryptophan in the C-terminal helix and a three amino acid insert the N-terminal to the PCxxC motif [32, 61]. The presence of three additional α-helices (the fourth, shown in black, is characteristic for editing deaminases and called “structural signature of editing deaminases” [49], the fifth and sixth, shown in gray, are conserved between Tad and APOBEC/AID) distinguish the vertebrate editing deaminases from other deaminases, such as ADAR and Cdd1-like cytidine deaminases [62]. The closest relatives of the AID/APOBEC family were identified among TadA/Tad2p-like tRNA adenosine deaminases; all share two distinct helices and several critical residues. The TadA/Tad2p-like deaminases and AID/APOBEC proteins thus appear to interact with nucleic acid substrates via a common mechanism. The crystal structure of APOBEC2 provided novel information on the unusual arrangement of subunits in deaminase dimer, which differs substantially from that previously described for deaminases operating on monomeric bases [63]. Structures of APOBEC3G catalytic domains show a substantial overlap with corresponding parts of inactive APOBEC2 and TadA structures. An interesting feature of APOBEC3G is that the protein is self-duplicated and has two cytidine deaminase active sites, inactive N-terminal, and active C-terminal. This is observed despite the conservation of all amino acids important for catalysis between these two domains and may result in a special structure of an active antiviral protein [64].
A search for protein sequence homologs of human AID revealed a new group of deaminases in vertebrates called APOBEC4 [61]. A multiple alignment of all related sequences revealed a new subfamily with strong conservation of the deaminase zinc-coordinating motif (H/C)xE…PCx2–6C [65, 66]. The presence of six amino acids (instead of two) in the PCx2–6 stretch is specific for APOBEC4 is (Fig. 1). APOBEC4 has an α-helix forming insertion characteristic to RNA/DNA editing enzymes that is absent in CDAs. As judged by a maximum likelihood phylogenetic tree, the APOBEC4 family is close to APOBEC1 and AID [61]. The APOBEC4 is expressed primarily in the testis.
Further screening for novel members of the AID/APOBEC family in the GenBank database led to the discovery of a new family member, hereafter referred to as APOBEC5 [29] (Fig. 1). The gene for APOBEC5 (XOO2897, GI: 64624554) was found in the bacterium Xanthomonas oryzae. This bacterium causes blight in rice, one of the major diseases of this important agricultural plant. Sequence alignment of APOBEC5 with other deaminases showed that APOBEC5 has two features that are characteristic only for the AID/APOBEC family: helix-4 (in black in Fig. 1) and an insertion of three amino acids upstream from the PCxxC motif. The presence of highly conserved motifs HxE and PCxxC, which are characteristic for various deaminases, indicates that APOBEC5 may be a functional deaminase. The C-terminus of this protein is shorter than those of the known AID/APOBEC family members (Fig. 1).
Phylogenetic analysis with the use of various methods (for their description, see [32]) consistently classified APOBEC5 as a distant AID/APOBEC family member. The absence of other members of the APOBEC5 subfamily may indicate a relatively recent transfer of this gene to X. oryzae from the vertebrate genome. The AID/APOBEC family members of lamprey were not assigned to any of the known deaminase subfamilies of jawed vertebrates, forming a separate subfamily. The divergence between subfamilies APOBEC1, APOBEC4, APOBEC2, AID, and APOBEC3 have probably occurred after the splitting of jawed and jawless vertebrates. The family of RNA-editing enzymes Tad2p/TadA is now considered to be the closest to the AID/APOBEC family [32]. These proteins have a number of traits characteristic of AID/APOBEC, including helixes 4–6 (Fig. 1). It is proposed that the ancestral AID/APOBEC emerged prior to the vertebrate radiation, most likely descending from the Tad2p lineage [32]. Tad proteins that convert adenine to inosine in tRNA in trypanosome are able to deaminate cytosine to uracil in single-strand DNA [54], thus supporting the relationship between Tads and APOBECs.
The wide distribution of DNA/RNA-editing deaminases superfamily suggests that they are involved in the ancient mechanism of regulating phenotypic and genetic variation that appeared at the dawn of the vertebrate radiation. Plant chloroplasts possess tRNA editing adenine deaminase [67]. It still has to be determined what additional editing deaminases are present in plants [68].
In the present study, in order to get a better understanding of the mechanisms of deaminases activities and their functions and evolution, we systematically analyzed the mutagenic activity and specificity of known and predicted members of AID/APOBEC superfamily.
MATERIALS AND METHODS
Bacterial strains
We converted E. coli strains MC1061 [69] and its nfi::cam derivative, NR13509 (kindly constructed for us by Dr. R. M. Schaaper, NIEHS), to λDE3 lysogene using λDE3 lyzogenization kit from Novagen (USA) (Cat. No. 69734-3). Mutation ung152::Tn10 was transferred into the strain Rosetta(DE3) pLysS (Novagen, Cat. No. 70956) from the strain BD2328 [70]. We used XL Blue (Stratagene, USA) and TOP10 (Invitrogen, USA) strains for routine cloning and DNA manipulations.
Yeast strains
For mutagenesis experiments we used 1B-D770 (MATa ura3-4 leu2-3,112 trp1-289 his7-2 ade5-1 lys2-Tn5-13) [71] and its ung1::kanMX deletion derivative generated by one step gene disruption with PCR product on hygB template using the following oligonucleotides:
UNG1:KAN_DISL, 5′-CAAACTACGATCGAAGACTTCTTTGGTACAAAGAAAAGCACTAATG cgTAcgcTgcAggTcgAc | CGTACGCTGCAGGTCGAC};
UNG1:KAN_DISR, 5′-CGTTCCAGGAACAACACTCCAGTCTATCATTTTCTCTCCGCGGGTAAT cgATgAATTcgAgcTcg | CGATGAATTCGAGCTCG}.
These strains allow concomitant measurement of mutation rates at several loci. These include: i) the forward mutation rate at the CAN1 locus, where mutations reflect a variety of substitution, frameshift, and more complex events; ii) the rate of reversion of nonsense mutations: the trp1-289 (TAG) [72] and ade5-1 (TAA) [73], where mutations reflect base substitutions in the nonsense codon as well as in suppressor genes encoding tRNAs; and iii) reversion of the his7-2 mutant allele that occurs mainly via +1 frameshifts in a homopolymeric A·T run [74]. To test if the activity of APOBEC4 is prevented with unsuspected glycosylase, we examined its activity in FF18733 (MATa leu2-3,112 trp1-289 his7-2 ura3-52 lys1-1) and its derivative with the deletion of five yeast glycosylases DGD39 (same but ung1-Δntg1-Δntg2::kanMX6 ogg1::URA3 mag1::hphMX4) [75] kindly provided by Dr. E. Sage, Institute Curie, Paris, France. We also tested the effect of apurinic endonucleases with strain E134 (MATα ade5-1 lys2::InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52 and its isogenic apn1::kanMX apn2::h3G variant.
For the experiments with plasmids carrying the HIS3 selectable marker we used BY4742 (MATα his-Δ1 leu2-Δlys2-Δura3-Δ) (Invitrogen) and BY4742 ung1 (MATα his-Δ1 leu2-Δlys2- Δura3-Δung1::kanMX) (Invitrogen).
Media
Standard LB media was used for E. coli and YEPD and SC media for yeast Saccharomyces cerevisiae cultivation. For bacterial mutagenesis experiments, rifampicin was added to the LB plates to the final concentration of 100 µg/ml. Ampicillin and kanamycin were added in standard concentrations to the media for plasmid selection. For the induction of expression from the GAL promoter, glucose was substituted for galactose in the yeast media.
Plasmids
We created new hAIDSc and hAPOBEC3GSc genes with the DNA sequence characteristic of highly expressed yeast genes. The construct for hAID was described previously [76]. We used yeast codon usage data [77] with revision by [78] to construct a DNA sequences with the preferable yeast codons. The DNA corresponding to this sequence and encoding for the c-myc tag at the C-terminus (hAIDSc) and at the N-terminus (hAPOBEC3GSc) was custom-synthesized and cloned into BamHI and SalI cut | The DNA encoding for hAPOBEC3G with c-myc tag at the N-terminus (hAPOBEC3GSc) was custom-synthesized and cloned into Xho I and Nhe I sites of } pESC-LEU (Stratagene, USA) expression vector by the McLab Company (USA). In this construct, the deaminases genes were put under the control of the strong, galactose-inducible GAL1 promoter. We made a vector for galactose-inducible expression of native hAPOBEC4 in yeast using human cDNA as a template for PCR (We made a vector for galactose_inducible expression of native hAPOBEC4 in yeast using human cDNA as a template for PCR (Human Kidney cDNA library, Stratagene #937250). Because hAPOBEC4 possess a long C-terminal extension, we made a truncated version. We cloned the PCR products derived from the two sets of primers into BglII-SacI sites of pESC-LEU. They direct the production of the full-length protein 367 amino acids long and the truncated protein with amino acids 1–195.
For expression in E. coli, the APOBEC4 gene was inserted into pETBlue2 vector via NcoI-NotI sites (full-length) and into pET24b vector via BamHI-XhoI sites (truncation of two C-terminal amino acids). APOBEC4 tagged by six histidines on the C-terminus is produced from this construct. We constructed a vector for constitutive production of a fusion protein of APOBEC4 and GFP. We used the plasmid pTSK241 [79] encoding for yeast nuclear protein Ace1 fused to triple GFP. We substituted the ACE1 with hAPOBEC4 open reading frame (ORF) cloned into the BamHI-NotI sites, resulting in a pTSK241-APOBEC4 construct.
The pTSK241-GAL-hAIDSc plasmid, encoding for hAIDSc-GFP fusion under the control of a galactose-inducible GAL1 promoter, was constructed in two steps. First, the CAP promoter was substituted for the GAL1 promoter in the pTSK241 plasmid using XhoI and BamHI sites. Second, ACE1 ORF was substituted for hAIDSc ORF using BamHI and NotI sites.
We cloned S. cerevisiae TAD2 ORF and cDNA of TAD3 into pCOLA-Duet-1 (Novagen) bacterial expression vector. The TAD2 gene was amplified by PCR using yeast genomic DNA (strain GT109) [80] as a template and cloned using BamHI and SacI sites. We used RT-PCR to obtain cDNA of TAD3, because TAD3 is one of the rare yeast genes that contain introns. The TAD3 coding sequence was cloned using BglII and KpnI sites. The resulting construct allows for an IPTG-inducible production of Tad2/Tad3 dimer, with Tad2 possessing N-terminal His6-tag and Tad3 possessing C-terminal S-tag. Interestingly, the coding sequences of both TAD2 and TAD3 genes obtained from the GT109 strain differ from corresponding GenBank entries. There were four synonymous (T150C, T273C, A579G, and T594C) and three non-synonymous substitutions (A296G, G608A, A722G, leading to the amino acid changes D99G, R203K, and K241R, respectively) in the TAD2 ORF obtained from the GT109 strain. The TAD3 coding sequence from the same strain contained five synonymous (C246T, T264C, T291C, C630T, T690C) and four non-synonymous (G196A, G229A, G314A and T315C, T351G, leading to V66I, D77N, S105N, and D118E amino acid changes, respectively) substitutions. These substitutions are not due to cloning errors because independent clones of plasmid constructs contain them.
The APOBEC5 gene was PCR-amplified from the X. oryzae genomic DNA (ATCC) and cloned into BamHI and XhoI sites of the pET24b bacterial expression vector. APOBEC5 that is produced from this construct has a C-terminal His6 tag.
The sequence of the Micromonas sp. gene MICPUN_56782 was optimized for E. coli and yeast expression, custom-synthesized, and cloned into pUC57 vector by the GenScript company. We inserted this gene into the pET24b vector using BamHI and HindIII sites introduced into pUC57-based construct. The MICPUN_56782 protein is also produced from this construct as a fusion with the His6-tag on the C-terminus.
Plasmids pGD309 (pYES2.0 expression vector with rAPOBEC1), pGD307 (same but with a sequence encoding for the NLS signal at N-terminus of rAPOBEC1), and pGD313 (pYES2.0 with yeast CDD1) [81, 82] were kindly sent by Dr. H. Smith of the University of Rochester. All proteins are produced with His6-HA tags at the N-terminus. YES2.0 | pYES2.0} (Invitrogen) was used as a control. Plasmid pET24b-PmCDA1 was described earlier [32]. Plasmid pET30-His6-APOBEC3G was kindly provided by Dr. S. Petersen-Mahrt (Clare Hall Laboratories, UK Cancer Research, England).
Nucleotide sequences of all primers used for constructions are available upon request.
Protein production in E. coli and yeast
Escherichia coli cultures transformed with respective plasmids were grown in liquid LB media containing appropriate antibiotics. Protein expression was induced according to the manufacturer’s recommendations (pET system manual from EMD, formerly Novagen). Cells were lysed by sonication or using EmulsiFlex, or with BugBuster reagent (Novagen). Lysates were cleared by 10,000g at 4°C, and supernatant and pellet fractions were mixed with LDS-sample buffer, boiled for 5 min at 100°C, and loaded on SDS-polyacrylamide gel. Gels were stained with Coomassie G-250.
Protein production in yeast was verified by Western blot as described earlier [76]. The Western Breeze Kit (Invitrogen) was used for detection of the protein in yeast extracts.
In vitro deamination assay
Deamination reactions were based on published protocol [83] and contained 25 mM Tris-HCl, pH 8.0, 50 mM NaCl, 5 mM EDTA, 0.2 µM oligo (5′-Cy5-TTTTTTTTTTTTTTTATCTTTTTTTTTTTACTTTTTTTTTTAAACCCAAATTTTTTTTTTT TTTTTTTTTTTTTTTTTTTTTTTT-3′-Bio), 2 U of uracil-DNA-glycosylase (UDG) (New England Biolabs, USA), 0.1 mg/ml RNase A (Qiagen, USA), and 200 nM deaminase. The hAID was purchased from Enzymax (USA), and hAPOBEC3G purified from E. coli was a gift from Dr. R. Harris (University of Minnesota, USA). After incubation in 37°C for 30 min, NaOH was added to the reaction to the final concentration of 0.2 M, and the samples were heated at 95°C for 5 min. Then an equal volume of formamide was added to the reaction mix, and the samples were heated again at 95°C for 5 min. Reaction products were separated on 16% polyacrylamide gel containing 8 M urea, and the gels were scanned using the Typhoon 9410 imaging system (GE Healthcare, USA).
Measurement of mutations rates, isolation, and sequencing of can1 mutants
Mutation rates were determined by fluctuation analysis as described earlier [74, 76, 84]. Independent transformants of the wild-type and ung1 derivatives of our basic strain were grown in a complete minimal medium lacking an ingredient to select for the plasmid, and containing galactose instead of glucose, to induce expression of deaminase genes.
Patches of yeast transformants originating from single colonies (64 per plate) were replica-plated onto galactose-containing medium without leucine. After two days, they were replicaplated onto canavanine-containing medium to select for can1 mutants. After five days of incubation, independent Canr colonies were colony-purified on canavanine-containing medium. Chromosomal DNA from cells originating from single colonies of these can1 mutants was isolated using a Yeast DNA Extraction Kit (Epicentre, USA). Subsequent PCR amplification and sequencing was performed as described earlier [76].
Comparison of mutation spectra
The MOTIFN program [15] was used for prediction of mutable motifs in the APOBEC3G and APOBEC1-induced mutation spectra. The significance of correlations between the distribution of mutable motifs and mutations along a target sequence was measured by a Monte Carlo procedure (the CONSEN program) [15, 85].
RESULTS
Mutagenic and nonmutagenic deaminases
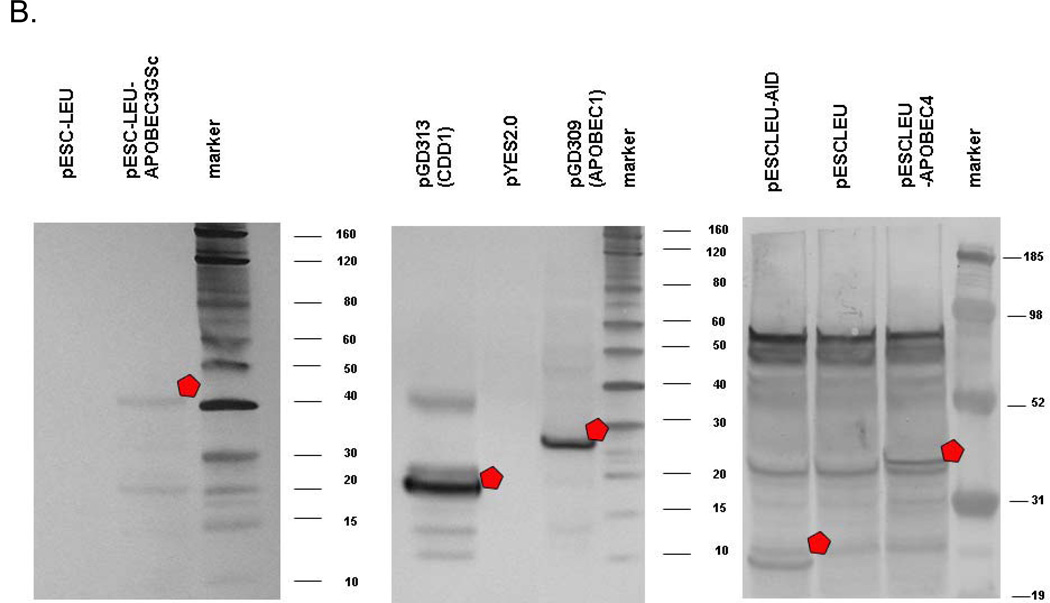
The summary of the results of the mutagenesis tests accomplished up to the present moment with bacteria and yeast expressing various deaminase genes is presented in Table 1. APOBEC1, AID, APOBEC3G, lamprey CDA1 are mutagenic in both hosts (Table 1, upper half). Abolishing the activity of uracil DNA glycosylase in the ung strains led to a major increase in the mutator effect, suggesting that the initial trigger was the deamination of cytosine. The presence of a small tag (or even larger GFP in AID) at the C-terminus or the N-terminus of deaminases (see “Materials and Methods”) or NLS in APOBEC1 did not affect their activity. Several other, novel, deaminases presented in Fig. 1 as well as Cdd1 and Tad2/Tad3 are not mutagenic. The results with these deaminases are summarized in the bottom part of Table 1. All deaminases were tested in ung1 strains and, in some cases, in additional genetically sensitized strains. The effect of the expression of the TAD2/TAD3 and APOBEC4 in bacteria were examined in the nfi strain defective in endonuclease V. This enzyme repairs DNA containing hypoxanthine, a product of adenine deamination. The effect of expression of APOBEC4 in yeast was examined in the yeast strain lacking all five known yeast glycosylases and in the strain defective in Apn1 and Apn2 apurinic/apyrimidinic endonucleases. Neither of these genetic backgrounds helped to reveal the mutagenic effects of these nonmutagenic deaminases. We conclude that they belong to the APOBEC2 group [35], deaminases that do not exhibit a mutator effect when produced in unnatural host. The difference in mutagenesis cannot be easily explained by the difference in the protein architecture because most members of the family retain conserved catalytic amino acids and exhibit a similar secondary structure (Fig. 1) with some minor variations. The additional helix instead of the beta strand right before the HAE motif in MICPUN_56782 can be explained by problems with the secondary structure prediction. The JHMM program [86] predicted the β-strand instead of the helix.
Table 1.
Mutagenic and nonmutagenic deaminases
| Mutator effect | Deaminase gene expressed |
E. coli strain* | Reference | S. cerevisiae strain* | Reference |
|---|---|---|---|---|---|
| Yes | rat APOBEC1 | [35] | ** | ||
| rat APOBEC1_NLS | ** | ||||
| human AID | [24, 35] | [76, 84]** | |||
| human AID-GTP | ** | ||||
| human APOBEC3G | [35] | [92]** | |||
| lamprey CDA1 | [32]** | [32]** | |||
| No | yeast CDD1 | wt. ung1 | ** | ||
| human APOBEC2 | wt, ung− | [35] | |||
| human APOBEC4 | wt, ung−, nfi− | ** | wt, ung1, Δ5G, Δapn1, Δapn2 | ** | |
| human APOBEC4*** | wt, ung1 | ** | |||
| X.oryzae APOBEC5 | wt, ung | ** | |||
| yeast TAD2/TAD3 | wt, ung−, nfi− | ** | |||
| M.sp MICPUN_56782 | wt, ung− | ** | |||
Height of letter in the column is proportional to the magnitude of mutahenic effect. Empty cells in the table mean that experiments were not done.
This work.
Truncated.
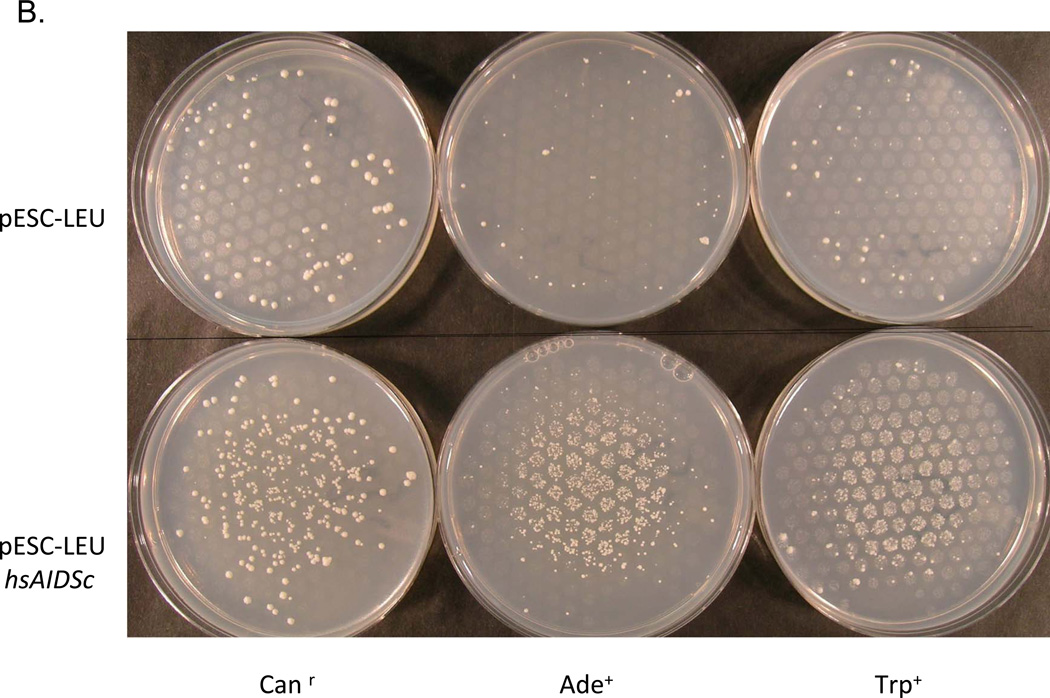
We present the examples of qualitative assay for mutagenic activity in spot test with bacteria in Fig. 2a and with yeast in Fig. 2b. In the test with the Rosetta strain, where the deaminase gene is under the T7 promoter, we spotted the inducer of T7 RNA polymerase in Rosetta, IPTG, in the center of the plates with bacteria as described in the legend to Fig. 2a. Rosetta with control vector yields a few rifr colonies (left column of plates, Fig. 2a). Knockout of the ung1 gene leads to a significant elevation of the number of resistant mutants. The absence of mutants in the central zone is explained by toxicity of IPTG. PmCDA1 production led to two circles of rifampicin-resistant colonies (middle column of plates, Fig. 2a). We explain the observation by interference of mutagenesis and survival curves dependent on the diffusion of ITPG on the plate. In the center, the induction of mutants is so magnificent that even a strong reduction of cell survival does not mask the mutator effect. Farther from the center, mutant frequency drops and low cell survival leads to the absence of resistant colonies. The next circle of mutants occurs when the survival is high and mutation induction is still much above the spontaneous level. The ung1 strain is much more sensitive to the mutator effect of the expression of pmCDA1. The expression of the MICPUN_56782 from Micromonas is clearly nonmutagenic.
Fig. 2.
Mutator effects of deaminases in spot test. a) Mutator effect of expression of lamprey CDA1 in bacteria in spot test. Rosetta or Rosetta ung1− were transformed by control pET24b vector or its derivatives with cloned deaminase genes from lamprey or from Micromonas. IPTG (20 µl of 100 mM solution) was added into the center of LB + kan plate with plated 108 bacteria. The bacteria were allowed to grow overnight and then were replica-plated on selective medium with rifampicin. Visible resistant colonies were scored after additional overnight incubation. b) Mutator effect of the expression of human AID in yeast in spot test. Yeast strain 1B-D770 ung1 was transformed by control pESC-LEU vector or its derivative with recoded human AID gene (“Materials and Methods”). Galactose (20 µl of 20% solution) was added into the center of the complete minimal plate selective for the plasmid marker and containing raffinose with plated ~107 yeast cells. Yeast were allowed to grow for two days and were replica-plated on the three types of selective media to score forward Canr mutations and reversion of the two auxotrophic markers, ade2-1 and trp1-289. Visible colonies of mutants or revertants were scored after four days of incubation.
In the spot tests with yeast we put a drop of the inducer of the GAL1/GAL10 promoter, galactose, into the center of the plates with yeast as described in the legend to Fig. 2b. Galactose apparently became mutagenic for strains capable of expressing human AID. The induction of forward mutation for canavanine resistance was moderate, while the mutagenic effect was very strong for the reversion of nonsense mutations. We did a more precise analysis of the response of different yeast markers to the expression of mutator deaminases in quantitative tests (Table 2). Only lamprey CDA1 was strongly mutagenic in the Ung+ background, leading to a 32-144-fold increase of mutant frequencies depending on the reporter. Other deaminases were only moderately active when uracil glycosylase was intact. The hAIDSc expression lead to an eight-fold increase in forward mutation and a three to six-fold increase in reversion of nonsense mutations; the expression of hAPOBEC3G and APOBEC1 resulted in a four to six-fold increase in forward mutation and did not markedly increase nonsense reversion. The ung1 mutation led to approximately a five to 30-fold increase of mutation rates. In the Ung− strain, all deaminases were strongly mutagenic for forward mutations, but again the pmCDA1 expression led to the largest effect. The mutator effect was multiplicative for Canr forward mutations (a 82-fold increase over the wild-type strain expressing hAIDSc) and synergistic for nonsense mutation reversion (a 410-to-1300-fold increase over the wild-type). When hAPOBEC3GSc was expressed in the ung1 strain, the mutator effect was multiplicative for Canr forward mutations (a 52-fold increase over the wild-type strain) and synergistic for TAG nonsense mutation reversion (a 185-fold increase over the wild-type). The hAPOBEC3G did not affect the reversion of the TAA nonsense codon (the ade5-1 reporter, reverts mostly by suppressors [73]). The expression of APOBEC1 led to a 75-fold increase of forward mutations and, quite opposite to APOBEC3G, a strong increase of Ade+ reversion (330-fold) but did not induce Trp+ reversion. The expression of the pmCDA1 exerted the most dramatic effects. It was exceptionally mutagenic for three different reporters, with a maximum increase of reversion by 3500-fold.
Table 2.
Mutator phenotypes resulting from hAIDSc, hAPOBEC3G, rat rAPOBEC1, lamprey CDA1, and yeast CDD1 expression in UNG1+ and ung1− haploid yeast
| Relevant genotype | Deaminase expression system | Mutation rates (95% confidence limits)* | ||
|---|---|---|---|---|
| forward mutations Canr × 10−7 |
TAA nonsense mutation reversion Ade+ × 10−8 |
TAG nonsense mutation reversion Trp+ × 10−8 |
||
| UNG1+ | vector | 2.5 (1.2–6.5) | 24 (21–34) | 4.1 (1.3–14) |
| hAIDSc | 19 (14–25) | 72 (60–124) | 24 (21–38) | |
| haPOBEC3GSc | 9.8 (7.7–16) | 21 (8.7–32) | 35 (21–6.7) | |
| rAPOBEC1 | 16 (14–21) | 48 (40–82) | 5.0 (3.7–5.9) | |
| pmCDA1 [32] | 360 (230–1300) | 2200 (1500–2800) | 130 (70–140) | |
| yCDD1 | 1.8 (1.2–3.2) | 16 (11–22) | 6.5 (5.2–13) | |
| ung1 | vector | 13 (10–31) | 210 (190–290) | 140 (110–270) |
| hAIDSc | 205 (170–220) | 9700 (7500–12 600) | 5300 (4400–6600) | |
| hAPOBEC3GSc | 130 (50–190) | 170 (140–240) | 760 (430–1300) | |
| rAPOBEC1 | 230 (104–280) | 7600 (5900–15 000) | 240 (210–350) | |
| pmCDA1 [32] | 430 (290–610) | 29 600 (18 000–34 000) | 19 000 (9000–28 000) | |
| yCDD1 | 8.3 (4.1–12) | 140 (100–170) | 130 (85–200) | |
Mutation frequencies statistically higher than corresponding wild-type are in bold.
The expression of the yCDD1 was completely nonmutagenic (Table 2) in this test as in spot tests described earlier.
Mutagenic specificity of deaminases
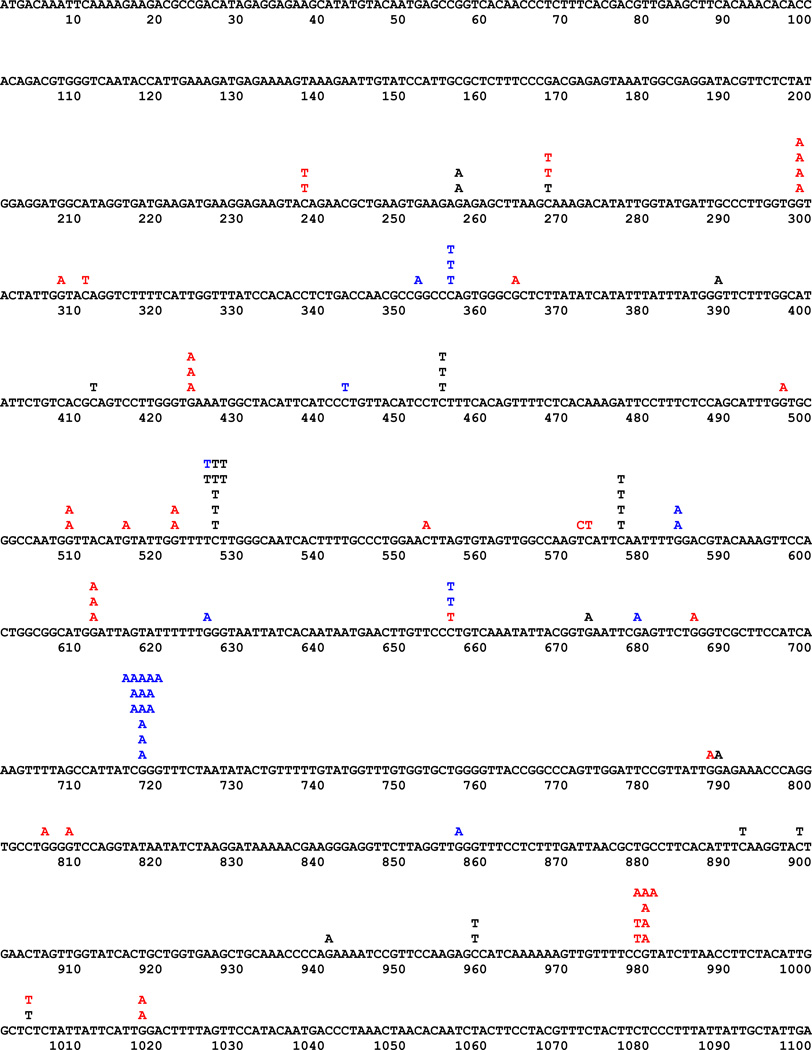
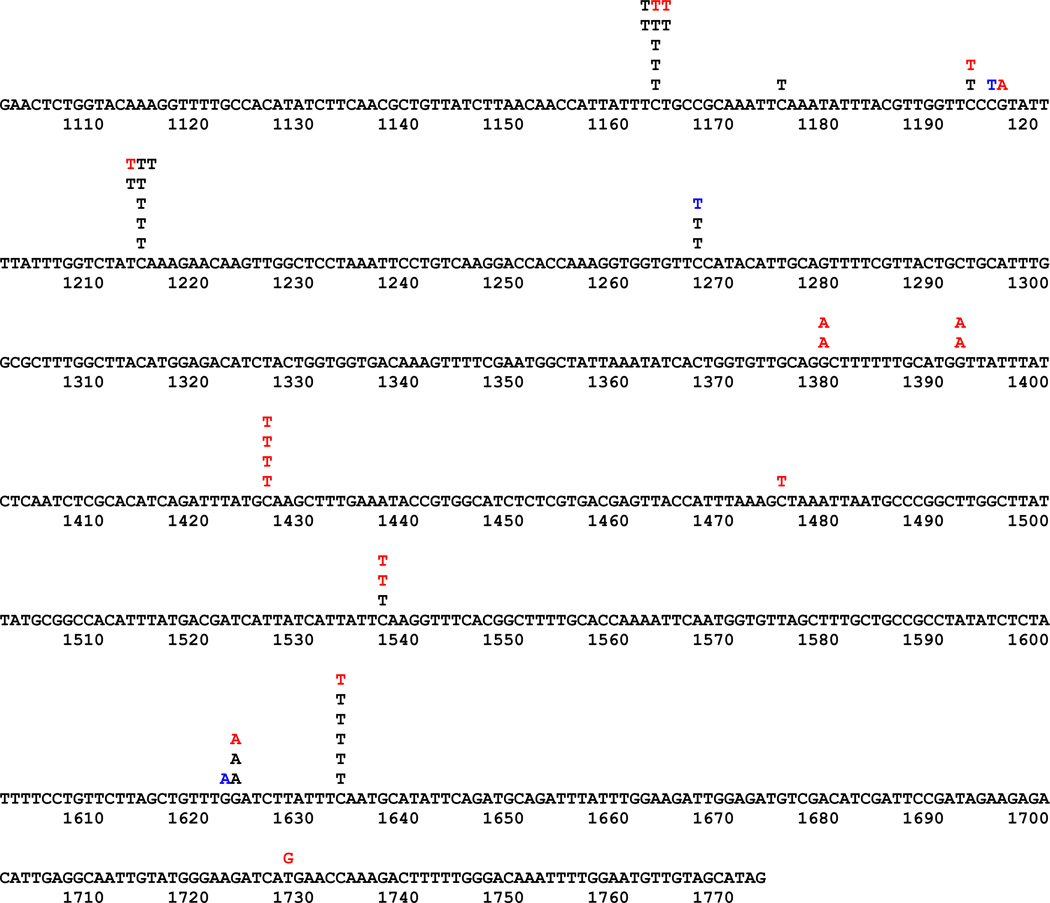
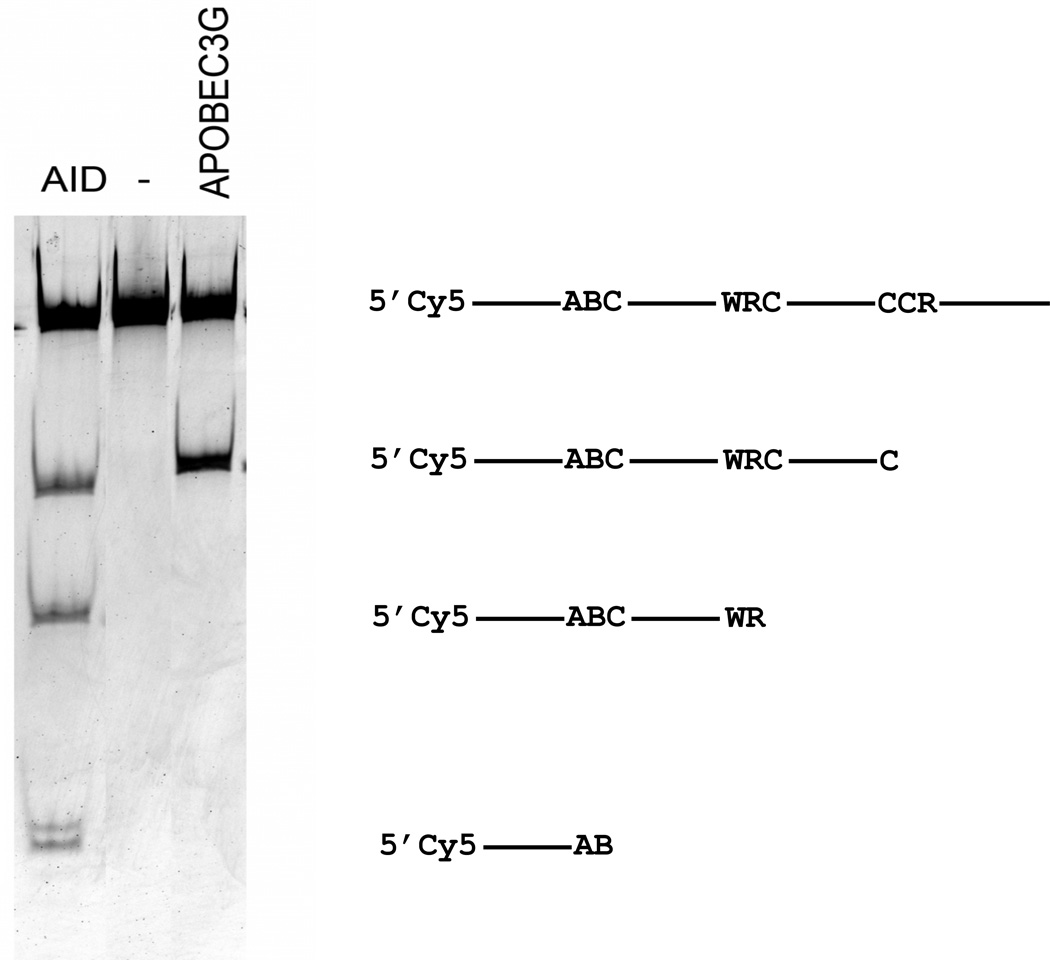
To get insights into why different markers responded so differently, we compared forward CAN1 mutation spectra for the four deaminases. For this analysis, we studied independent Canr mutants obtained under conditions of hAIDSc expression in the wild-type and the ung1 strain and in the ung1 strain only for other deaminases (Fig. 3 and Table 3). It is worth mentioning that every single mutant had only one single base substitution. The dependence of mutagenic effects of the expression of mutator deaminase genes on the status of uracil DNA glycosylase unequivocally means that cytidine to uracil deamination is the major cause of the mutator effect. Indeed, the vast majority of mutations were transitions of the C/G pair to the T/A pair as seen before [32, 76]. Quite amazingly, the distribution of mutations induced by different deaminases was completely different (Fig. 3) as well as extracted hotspot motifs (Table 3). Mutations in the ung1 strain, representing deamination proclivity of AID before repair, occur at a higher rate on the transcribed strand. This is different from the effect of expression of AID observed in the E. coli selective system [87]. In the wild-type, there is some prevalence of mutations due to putative non-transcribed strand deaminations, suggesting the possibility that, in our system, the repair of uracil in the transcribed strand is more efficient. APOBEC1 and pmCDA1 induced more mutations in the non-transcribed DNA strand in the ung1 strain (Table 3). It is interesting to note that the clarity of the predicted mutable motif was also dependent on the strand analyzed. The stringency of mutational specificity was higher for hAPOBEC3G (specificity index around 8, Table 3) than for hAID (specificity index 3). It is interesting that this in vivo specificity correlated with the activity of the enzymes in vitro. When we analyzed the specificity of the two enzymes biochemically with oligonucleotide with three different hotspot motifs, hAPOBEC3G also demonstrated high stringency for deamination of only CCC motif (Fig. 4).
Fig. 3.
Spectra of deaminase-induced forward mutations in yeast CAN1 gene. Letters above the sequence of CAN1 are changes observed. Black on white, AID as a reference, taken from [76]; white on black, APOBEC1; black on gray, APOBEC3G.
Table 3.
Comparison of the types of DNA sequence changes induced in yeast by expression of human AID, rat APOBEC1, human APOBEC3G, and pmCDA1
| Strain, plasmid, reference |
Total mutations* |
Transitions at G-C base pairs |
c→T** | G→A changes*** |
Preference for mutable motifs**** |
|---|---|---|---|---|---|
| WT, hAIDSc [76] | 64 | 56 | 37 | 19 | WRC/GYW 2.2/6.2 |
| ung1, hAIDSc [76] | 62 | 61 | 24 | 37 | WRC/GYW 3.8/2.8 |
| ung1, rAPOBEC1 | 49 | 49 | 42 | 7 | TCW/WGA 15.4/1.5 |
| ung1, hAPOBEC3GSc | 62 | 58 | 21 | 33 | CCR/YGG 9.0/7.4 |
| ung1, pmCDA1 [32] | 104 | 102 | 68 | 34 | ABC/GVT 4.5/1.6 |
All mutants had only one nucleotide change in the CAN1 ORF.
Deamination of cytosine in the non-transcribed strand.
Deamination of cytosine in the transcribed strand.
Mutated base is underlined. W stand for weak A–T pair, R – purine, Y – pyrimidine, B – any base but A, V is any base but T. The values listed represent the fold increase in occurrence of mutations at a mutable site above the average occurrence of mutations at non-hotspot G:C sites (specificity index). Mutable sites match with mutable motifs that are defined elsewhere [28, 91]. Underlined values represent a statistically significant correlation (p < 0.05) between a mutable motif and the distribution of mutations, as revealed by using a Monte Carlo procedure.
Fig. 4.
Preference of hAID and hAPOBEC3G for specific motifs as judged by in vitro deamination assay. Activity of the two enzymes was compared with the use of Cy5 labeled oligonucleotide containing three sequence motifs (“Materials and Methods”) matching hotspot motifs for cytosine deamination (Table 3). The gel image to the left is the result of separation of processed deamination reaction products on denaturing polyacrylamide gel. The diagram to the right explains what sites were deaminated to generate the observed bands.
Production and intracellular distribution of mutagenic and nonmutagenic deaminases
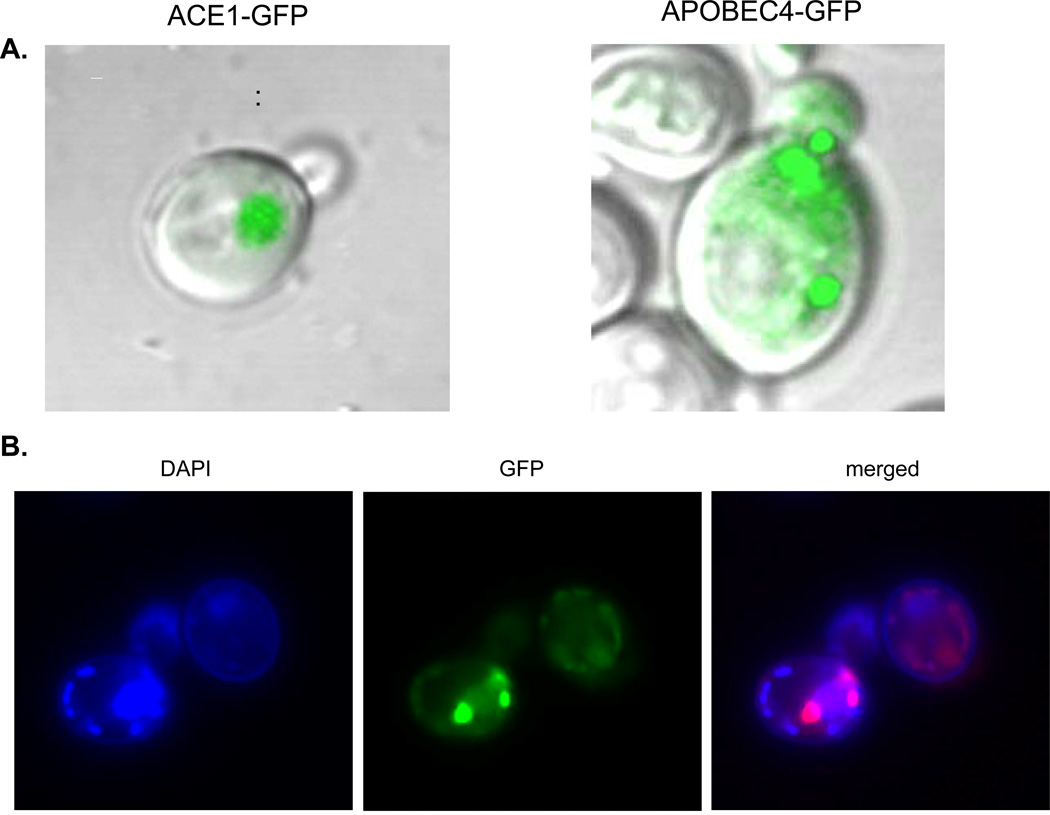
The lack of mutagenic effect of deaminases in bacteria and yeast can be due to various reasons, and we explored several trivial explanations. We found that all, nonmutagenic and mutagenic, deaminases were found largely in the pellet fraction of cell extracts, and therefore, were largely insoluble when produced in bacteria (Fig. 5a). This corresponds to published data [88, 89]. We conclude that insolubility by itself can not explain the absence of the effect of nonmutagenic deaminases. On the other hand, we have shown that nonmutagenic CDD1 and APOBEC4 are detected in a soluble fraction of yeast extracts, even better than in highly mutagenic deaminases APOBEC1, APOBEC3, and AID (Fig. 5b). Together with the observation that APOBEC2 is soluble in E. coli ([49] and our data), these results indicate that there is no simple correlation between solubility and mutagenic activity. We demonstrated that GFP-fused hAID and APOBEC4 do not localize in the nucleus in yeast cells (Fig. 6; see color insert). In Fig. 6a we compare the distribution of GFP tagged transcription factor Ace1 that has nuclear localization (left photo) with APOBEC4-GFP. The latter has an irregular pattern of distribution in the cell. In Fig. 6b we demonstrate that AID-GFP is localized primarily outside the nucleus (stained by DAPI in the left photo). In the lower cell there are bright spots of aggregates of AID-GFP outside the nucleus with some weak signal in the nucleus. In the upper cell most of the GFP signal is in the cell compartment located opposite to the nucleus. It is likely that even a small amount of AID available in the nucleus is sufficient to drive hypermutagenesis (Table 1).
Fig. 5.
Detection of AID/CDD1/APOBEC proteins in extracts. a) Production of deaminases in bacteria. Proteins in bacterial extracts were separated on 12% polyacrylamide gels and stained by Coomassie G-450. Lanes: M, molecular weight marker; S, supernatant of the protein extract; P, protein extract pellet prepared as described in “Materials and Methods”. Predicted molecular weights are for 27.2 kDa for PmCDA1-His6, 47.2 kDa for APOBEC3G, 17.7 kDa for APOBEC5-His6, 29.4 kDa for His6-Tad2, and 39.4 kDa for Tad3-S-tag. b) Production of deaminases in yeast. The results of Western blots are shown. Proteins of correct size are marked by a white pentagon. Protein in yeast extracts of appropriate strains were separated using 4–12% gradient polyacrylamide gel (Invitrogen). Transfer to PVDF membrane and reaction with appropriate primary antibodies from mouse and then secondary antibodies from goat was accomplished as suggested by the vendor (Western Breeze kit; Invitrogen). Primary antibodies were mouse anti c-myc for tagged hApobec3G, anti-FLAG for tagged APOBEC4, monoclonal anti-HA for tagged APOBEC1 and CDD1.
Fig. 6.
Intracellular distribution of GFP-fused deaminases in yeast. a) Confocal LSM images of cells of BY4742 ung1 producing nuclear ACE1-GFP or APOBEC4-GFP. Live cells were imaged at UNMC core Confocal microscopy facility by Zeiss LSM 410 confocal laser microscope. b) Intracellular distribution of hAIDSc-GFP in BY4742 ung1. Cells were imaged with Olympus 100 × 1.35 NA oil immersion objective on the Delta Vision microscopy system (Applied Precision, USA) consisting of an Olympus IX70 microscope (Olympus America, USA) and a CoolSnap HQ 12-bit camera (Photometrics/Roper Scientific, USA) and controlled by SoftWoRx software.
DISCUSSION
Many deaminases of the APOBEC1/AID/TAD superfamily possess enzymatic activity on polynucleotides and exert genome-wide mutagenic effects when produced in the heterologous hosts. They are involved, as we discussed in the introduction, in different biological transactions. In the current paper we summarize our studies of activity and specificity of deaminases with known mutagenic effects and examine the properties of several new deaminases. We have used two model organisms, bacterium E. coli and unicellular eukaryote yeast S. cerevisiae, to study the in vivo biological activity of deaminases and to purify recombinant deaminases for in vitro assays.
Mutator proteins produced in host microorganisms led to the apparent conversion of nonmutagenic, harmless compounds to hypermutagens by indirect action, because IPTG or galactose induced the production of these proteins. AID induces mutations when expressed in E. coli [24, 90] and in yeast [76, 84]. It is amazing and significant that these mutations occur in DNA sequence motifs similar to mutations during SHM [24, 76, 91]. The mutator effects are enhanced in uracil DNA glycosylase-deficient ung1− strains, which are unable to repair uracil in DNA, suggesting that the deamination of cytosine to uracil in DNA is the cause of these mutations [24, 84]. It was found that the expression of two other homologous deaminases, APOBEC1 and APOBEC3G, is highly mutagenic in bacteria [35] and yeast [92]. Almost all mutations arising under the expression of deaminases in prokaryotes or in yeast were G→C to A→T transitions. Mutations at the A-T base are not observed, implying that deamination was restricted to cytosines [32, 44, 76, 84, 87]. Expression of AID was also recombinogenic in Ung1+ yeast, suggesting that nicks during repair of deaminated cytosine trigger recombination [84]. Expression of APOBEC3G in yeast also inhibited Ty1 retrotransposition [92, 93]. CDA1 from lamprey was a potent mutagen and inducer of recombination in yeast [32]. In all cases the mutagenic signature of deaminases produced in microorganisms resembled the signature attributable to the particular deaminase in vivo in the natural host.
We have extended the studies and compared the activity and specificity of these four enzymes. The most powerful mutagenic effect was detected for lamprey CDA1, both in bacteria and yeast (Fig. 2a and Tables 1 and 2). The effect of pmCDA1 expression was generally greatly elevated in the ung1− background but not for all markers. The lack of influence of ung1 was most striking for forward mutations in yeast (Table 2). We interpret this finding as evidence for hotspot motifs of deaminations in the CAN1 gene that are not corrected by uracil glycosylase. The analysis of the context can1 mutations induced by pmCDA1 revealed that it is very different from the context of mutations induced by other deaminases (Table 3).
The next strongest mutagenic effect in bacteria was for APOBEC1 produced in bacteria and AID produced in yeast. The difference could be caused by differences in levels of the active protein in the cell or the difference in reporter response or both. The APOBEC3G mutagenic effect in yeast was the most modest (Table 2). It is likely caused by a unique mutation signature involving runs of cytosines (Table 3), which are less abundant in yeast, generally an AT-rich organism.
The effects of deaminases in reversion tests were strongly dependent on a particular enzyme. The expression of the hAID or lamprey CDA1 strongly increased the reversion of both nonsense mutations ade5-1 and trp1-289. The expression of the hAPOBEC3G was nonmutagenic for the ade5-1 reversion and strongly mutagenic in the trp1-289 reversion. Effects on the two markers were reversed for the expression of the rAPOBEC1. Therefore, the tester yeast strain 1B-D770 ung1 can be used for the express-analysis of the specificity of deaminases (Table 2).
It is interesting that in the collection of mutants induced by the expression of deaminases in yeast there were generally only one base substitution per mutant. Quite contrary, it is well documented that in vitro on single-stranded DNA both AID and APOBEC3G produce bursts of multiple deaminations in a processive manner [28, 39, 94, 95]. Multiple mutations are also seen in HIV or retroelements in yeast surviving after APOBEC3 restriction deamination [36, 92, 96, 97]. Apparently, chromosomal DNA in living cells is protected within the cell from the processive action of deaminases. This mechanism is missing in vitro and is somehow disrupted during the viral cycle or retrotransposition.
In summary for this section, the expression of hAIDSc, hAPOBEC3G, rAPOBEC1, and pmCDA1 is highly mutagenic in yeast, due to genome-wide cytosine to uracil deamination. The specificity of the mutator effects is very distinct for every deaminase, which could be partially responsible for the divergence of functions of deaminases. This observation suggests that the current reporters may be used for screening and estimation of specificity of new deaminase variants.
Yeast Cdd1, cytidine/deoxycytidine deaminase, was nonmutagenic. It has been suggested that yeast Cdd1, a classical cytidine/deoxycytidine deaminase, can perform RNA editing in yeast. It was named an orphan editase, a prototype for DNA/RNA editing enzymes similar to APOBEC1 [81]. A lack of mutagenesis does not support the proposal on the similarity of the two enzymes. The same conclusion comes from the analysis of the general structural plan of Cdd1 and editing deaminases. Cdd1 lacks three helices that have been implicated in the ability of deaminases to act on polynucleotides (Fig. 1) [32]. Most likely, deaminases operating in nucleotide pools do not possess the ability to edit DNA and are thus nonmutagenic.
Tad2/Tad3 was also nonmutagenic, despite the closer relationship to other editing deaminases (Fig. 1) and the ability to deaminate specific RNAs. It is possible that they recognize only tRNA.
Other new deaminases we have tested were nonmutagenic in yeast or bacteria (Table 1). Along with the known fact of the nonmutagenicity of the expression of APOBEC2 [35], this finding leads to the conclusion that the mutagenic effect in a heterologous host is a relatively rare observation. The absence of the mutagenic effect is probably caused by the intrinsic properties of deaminases but not by the unavailability of their active forms in the foreign hosts, because they were robustly produced in bacteria and yeast and their intracellular distribution was not different from mutagenic deaminases (Figs. 5 and 6). The presence of a nuclear localization signal in APOBEC1 fusion protein, on the other hand, had no effect on the already high mutagenic potential of this protein. In theory, it is possible that their activity in natural hosts depends on auxiliary factors. For example, specific RNA editing by APOBEC1 requires complementing factors [82]. However, APOBEC1 is highly mutagenic when produced in bacteria and yeast, where these factors are apparently missing.
Structural features of deaminases do not immediately help to understand the critical differences between mutagenic and nonmutagenic deaminases. APOBEC4 has an insert of four amino acids between catalytic residues PC and C (Fig. 1) [61]. Does this disrupt any predicted helices/sheets? APOBEC5 does not possess α-helices 5 and 6; however, highly conserved motifs HxE and PCxxC are present. More closely related AID, APOBEC1, APOBEC3G, and APOBEC4 have the xxLRxL motif. In APOBEC2 the R is changed to K, and in Tad2 enzymes two leucines are changed to valines, while in APOBEC5 this motif is beyond recognition. Further studies are required to find how these differences affect deamination ability and the function of deaminases. We propose that nonmutagenic deaminases perform a very specific DNA/RNA editing due to unique structural features, and such specific events are not detected in the relatively rough detection system of heterologous expression. It is known that the specificity of a deaminase could be altered very efficiently by single amino acid changes [98]. Further work is needed to find the functions of nonmutagenic deaminases.
It is also possible that some deaminases do not possess their original deaminase activity and have acquire another function. Evidence is accumulating that inactive enzymes or protein domains comprise around 10% of all proteins. They are found in virtually all enzyme families and are stably maintained during evolution [99]. Inactive domains were thought to be rare among DNA replication and other enzymes of DNA metabolism. However, a class of inactive DNA polymerases has been discovered in archae [100, 101]. The second subunit of human DNA polymerase δ possesses an inactivated phosphodiesterase domain [102]. An essential replicative catalytic subunit of DNA polymerase ε polypeptide consists of two polymerases belonging to different types of DNA polymerases of the B-family, inactive N-terminal domain polymerase and active C-terminal domain polymerase [103]. This is similar to APOBEC3G, possessing inactive and active domains in the same polypeptide.
Acknowledgments
We are grateful to V. G. Liston for expert technical assistance and to rotation students S. J. Wingett, P. S. Seymour, and E. Worrall for participation in the experiments. We acknowledge the UNMC confocal microscopy facility for help with imaging in the yeast cells producing APOBEC1-GFP fusions and the UNMC Structural Biology facility which is funded in part by the Eppley Cancer Center and the Nebraska Research Initiative for help with protein purification. We thank Drs. E. Sage, H. Smith, S. Petersen-Mahrt, R. Harris, and R. Schaaper for sending us strains and plasmids.
This work has been supported, in part, by a UNMC pilot grant awarded in 2008 (YIP, Co-PI) and National Cancer Institute Eppley Cancer Center Support Grant P30CA036727. IBR was supported in part by the Intramural Research Program of the National Library of Medicine at the National Institutes of Health/DHHS. LEGENDS TO FIGURES.
Abbreviations
- ADA
adenosine deaminase
- AID
activation induced deaminase
- Apn
apurinic/apyrimidinic nuclease
- APOBEC
apolipoprotein B editase complex related enzyme
- AP site
apurinic/apyrimidinic site
- CDA or CDD
cytidine deaminase or various organisms
- CSR
class switch recombination
- GC
gene conversion
- GFP
green fluorescent protein
- HIV
human immunodeficiency virus
- IPTG
isopropyl-β-D-1-thiogalactopyranoside
- LB
Luria–Bertani medium
- LDS
sodium lauryl sulfate, leucine-rich repeats
- Nfi
endonuclease five
- NLS
nuclear localization signal
- ORF
open reading frame
- PmCDA
Pteromyzon marinus cytidine deaminase
- SC
synthetic medium for yeast cultivation
- SDS
sodium dodecyl sulfate
- SHM
somatic hypermutation
- Tad
tRNA adenosine deaminase
- UNG or UDG
uracil-DNA-glycosylase
- Vif
virus infectivity factor
- VLR
variable lymphocyte receptors
- YEPD
complete medium for yeast cultivation
Contributor Information
A. G. Lada, Email: lada@unmc.edu.
C. Frahm Krick, Email: cfrahmkrick@benchmarkbiolabs.com.
S. G. Kozmin, Email: stanislav.kozmin@duke.edu.
V. I. Mayorov, Email: mayorov_vi@mercer.edu.
T. S. Karpova, Email: karpovat@mail.nih.gov.
I. B. Rogozin, Email: rogozin@ncbi.nlm.nih.gov.
Y. I. Pavlov, Email: ypavlov@unmc.edu.
REFERENCES
- 1.Hayaishi O, Kornberg A. J. Biol. Chem. 1952;197:717–732. [PubMed] [Google Scholar]
- 2.Kornberg A, Baker TA. DNA Replication. New York: W.H, Freeman; 1992. [Google Scholar]
- 3.Sugiyama E, Lee SJ, Lee SS, Kim WY, Kim SR, Tohkin M, Hasegawa R, Okuda H, Kawamoto M, Kamatani N, Sawada J, Kaniwa N, Saito Y, Shin JG. Drug Metab. Pharmacokinet. 2009;24:553–556. doi: 10.2133/dmpk.24.553. [DOI] [PubMed] [Google Scholar]
- 4.Sauer AV, Aiuti A. Curr. Opin. Allergy Clin. Immunol. 2009;9:496–502. doi: 10.1097/ACI.0b013e3283327da5. [DOI] [PubMed] [Google Scholar]
- 5.Pankratova EV, Stepchenko AG. Genetika. 2010;46:5–13. [PubMed] [Google Scholar]
- 6.Neuberger MS, Harris RS, Di Noia J, Petersen-Mahrt SK. Trends Biochem. Sci. 2003;28:305–312. doi: 10.1016/S0968-0004(03)00111-7. [DOI] [PubMed] [Google Scholar]
- 7.Harris RS, Liddament MT. Nat. Rev. Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 8.Samaranayake M, Bujnicki JM, Carpenter M, Bhagwat AS. Chem. Rev. 2006;106:700–719. doi: 10.1021/cr040496t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng W. Bioessays. 2010;32:385–387. doi: 10.1002/bies.201000014. [DOI] [PubMed] [Google Scholar]
- 10.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teng B, Burant CF, Davidson NO. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 12.Morrison JR, Paszty C, Stevens ME, Hughes SD, Forte T, Scott J, Rubin EM. Proc. Natl. Acad. Sci. USA. 1996;93:7154–7159. doi: 10.1073/pnas.93.14.7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milstein C, Rada C. The Maturation of Immune Response. London: Academic Press; 1995. [Google Scholar]
- 14.Kinoshita K, Honjo T. Nat. Rev. Mol. Cell Biol. 2001;2:493–503. doi: 10.1038/35080033. [DOI] [PubMed] [Google Scholar]
- 15.Rogozin IB, Kolchanov NA. Biochim. Biophys. Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 16.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Nat. Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 17.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 18.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 19.Durandy A, Honjo T. Curr. Opin. Immunol. 2001;13:543–548. doi: 10.1016/s0952-7915(00)00256-9. [DOI] [PubMed] [Google Scholar]
- 20.Yoshikawa K, Okazaki IM, Eto T, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. Science. 2002;296:2033–2036. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
- 21.Okazaki IM, Kinoshita K, Muramatsu M, Yoshikawa K, Honjo T. Nature. 2002;416:340–345. doi: 10.1038/nature727. [DOI] [PubMed] [Google Scholar]
- 22.Arakawa H, Hauschild J, Buerstedde JM. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 23.Di Noia JM, Neuberger MS. Eur. J. Immunol. 2004;34:504–508. doi: 10.1002/eji.200324631. [DOI] [PubMed] [Google Scholar]
- 24.Petersen-Mahrt SK, Harris RS, Neuberger MS. Nature. 2002;418:99–103. [PubMed] [Google Scholar]
- 25.Poltoratsky V, Goodman MF, Scharff MD. J. Exp. Med. 2000;192:F27–F30. doi: 10.1084/jem.192.10.f27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pavlov YI, Rogozin IB, Galkin AP, Aksenova AY, Hanaoka F, Rada C, Kunkel TA. Proc. Natl. Acad. Sci. USA. 2002;99:9954–9959. doi: 10.1073/pnas.152126799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Curr. Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 28.Pham P, Bransteitter R, Petruska J, Goodman MF. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 29.Lada AG, Iyer LM, Rogozin IB, Aravind L, Pavlov IuI. Genetika. 2007;43:1311–1327. [PubMed] [Google Scholar]
- 30.Pancer Z, Amemiya CT, Ehrhardt GR, Ceitlin J, Gartland GL, Cooper MD. Nature. 2004;430:174–180. [PubMed] [Google Scholar]
- 31.Alder MN, Rogozin IB, Iyer LM, Glazko GV, Cooper MD, Pancer Z. Science. 2005;310:1970–1973. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- 32.Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, Pavlov YI, Aravind L, Pancer Z. Nat. Immunol. 2007;8:647–656. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- 33.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 34.Sheehy AM, Gaddis NC, Malim MH. Nat. Med. 2003;9:1404–1407. doi: 10.1038/nm945. Epub 2003 Oct 1405. [DOI] [PubMed] [Google Scholar]
- 35.Harris RS, Petersen-Mahrt SK, Neuberger MS. Mol. Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 36.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 37.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Nat. Struct. Mol. Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 38.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 39.Chelico L, Pham P, Calabrese P, Goodman MF. Nat. Struct. Mol. Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- 40.Coker HA, Petersen-Mahrt SK. DNA Repair (Amst) 2007;6:235–243. doi: 10.1016/j.dnarep.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Stenglein MD, Harris RS. J. Biol. Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 42.Chiu YL, Greene WC. J. Biol. Chem. 2006;281:8309–8312. doi: 10.1074/jbc.R500021200. [DOI] [PubMed] [Google Scholar]
- 43.Yu K, Huang FT, Lieber MR. J. Biol. Chem. 2004;279:6496–6500. doi: 10.1074/jbc.M311616200. Epub 2003 Nov 6425. [DOI] [PubMed] [Google Scholar]
- 44.Beale RC, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS. J. Mol. Biol. 2004;337:585–596. doi: 10.1016/j.jmb.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 45.Rausch JW, Chelico L, Goodman MF, Le Grice SF. J. Biol. Chem. 2009;284:7047–7058. doi: 10.1074/jbc.M807258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carpenter MA, Rajagurubandara E, Wijesinghe P, Bhagwat AS. DNA Repair (Amst) 2010;9:579–587. doi: 10.1016/j.dnarep.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harjes E, Gross PJ, Chen KM, Lu Y, Shindo K, Nowarski R, Gross JD, Kotler M, Harris RS, Matsuo H. J. Mol. Biol. 2009;389:819–832. doi: 10.1016/j.jmb.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. Nature. 2007;445:447–451. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- 50.Autore F, Bergeron JR, Malim MH, Fraternali F, Huthoff H. PLoS One. 2010;5:e11515. doi: 10.1371/journal.pone.0011515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato Y, Probst HC, Tatsumi R, Ikeuchi Y, Neuberger MS, Rada C. J. Biol. Chem. 2010;285:7111–7118. doi: 10.1074/jbc.M109.052977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Etard C, Roostalu U, Strahle U. J. Cell Biol. 2010;189:527–539. doi: 10.1083/jcb.200912125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rubio MA, Pastar I, Gaston KW, Ragone FL, Janzen CJ, Cross GA, Papavasiliou FN, Alfonzo JD. Proc. Natl. Acad. Sci. USA. 2007;104:7821–7826. doi: 10.1073/pnas.0702394104. Epub 2007 May 7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerber AP, Keller W. Science. 1999;286:1146–1149. doi: 10.1126/science.286.5442.1146. [DOI] [PubMed] [Google Scholar]
- 56.Johansson E, Mejlhede N, Neuhard J, Larsen S. Biochemistry. 2002;41:2563–2570. doi: 10.1021/bi011849a. [DOI] [PubMed] [Google Scholar]
- 57.Huthoff H, Malim MH. Virology. 2005;334:147–153. doi: 10.1016/j.virol.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 58.Kuratani M, Ishii R, Bessho Y, Fukunaga R, Sengoku T, Shirouzu M, Sekine S, Yokoyama S. J. Biol. Chem. 2005;280:16002–16008. doi: 10.1074/jbc.M414541200. [DOI] [PubMed] [Google Scholar]
- 59.Losey HC, Ruthenburg AJ, Verdine GL. Nat. Struct. Mol. Biol. 2006;13:153–159. doi: 10.1038/nsmb1047. [DOI] [PubMed] [Google Scholar]
- 60.Chen KM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, Harris RS, Matsuo H. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 61.Rogozin IB, Basu MK, Jordan IK, Pavlov YI, Koonin EV. Cell Cycle. 2005;4:1281–1285. doi: 10.4161/cc.4.9.1994. [DOI] [PubMed] [Google Scholar]
- 62.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Mol. Biol. Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. Epub 2004 Oct 2020. [DOI] [PubMed] [Google Scholar]
- 63.Conticello SG, Langlois MA, Neuberger MS. Nat. Struct. Mol. Biol. 2007;14:7–9. doi: 10.1038/nsmb0107-7. [DOI] [PubMed] [Google Scholar]
- 64.Zhang KL, Mangeat B, Ortiz M, Zoete V, Trono D, Telenti A, Michielin O. PLoS One. 2007;2:e378. doi: 10.1371/journal.pone.0000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Betts L, Xiang S, Short SA, Wolfenden R, Carter CW., Jr J. Mol. Biol. 1994;235:635–656. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- 66.Mejlhede N, Neuhard J. Biochemistry. 2000;39:7984–7989. doi: 10.1021/bi000542t. [DOI] [PubMed] [Google Scholar]
- 67.Karcher D, Bock R. RNA. 2009;15:1251–1257. doi: 10.1261/rna.1600609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salone V, Rudinger M, Polsakiewicz M, Hoffmann B, Groth-Malonek M, Szurek B, Small I, Knoop V, Lurin C. FEBS Lett. 2007;581:4132–4138. doi: 10.1016/j.febslet.2007.07.075. [DOI] [PubMed] [Google Scholar]
- 69.Casadaban MJ, Cohen SN. J. Mol. Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 70.Duncan BK. J. Bacteriol. 1985;164:689–695. doi: 10.1128/jb.164.2.689-695.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shcherbakova PV, Pavlov YI. Genetics. 1996;142:717–726. doi: 10.1093/genetics/142.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Calderon IL, Contopoulou CR, Mortimer RK. Gene. 1984;29:69–76. doi: 10.1016/0378-1119(84)90167-7. [DOI] [PubMed] [Google Scholar]
- 73.Achilli A, Matmati N, Casalone E, Morpurgo G, Lucaccioni A, Pavlov YI, Babudri N. BMC Genet. 2004;5:34. doi: 10.1186/1471-2156-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shcherbakova PV, Kunkel TA. Mol. Cell Biol. 1999;19:3177–3183. doi: 10.1128/mcb.19.4.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kozmin SG, Sedletska Y, Reynaud-Angelin A, Gasparutto D, Sage E. Nucleic Acids Res. 2009;37:1767–1777. doi: 10.1093/nar/gkp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mayorov VI, Rogozin IB, Adkison LR, Frahm C, Kunkel TA, Pavlov YI. BMC Immunol. 2005;6:10. doi: 10.1186/1471-2172-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bennetzen JL, Hall BD. J. Biol. Chem. 1982;257:3026–3031. [PubMed] [Google Scholar]
- 78.Jansen R, Bussemaker HJ, Gerstein M. Nucleic Acids Res. 2003;31:2242–2251. doi: 10.1093/nar/gkg306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karpova TS, Kim MJ, Spriet C, Nalley K, Stasevich TJ, Kherrouche Z, Heliot L, McNally JG. Science. 2008;319:466–469. doi: 10.1126/science.1150559. [DOI] [PubMed] [Google Scholar]
- 80.Chernoff YO, Galkin AP, Lewitin E, Chernova TA, Newnam GP, Belenkiy SM. Mol. Microbiol. 2000;35:865–876. doi: 10.1046/j.1365-2958.2000.01761.x. [DOI] [PubMed] [Google Scholar]
- 81.Dance GS, Beemiller P, Yang Y, Mater DV, Mian IS, Smith HC. Nucleic Acids Res. 2001;29:1772–1780. doi: 10.1093/nar/29.8.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dance GS, Sowden MP, Yang Y, Smith HC. Nucleic Acids Res. 2000;28:424–429. doi: 10.1093/nar/28.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bransteitter R, Pham P, Scharff MD, Goodman MF. Proc. Natl. Acad. Sci. USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Poltoratsky VP, Wilson SH, Kunkel TA, Pavlov YI. J. Immunol. 2004;172:4308–4313. doi: 10.4049/jimmunol.172.7.4308. [DOI] [PubMed] [Google Scholar]
- 85.Rogozin IB, Pavlov YI. Mutat. Res. 2003;544:65–85. doi: 10.1016/s1383-5742(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 86.Cole C, Barber JD, Barton GJ. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bhagwat AS. DNA Repair (Amst) 2004;3:85–89. doi: 10.1016/j.dnarep.2003.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coker HA, Morgan HD, Petersen-Mahrt SK. Meth. Enzymol. 2006;408:156–170. doi: 10.1016/S0076-6879(06)08010-4. [DOI] [PubMed] [Google Scholar]
- 89.Chen KM, Martemyanova N, Lu Y, Shindo K, Matsuo H, Harris RS. FEBS Lett. 2007;581:4761–4766. doi: 10.1016/j.febslet.2007.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sohail A, Klapacz J, Samaranayake M, Ullah A, Bhagwat AS. Nucleic Acids Res. 2003;31:2990–2994. doi: 10.1093/nar/gkg464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rogozin IB, Diaz M. J. Immunol. 2004;172:3382–3384. doi: 10.4049/jimmunol.172.6.3382. [DOI] [PubMed] [Google Scholar]
- 92.Schumacher AJ, Nissley DV, Harris RS. Proc. Natl. Acad. Sci. USA. 2005;102:9854–9859. doi: 10.1073/pnas.0501694102. Epub 2005 Jul 9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dutko JA, Schafer A, Kenny AE, Cullen BR, Curcio MJ. Curr. Biol. 2005;15:661–666. doi: 10.1016/j.cub.2005.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bransteitter R, Pham P, Calabrese P, Goodman MF. J. Biol. Chem. 2004;14:14. doi: 10.1074/jbc.M408135200. [DOI] [PubMed] [Google Scholar]
- 95.Nowarski R, Britan-Rosich E, Shiloach T, Kotler M. Nat. Struct. Mol. Biol. 2008;15:1059–1066. doi: 10.1038/nsmb.1495. [DOI] [PubMed] [Google Scholar]
- 96.Suspene R, Rusniok C, Vartanian JP, Wain-Hobson S. Nucleic Acids Res. 2006;34:4677–4684. doi: 10.1093/nar/gkl555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sato K, Izumi T, Misawa N, Kobayashi T, Yamashita Y, Ohmichi M, Ito M, Takaori-Kondo A, Koyanagi Y. J. Virol. 2010;84:9546–9556. doi: 10.1128/JVI.00823-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen Z, Eggerman TL, Bocharov AV, Baranova IN, Vishnyakova TG, Csako G, Patterson AP. RNA. 2010;16:1040–1052. doi: 10.1261/rna.1863010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pils B, Schultz J. J. Mol. Biol. 2004;340:399–404. doi: 10.1016/j.jmb.2004.04.063. [DOI] [PubMed] [Google Scholar]
- 100.Edgell DR, Klenk HP, Doolittle WF. J. Bacteriol. 1997;179:2632–2640. doi: 10.1128/jb.179.8.2632-2640.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rogozin IB, Makarova KS, Pavlov YI, Koonin EV. Biol. Direct. 2008;3:32. doi: 10.1186/1745-6150-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Baranovskiy AG, Babayeva ND, Liston VG, Rogozin IB, Koonin EV, Pavlov YI, Vassylyev DG, Tahirov TH. Cell Cycle. 2008;7:3026–3036. doi: 10.4161/cc.7.19.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tahirov TH, Makarova KS, Rogozin IB, Pavlov YI, Koonin EV. Biol. Direct. 2009;4:11. doi: 10.1186/1745-6150-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]